Abstract
cDNA microarray technology has been increasingly used to monitor global gene expression patterns in various tissues and cell types. However, applications to mammalian development have been hampered by the lack of appropriate cDNA collections, particularly for early developmental stages. To overcome this problem, a PCR-based cDNA library construction method was used to derive 52,374 expressed sequence tags from pre- and peri-implantation embryos, embryonic day (E) 12.5 female gonad/mesonephros, and newborn ovary. From these cDNA collections, a microarray representing 15,264 unique genes (78% novel and 22% known) was assembled. In initial applications, the divergence of placental and embryonic gene expression profiles was assessed. At stage E12.5 of development, based on triplicate experiments, 720 genes (6.5%) displayed statistically significant differences in expression between placenta and embryo. Among 289 more highly expressed in placenta, 61 placenta-specific genes encoded, for example, a novel prolactin-like protein. The number of genes highly expressed (and frequently specific) for placenta has thereby been increased 5-fold over the total previously reported, illustrating the potential of the microarrays for tissue-specific gene discovery and analysis of mammalian developmental programs.
Systematic studies of gene expression patterns by using cDNA microarrays provide a powerful approach to molecular dissection of cells and tissues by comparing expression levels of tens of thousands of genes at a time (reviewed in refs. 1 and 2). Successful applications have been done with the full gene cohort of yeast (3–6). In mammalian systems, analyses of human fibroblast activation (7) and molecular classification of tumors (8–14) have successfully built on the use of a relatively small microarray containing mainly well-characterized genes. In those instances, the absence of particular genes in the microarray does not affect overall conclusions.
In contrast, inclusion of as many genes as possible in microarrays is desirable for exploratory approaches in which critical genes for biological processes are sought without prior knowledge (15). The problem of maximum coverage is notable for studies of early development, because a recent mouse embryo expressed sequence tag (EST) project (16, 17) has revealed that a significant fraction of the genes expressed in early life are not represented in the collections of genes previously available. We have therefore been assembling a cohort of developmentally active genes to supplement microarray efforts.
Mid-gestation placenta was chosen for a test of the utility of the microarray for several reasons. Methodologically, the placenta can be cleanly separated from the embryo proper, and substantial amounts of RNA can be easily recovered. Biologically, studies of human pregnancy loss and of mouse mutants that render embryos nonviable have motivated widespread interest in the placenta as the indispensable organ for embryo survival (18–20). Furthermore, the extraembryonic lineage that develops into placenta is the first cell type to differentiate from fertilized eggs, and it has therefore been studied as the first cell differentiation that is unique to mammalian species. Yet the number of genes identified as placenta-specific (or highly expressed in placenta) is very limited, and the mechanisms of placenta development and maintenance are still largely unknown. The assembly and use of the microarray to identify and profile genes expressed in early placenta and embryo are reported here.
Materials and Methods
cDNA Libraries, EST Collections, and Assembly of Unique Gene Set.
Procedures for cDNA library construction and EST projects are essentially the same as described previously (16, 17). Sequence information is available at dbEST (http://www.ncbi.nlm.nih.gov/) and at the National Institute on Aging web site (http://lgsun.grc.nia.nih.gov/). All cDNA clones are available from American Type Culture Collection (ATCC; http://www.atcc.org/). Selection of the unique gene set was done by all-against-all similarity searches with blast 1.4 algorithm (21) with a cut-off score of 800 to group ESTs into unique genes. A total of 15,264 unique genes were rearrayed in 96-well microtiter plates.
Preparation of Microarray.
Plasmid DNAs were prepared in 96-well format (R.E.A.L. Prep 96 plasmid kit; Qiagen, Chatsworth, CA). To amplify cDNA inserts, PCR primers (5′-CCAGTCACGACGTTGTAAAACGAC-3′; 5′-GTGTGGAATTGTGAGCGGATAACAA-3′) were designed from sequences flanking the multiple cloning site of pSPORT1 plasmid vector (Life Technologies, Rockville, MD), which was used for cDNA cloning. For each cDNA clone, about 10 ng of plasmid DNA was used in 100 μl PCR reaction containing 10 mM Tris⋅HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.5 μM each primer, 0.8 mM dNTPs, and 3.25 units of Taq polymerase (PE Biosystems, Foster City, CA). Amplification was done in a Peltier Thermal Cycler (MJ Research, Cambridge, MA) under the following conditions: initial denaturation at 95°C for 1 min; 38 cycles of denaturation at 94°C for 30 s, annealing at 65°C for 45 s, and extension at 72°C for 3.5 min; and a final extension at 72°C for 3 min. PCR products were precipitated with 50% isopropanol, washed in 70% ethanol, and resuspended in 6.5 μl TE buffer (10 mM Tris/1 mM EDTA, pH 8.0). To examine the quality and quantity of PCR products, they were run on 1% agarose gels (Embi Tec, San Diego, CA). Greater than 90% of the clones gave single PCR products. PCR products were denatured with 0.1 M NaOH and spotted onto nylon membranes (Schleicher and Schüll, Dassel, Germany) by using a GMS417 Microarrayer (Genetic Microsystems, Woburn, MA). Genes (2,304) were spotted on each membrane with 2.5 × 7.5 cm dimension at 665 μm distance between spots. Diameter of the spotting pin was 300 μm.
Probe Preparation.
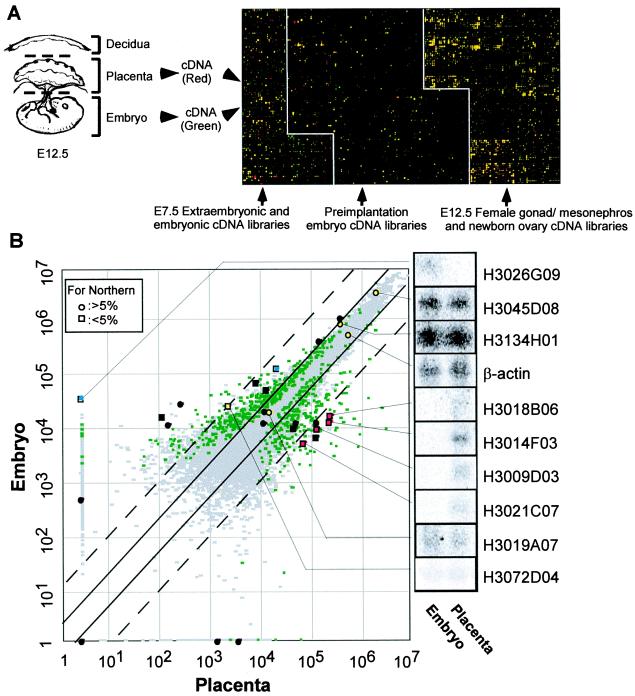
Hybridization of the microarray with a 33P-labeled 260-bp vector probe showed relatively uniform signal intensities across individual membranes. Placenta (including yolk sac and amnion) and embryo were dissected out from C57BL/6J at 12.5 days postcoitum [embryonic day (E) 12.5]. Maternal part of the placenta (i.e., decidua and uterus) was stripped away, and the conceptus-derived part of the placenta was used for experiments (see Fig. 2A). Total RNA was extracted by Trizol reagent (Life Technologies); 100–300 μg of total RNA was used as a template to incorporate [α-33P]dCTP (Amersham Pharmacia, Piscataway, NJ) during cDNA synthesis using SuperScript II reverse transcriptase (SSTII; Life Technologies) with oligo(dT) primer (Amersham Pharmacia). Each labeling reaction contained 10–13 μg of a heat-denatured template RNA, 1× first strand reaction buffer, 0.01 M DTT, 0.5 mM each of dATP, dGTP, and dTTP (Amersham Pharmacia), 40 units RNAsin (Boehringer Mannheim) and 50 μCi/mmol [α- 33P]dCTP. The reaction mixture was heated at 65°C for 10 min and then cooled down gradually to 42°C, where 400 units of SSTII was added for 2 h. After a second hour incubation with 400 units more enzyme, template RNA was degraded with 0.2 M NaOH at 65°C for 15 min, and the mixture was then neutralized with 0.3 M Tris⋅HCl (pH 7.5).
Figure 2.
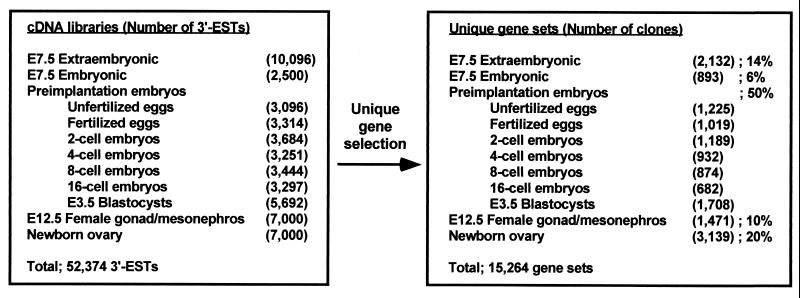
Expression profiling of 15K genes between embryo and placenta at E12.5. (A) Schematic representation of experimental procedures and pseudocolored image for a typical hybridization. After removal of maternal tissues, embryo and placenta including yolk sac and amnion were dissected out and used for RNA extraction. Radiolabeled cDNAs from each sample were hybridized separately on the 15K microarray. Hybridization was repeated three times independently for each sample. Gene expression levels in placenta were represented as a gradation of red, and those in embryo were represented as a gradation of green. Genes expressed commonly in two samples appear yellow. (B) For each gene, average expression levels (in arbitrary units) were calculated from three independent hybridizations for placenta and embryo, respectively, and displayed on a scatter plot. Genes that show significantly different expression levels between embryo and placenta at the 5% significance level (P < 0.05) are displayed as green spots, and the other genes are displayed as gray spots. Solid lines indicate 2-fold expression differences between embryo and placenta; dotted lines indicate 10-fold expression differences. Results of Northern blot analyses of selected genes are shown with coordinates in the scatter plot. A total 28 clones were examined, and Northern blot for 10 clones are shown. Spots were color coded according to their expression patterns on Northern blot: placenta-specific expression (red), embryo-specific expression (blue), commonly expressed genes (yellow), and no detectable expression in both placenta and embryo (black). Genes that show statistically significant differences are represented as squares, and ones that do not are represented as circles.
Microarray Hybridization and Data Analyses.
Prehybridization was done at 65°C for 4 h with MicroHyb solution (Research Genetics, Huntsville, AL) supplemented with yeast tRNA (1 mg/ml; Sigma) and poly(A) RNA (1 mg/ml; Amersham Pharmacia). Then hybridization was done with purified probe 10% dextran sulfate (Sigma), and mouse Cot1 DNA (70 μg/ml; Life Technologies) added to the prehybridization solution at 65°C for overnight incubation. Membranes were washed twice in 2× SSC/0.1% SDS for 20 min at room temperature, followed by two washings in 2× SSC/0.1% SDS at 65°C for 30 min. After a final wash with 0.1× SSC/1% SDS for 1 h at 65°C, membranes were exposed to Phosphorscreen (Amersham Pharmacia) for 10 days at room temperature; then the screen was scanned by Storm 860 (Amersham Pharmacia) at a maximum resolution of 50 μm/pixel. Results from three independent hybridizations were obtained for each probe. Images were analyzed by imagequant 5.0 (Amersham Pharmacia). Each image was overlaid with grids so that signal intensities of individual spots could be assessed. Local background for each membrane was calculated on the basis of 18 positions with no DNA spotted area. Expression levels of individual genes are represented in arbitrary units after subtracting background. All of the expression data are available at the National Institute on Aging web site (http://lgsun.grc.nia.nih.gov/array/data1.html).
Gene Expression Analyses by Northern Blot and in Situ Hybridization.
For Northern blot analysis, 20 μg of total RNA from E12.5 embryo and placenta were size fractionated on 1% formaldehyde agarose gel and transferred to nylon membranes (Oncor, Gaithersburg, MD). cDNA inserts from enzyme-digested plasmid DNAs were purified and used as probes for hybridization. In situ hybridization for one clone, H3018B06, was performed as described (22). Digoxigenin-labeled antisense probe was hybridized on the section of E13.5 embryos with placenta. As a marker for placenta-specific hybridization, 4311 (23) was also used as a probe.
Results and Discussion
Assembly of a 15K Mouse Developmental cDNA Microarray.
To collect ESTs from early mouse embryos, a PCR-based cDNA library construction method was used to derive ESTs on a large scale from E7.5 extraembryonic tissue (16) and preimplantation embryos (17). Additional ESTs were generated for this study from E7.5 extraembryonic cDNA libraries (8,088 cDNAs, GenBank nos. AW537829–AW545916), E7.5 embryonic cDNA libraries (1,589 cDNAs, GenBank nos. AW536144–AW537732), and other libraries (13,241 cDNAs, GenBank nos. AW545922–AW559162) (Fig. 1). All of the ESTs were derived from the 3′-ends of cDNAs. All-against-all similarity searches by blast program (21) classified the total of 52,374 3′-ESTs into 15,264 unique genes, which were rearrayed in a set of 96-well microtiter plates (Fig. 1). This 15K clone set contains cDNA inserts of average 1.5 kb and has been sequenced from both 5′- and 3′-ends to verify clone identity and further characterize individual genes (G.J.K., D. B. Dudekula, Y. Qian, and M.S.H.K. unpublished work). About 3,400 represent known genes, suggesting that one-third to one-half of well-characterized mammalian genes are included. Because the majority of the cDNAs are derived from early mouse embryos and 78% are either completely novel (55%) or are similar but not identical to known genes (23%), the collection provides a unique source for studies of developmental biology and is complementary to other mouse/human EST cohorts.
Figure 1.
Construction of 15K mouse cDNA microarray. cDNA libraries used for EST generations and the number of ESTs collected from each library are shown (Left). ESTs were clustered by their sequence similarity, and representative cDNA clones were selected from each gene cluster. Numbers and sources of cDNA clones in the 15,264 (15K) unique gene set are shown (Right).
Expression Profiling of Placenta and Embryo.
The global gene expression pattern of E12.5 placenta was compared with that of E12.5 embryo (Fig. 2A). Signal intensities of individual genes were converted to color intensities and superimposed (Fig. 2A). A number of “placenta-specific” (red in Fig. 2A) and “embryo-specific” genes (green in Fig. 2A) are apparent. Rather than applying a current standard procedure, which directly uses ratios of values for each gene (or an arbitrary 2- or 10-fold expression difference) to characterize “differences” in gene expression levels in a single determination, three independent hybridization experiments were first performed for each probe. The multiple probings permit an analysis that satisfies conservative statistical criteria (24) and allows one to identify 720 genes that are expressed differentially between two samples at the 5% significance level by t test (P value < 0.05). These genes are shown as spots in a scatter plot of the average signal intensity of each gene (Fig. 2B).
Next, the conventional criteria of 2-fold and 10-fold differences were used to subclassify the significantly different genes into six categories. Sixty-one genes that show 10-fold higher expression in placenta than in embryo are classified as placenta-specific genes; 166 genes with expression levels between 2-fold and 10-fold higher in placenta are classified as predominantly expressed in placenta. Similarly, 57 genes with 10-fold greater expression in embryo are classified as embryo-specific, and 253 whose expression levels are between 2-fold and 10-fold higher in embryo are classified as predominantly expressed in embryo. Expression differences of less than 2-fold have usually been considered at the limit of detection in previous analyses; but, by the statistical criteria applied here, significant differences are reproducibly seen for many differences in the range of 1.2- to 2-fold. The range showing statistically significant differences in less than 2-fold includes 62 genes whose expression is higher in placenta and 121 genes whose expression is higher in embryo. The statistically supported indications are particularly promising for the discovery and analysis of genes that are dosage controlled. Frequently important in early development, they include genes that are imprinted or X inactivated.
On the other hand, even genes showing more than 2-fold differences in their average expression levels sometimes fall into the statistically nonsignificant category (shown in gray in Fig. 2B), because of unknown variables that affect the relative signal from one or another probe. The statistical test of triplicate hybridizations discriminates the likely dependable values from false positives or negatives.
Verification of cDNA Microarray Data.
The rankings of gene expression levels are supported by Northern blot analyses of selected genes. Results for 27 genes examined correlated well with cDNA microarray intensities and subclassification (Fig. 2B). Beta-actin, for example, displayed equivalent expression levels on the scatter plot, consistent with Northern blot data. Notably, a 28th gene (H3010F03) again showed the utility of the statistical test. Although it seemed placenta specific, based on average expression levels, it was judged as not significant by the statistical test and indeed showed only very faint signals in both placenta and embryo in Northern blot hybridization tests. The results also illustrated the greater sensitivity of microarray analyses compared with Northern blots. Expression levels 100-fold below the level detected on Northern blots were reproducibly observed in the microarray analyses (Fig. 2B).
Identification of Individual Genes.
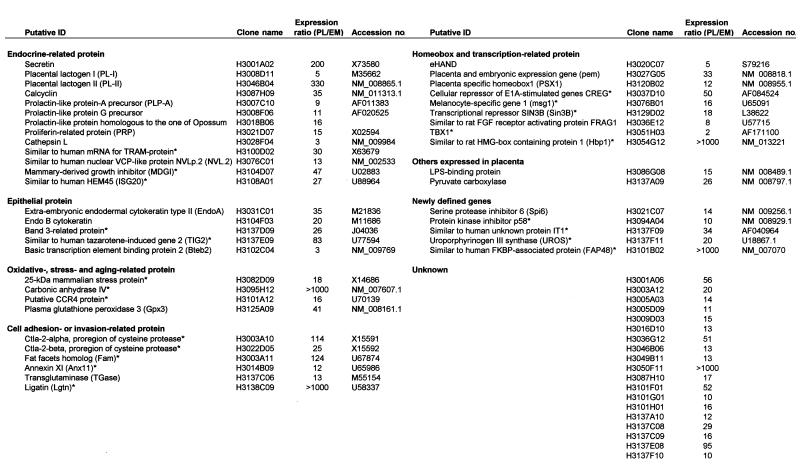
Details about some of the placenta-specific genes, including putative identifications and functional groupings, are shown in Table 1. Based on current gene classifications, they fall into three categories. One comprises 30 genes that are already known and well-described as placenta-specific genes (19, 20), which include placental lactogen II (25), extraembryonic endodermal cytokeratin type II [endoB (26)], Psx (27), proliferin-related protein, prolactin (PRL)-like protein G precursor, and PRL-like protein A precursor (reviewed in refs. 25 and 28). These results further support the validity of the cDNA microarray-based analyses for identifying placenta-specific genes.
Table 1.
List of genes expressed highly in placenta
Presence of these genes in placenta has not been previously reported. PL, Average expression level of genes with placenta probes; EM, average expression level of genes with embryo probes.
A second category includes 78 genes that are already known, but for which expression in placenta had not been determined. This category includes cytotoxic T lymphocyte-associated protein 2 (Ctla-2α, Ctla-2β) (29), and fat facets (Faf). Although Ctla-2 genes may originate from circulating blood cells in placenta, they might also function there. Faf is a deubiquitination enzyme specifically required for normal eye development in Drosophila; but its mouse homologue, Fam, was cloned (30) and implicated in cell–cell adhesion (31), suggesting a possible involvement of this gene in the implantation process.
The third category comprises 181 genes that are new and, together with genes in the second category, will provide a source of discovery of genes with a previously unrecognized important involvement in implantation and placenta development. In fact, the number of genes highly expressed or specific for the placenta are thus increased about 5-fold over the total reported previously.
Global Characteristics of Placenta.
The list of putative placenta-specific genes elucidates some characteristic features of placenta (Table 1). Many genes involved in secretion stand out in the list and can be correlated with the status of the placenta as a primary endocrine organ. As a derivative of trophectoderm with epithelial character, mid-gestation placenta also actively synthesizes proteins characteristic of epithelia. The presence of a number of cell adhesion and invasion-related proteins is consistent with trophoblast function as the first-line invasion front for the embryo into the maternal uterine epithelia. Interestingly, the additional presence of a number of stress- and aging-related proteins may support the concept of the placenta as a prematurely aging organ (32).
Example of Newly Identified Genes.
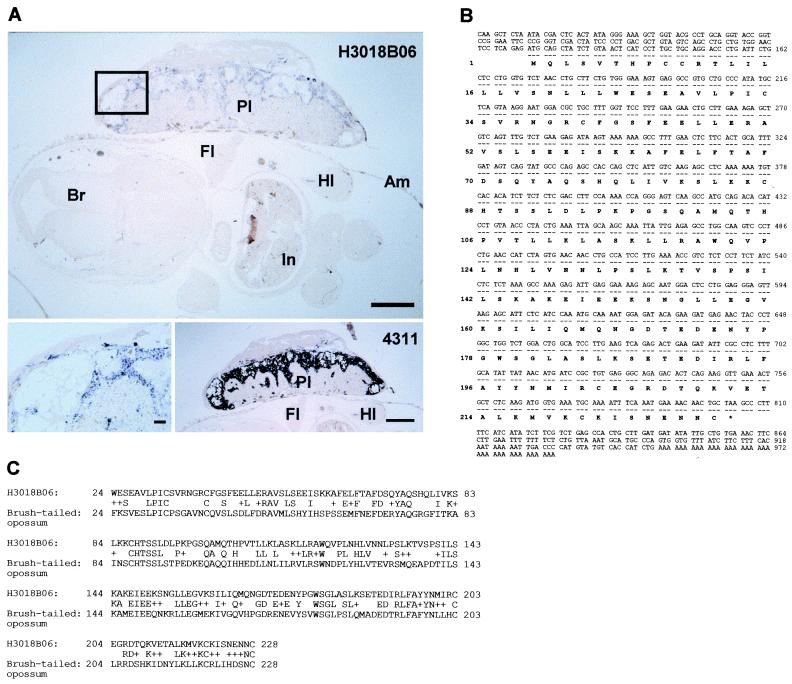
In an example of further gene characterization based on the microarray data, one new species that shows placenta-specific expression (H3018B06) was further characterized. Northern blot showed strong placenta-specific expression (Fig. 2B), and in situ hybridization (Fig. 3A) revealed expression exclusively in spongiotrophoblast and trophoblast giant cells, with no detectable signal in the embryo. This expression pattern is the same as that of a well-known trophoblast-specific marker gene, clone 4311 (23) (Fig. 3A). The cDNA clone has a 987-bp insert, which suggests that it is essentially full-length (the 1.1-kb transcript observed in Northern blot includes a poly(A) tract of several hundred residues). The clone was completely sequenced (Fig. 3B, GenBank no, AF272368). Computer-assisted searches detected significant homology only to a PRL-like protein from opossum (Fig. 3C). Thus, this gene seems to be a new member of the placental growth-related hormone family (25, 28).
Figure 3.
New placenta-specific gene. (A, Top) In situ hybridization of H3018B06 gene. Transverse section of E13.5 embryo with placenta was probed with digoxigenin-labeled RNA transcribed from H3018B06 (bar = 1 mm). (Bottom Left) Magnified view of the region marked with a rectangle in the top panel (bar = 100 μm). (Bottom Right) Control in situ hybridization with a trophoblast-specific gene, 4311 (bar = 1 mm). Am, amnion; Br, brain; Fl, forelimb; Hl, hindlimb; In, intestine; Pl, placenta. (B) Nucleotide and putative amino acid sequence of H3018B06. (C) Amino acid sequence alignment between mouse H3018B06 gene and opossum PRL gene.
Similarity of Global Gene Expression Patterns Between Placenta and Embryo.
Although we have emphasized the 5% of the genes that showed statistically significant differences between the placenta and developing embryo, it is striking that global gene expression patterns are overall rather similar for most genes in these very distinctive portions of the embryo. These results may rationalize the unexpected findings of placental failure in a significant fraction of mice in which nonplacental “tissue-specific” genes had been disrupted (reviewed in ref. 18). It may be relevant that, at E12.5, more than 70% of placental cells are trophectoderm-derived trophoblast cells (33), and it is thus very likely that the expression patterns in placenta in this experiment represent trophoblasts. Trophoblast cells first appear in mammals, likely between the Jurassic [≈150 million years (Myr)] and Cretaceous (≈130 Myr) periods, after which the explosive diversification of mammalian species (“mammalian radiation”) took place (34). To create a functional placenta in such a short evolutionary span, it may be that genetic pathways and networks were taken directly from ones already extant for basic body plan, with minimum modifications.
Possible Applications of the 15K Mouse Developmental cDNA Microarray.
Determining the degree of divergence of placenta from embryo at a particular stage of development is an example of the use of the microarray. Because it includes a large number of genes from pre- and periimplantation mouse embryogenesis, it is adapted to comparable analyses of other stages and tissues during mouse development. For example, it should be possible to characterize genes that change expression as trophoblasts, which have a finite lifespan, form from embryonic cells with indefinite growth potential. Identification of downstream target genes in transgenic/knockout mice and global gene expression analysis of natural/induced mouse mutants are some of the obvious applications of this microarray. Recent findings indicate that some embryonic genes are reactivated in aging heart and muscle, suggesting the potential utility of the microarray for analyses of features of aging as well as growth control and tumorigenesis (35).
Note Added in Proof.
The 15K mouse cDNA clone set described in this paper has been made available (see details at http://lgsun.grc.nia.nih.gov).
Acknowledgments
We thank Dr. David Schlessinger for critical reading of the manuscript, Dr. Chris Morrell for the help in statistical analysis, and Dr. Janet Rossant for providing 4311 clone probe.
Abbreviations
- EST
expressed sequence tag
- En
embryonic day n
- PRL
prolactin
Footnotes
References
- 1.Ferea T L, Brown P O. Curr Opin Genet Dev. 1999;9:715–722. doi: 10.1016/s0959-437x(99)00033-7. [DOI] [PubMed] [Google Scholar]
- 2.Duggan D J, Bittner M, Chen Y, Meltzer P, Trent J M. Nat Genet. 1999;21:10–14. doi: 10.1038/4434. [DOI] [PubMed] [Google Scholar]
- 3.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 4.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown P O, Herskowitz I. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 5.Spellman P T, Sherlock G, Zhang M Q, Iyer V R, Anders K, Eisen M B, Brown P O, Botstein D, Futcher B. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts C J, Nelson B, Marton M J, Stoughton R, Meyer M R, Bennett H A, He Y D, Dai H, Walker W L, Hughes T R, Tyers M, Boone C, Friend S H. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- 7.Iyer V R, Eisen M B, Ross D T, Schuler G, Moore T, Lee J C F, Trent J M, Staudt L M, Hudson J, Jr, Boguski M S, Lashkari D, Shalon D, Botstein D, Brown P O. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 8.DeRisi J, Penland L, Brown P O, Bittner M L, Meltzer P S, Ray M, Chen Y, Su Y A, Trent J M. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 9.Pollack J R, Perou C M, Alizadeh A A, Eisen M B, Pergamenschikov A, Williams C F, Jeffrey S S, Botstein D, Brown P O. Nat Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 10.Khan J, Simon R, Bittner M, Chen Y, Leighton S B, Pohida T, Smith P D, Jiang Y, Gooden G C, Trent J M, Meltzer P S. Cancer Res. 1998;58:5009–5013. [PubMed] [Google Scholar]
- 11.Perou C M, Jeffrey S S, van de Rijn M, Rees C A, Eisen M B, Ross D T, Pergamenschikov A, Williams C F, Zhu S X, Lee J C, Lashkari D, Shalon D, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alizadeh A A, Eisen M B, Davis R E, Ma C, Lossos I S, Rosenwald A, Boldrick J C, Sabet H, Tran T, Yu X, et al. Nature (London) 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 13.Ross D T, Scherf U, Eisen M B, Perou C M, Rees C, Spellman P, Iyer V, Jeffrey S S, Van De Rijn M, Waltham M, et al. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 14.Scherf U, Ross D T, Waltham M, Smith L H, Lee J K, Tanabe L, Kohn K W, Reinhold W C, Myers T G, Andrews D T, et al. Nat Genet. 2000;24:236–244. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- 15.Khan J, Bittner M L, Saal L H, Teichmann U, Azorsa D O, Gooden G C, Pavan W J, Trent J M, Meltzer P S. Proc Natl Acad Sci USA. 1999;96:13264–13269. doi: 10.1073/pnas.96.23.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ko M S H, Threat T A, Wang X, Horton J H, Cui Y, Wang X, Pryor E, Paris J, Wells-Smith J, Kitchen J R, et al. Hum Mol Genet. 1998;7:1967–1978. doi: 10.1093/hmg/7.12.1967. [DOI] [PubMed] [Google Scholar]
- 17.Ko M S H, Kitchen J R, Wang X, Threat T A, Wang X, Hasegawa A, Sun T, Grahovac M J, Kargul G J, Lim M K, et al. Development. 2000;127:1737–1749. doi: 10.1242/dev.127.8.1737. [DOI] [PubMed] [Google Scholar]
- 18.Copp A J. Trends Genet. 1995;11:87–93. doi: 10.1016/S0168-9525(00)89008-3. [DOI] [PubMed] [Google Scholar]
- 19.Cross J C, Werb Z, Fisher S J. Science. 1994;266:1508–1518. doi: 10.1126/science.7985020. [DOI] [PubMed] [Google Scholar]
- 20.Rinkenberger J L, Cross J C, Werb Z. Dev Genet. 1997;21:6–20. doi: 10.1002/(SICI)1520-6408(1997)21:1<6::AID-DVG2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 21.Altschul S F, Gish W, Miller W, Myers E, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Wilkinson D G, Nieto M A. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- 23.Lescisin K R, Varmuza S, Rossant J. Genes Dev. 1988;2:1639–1646. doi: 10.1101/gad.2.12a.1639. [DOI] [PubMed] [Google Scholar]
- 24.Claverie J M. Hum Mol Genet. 1999;8:1821–1832. doi: 10.1093/hmg/8.10.1821. [DOI] [PubMed] [Google Scholar]
- 25.Linzer D I, Fisher S J. Mol Endocrinol. 1999;13:837–840. doi: 10.1210/mend.13.6.0286. [DOI] [PubMed] [Google Scholar]
- 26.Hashido K, Morita T, Matsushiro A, Nozaki M. Exp Cell Res. 1991;192:203–212. doi: 10.1016/0014-4827(91)90177-v. [DOI] [PubMed] [Google Scholar]
- 27.Chun J Y, Han Y J, Ahn K Y. Dev Dyn. 1999;216:257–266. doi: 10.1002/(SICI)1097-0177(199911)216:3<257::AID-DVDY4>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 28.Hamilton R T, Millis A J. Curr Top Dev Biol. 1990;24:193–218. doi: 10.1016/s0070-2153(08)60088-5. [DOI] [PubMed] [Google Scholar]
- 29.Denizot F, Brunet J F, Roustan P, Harper K, Suzan M, Luciani M F, Mattei M G, Golstein P. Eur J Immunol. 1989;19:631–635. doi: 10.1002/eji.1830190409. [DOI] [PubMed] [Google Scholar]
- 30.Wood S A, Pascoe W S, Ru K, Yamada T, Hirchenhain J, Kemler R, Mattick J S. Mech Dev. 1997;63:29–38. doi: 10.1016/s0925-4773(97)00672-2. [DOI] [PubMed] [Google Scholar]
- 31.Taya S, Yamamoto T, Kano K, Kawano Y, Iwamatsu A, Tsuchiya T, Tanaka K, Kanai-Azuma M, Wood S A, Mattick J S, et al. J Cell Biol. 1998;142:1053–1062. doi: 10.1083/jcb.142.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosso P. Curr Concepts Nutr. 1976;4:23–41. [PubMed] [Google Scholar]
- 33.Rossant J, Croy B A. J Embryol Exp Morphol. 1985;86:177–189. [PubMed] [Google Scholar]
- 34.Easteal S. Bioessays. 1999;21:1052–1058. doi: 10.1002/(SICI)1521-1878(199912)22:1<1052::AID-BIES9>3.0.CO;2-6. ; discussion 1059. [DOI] [PubMed] [Google Scholar]
- 35.Schlessinger D, Ko M S H. Genomics. 1998;52:113–118. [PubMed] [Google Scholar]