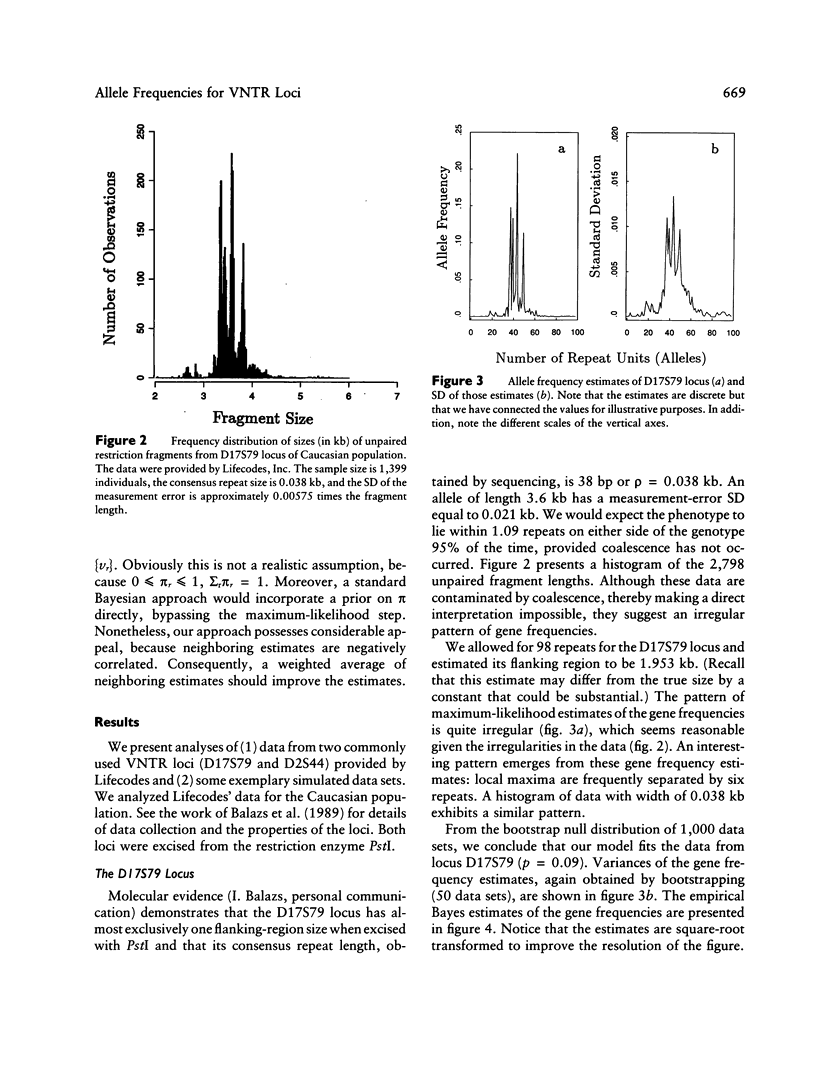
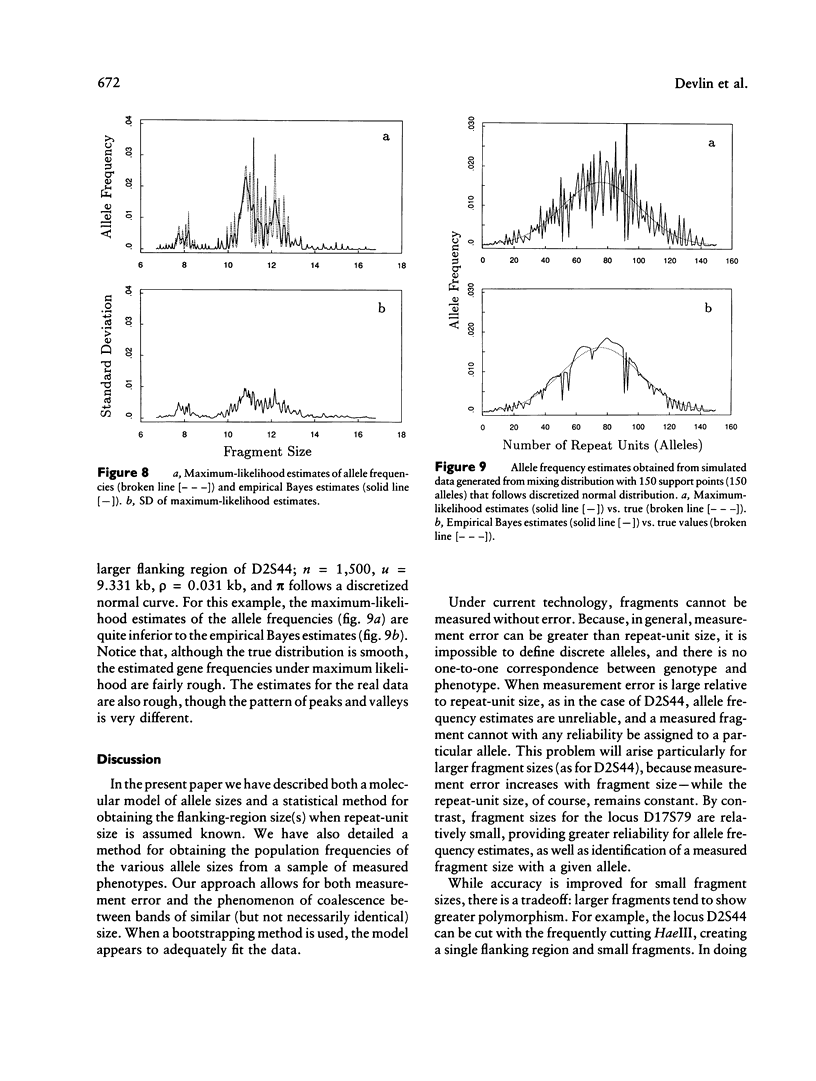
Abstract
VNTR loci provide valuable information for a number of fields of study involving human genetics, ranging from forensics (DNA fingerprinting and paternity testing) to linkage analysis and population genetics. Alleles of a VNTR locus are simply fragments obtained from a particular portion of the DNA molecule and are defined in terms of their length. The essential element of a VNTR fragment is the repeat, which is a short sequence of basepairs. The core of the fragment is composed of a variable number of identical repeats that are linked in tandem. A sample of fragments from a population of individuals exhibits substantial variation in length because of variation in the number of repeats. Each distinct fragment length defines an allele, but any given fragment is measured with error. Therefore the observed distribution of fragment lengths is not discrete but is continuous, and determination of distinct allele classes is not straightforward. A mixture model is the natural statistical method for estimating the allele frequencies of VNTR loci. In this article we develop nonparametric methods for obtaining the distribution of allele sizes and estimates of their frequencies. Methods for obtaining maximum-likelihood estimates are developed. In addition, we suggest an empirical Bayes method to improve the maximum-likelihood estimates of the gene frequencies; the empirical Bayes procedure effects a local smoothing. The latter method works particularly well when measurement error is large relative to the repeat size, because the estimated distribution of allele frequencies when maximum likelihood is used is unreliable because of an alternating pattern of over- and underestimation. We define alleles and estimate the allele frequencies for two VNTR loci from the human genome (D17S79 and D2S44), from data obtained from Lifecodes, Inc.
Full text
PDF












Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balazs I., Baird M., Clyne M., Meade E. Human population genetic studies of five hypervariable DNA loci. Am J Hum Genet. 1989 Feb;44(2):182–190. [PMC free article] [PubMed] [Google Scholar]
- Devlin B., Risch N., Roeder K. No excess of homozygosity at loci used for DNA fingerprinting. Science. 1990 Sep 21;249(4975):1416–1420. doi: 10.1126/science.2205919. [DOI] [PubMed] [Google Scholar]
- Giusti A., Baird M., Pasquale S., Balazs I., Glassberg J. Application of deoxyribonucleic acid (DNA) polymorphisms to the analysis of DNA recovered from sperm. J Forensic Sci. 1986 Apr;31(2):409–417. [PubMed] [Google Scholar]
- Ito H., Yasuda N., Matsumoto H. The probability of parentage exclusion based on restriction fragment length polymorphisms. Jinrui Idengaku Zasshi. 1985 Dec;30(4):261–269. doi: 10.1007/BF01907963. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Royle N. J., Wilson V., Wong Z. Spontaneous mutation rates to new length alleles at tandem-repetitive hypervariable loci in human DNA. Nature. 1988 Mar 17;332(6161):278–281. doi: 10.1038/332278a0. [DOI] [PubMed] [Google Scholar]
- Kanter E., Baird M., Shaler R., Balazs I. Analysis of restriction fragment length polymorphisms in deoxyribonucleic acid (DNA) recovered from dried bloodstains. J Forensic Sci. 1986 Apr;31(2):403–408. [PubMed] [Google Scholar]
- Lynch M. Estimation of relatedness by DNA fingerprinting. Mol Biol Evol. 1988 Sep;5(5):584–599. doi: 10.1093/oxfordjournals.molbev.a040518. [DOI] [PubMed] [Google Scholar]
- Mendell N. R., Simon G. A. A general expression for the variance-covariance matrix of estimates of gene frequency: the effects of departures from Hardy-Weinberg equilibrium. Ann Hum Genet. 1984 Jul;48(Pt 3):283–286. doi: 10.1111/j.1469-1809.1984.tb01026.x. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Leppert M., O'Connell P., Wolff R., Holm T., Culver M., Martin C., Fujimoto E., Hoff M., Kumlin E. Variable number of tandem repeat (VNTR) markers for human gene mapping. Science. 1987 Mar 27;235(4796):1616–1622. doi: 10.1126/science.3029872. [DOI] [PubMed] [Google Scholar]
- Risch N. Linkage strategies for genetically complex traits. III. The effect of marker polymorphism on analysis of affected relative pairs. Am J Hum Genet. 1990 Feb;46(2):242–253. [PMC free article] [PubMed] [Google Scholar]


