Abstract
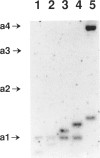
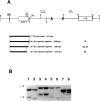
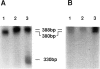
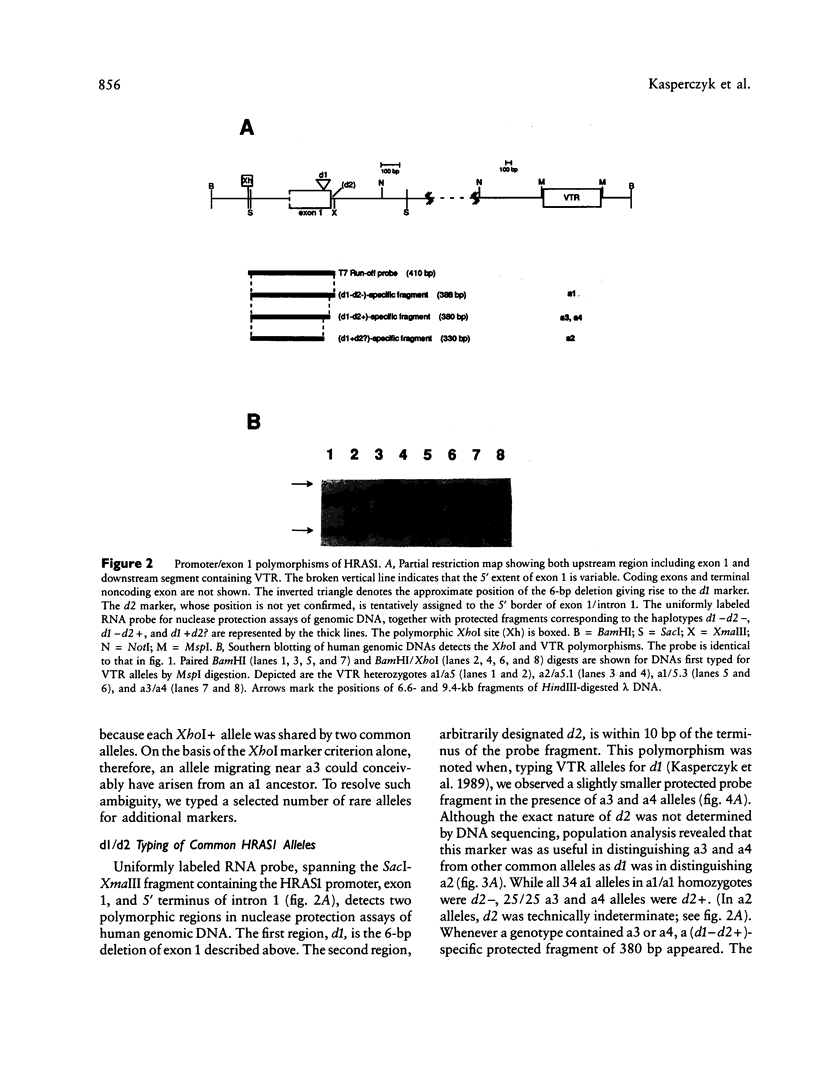
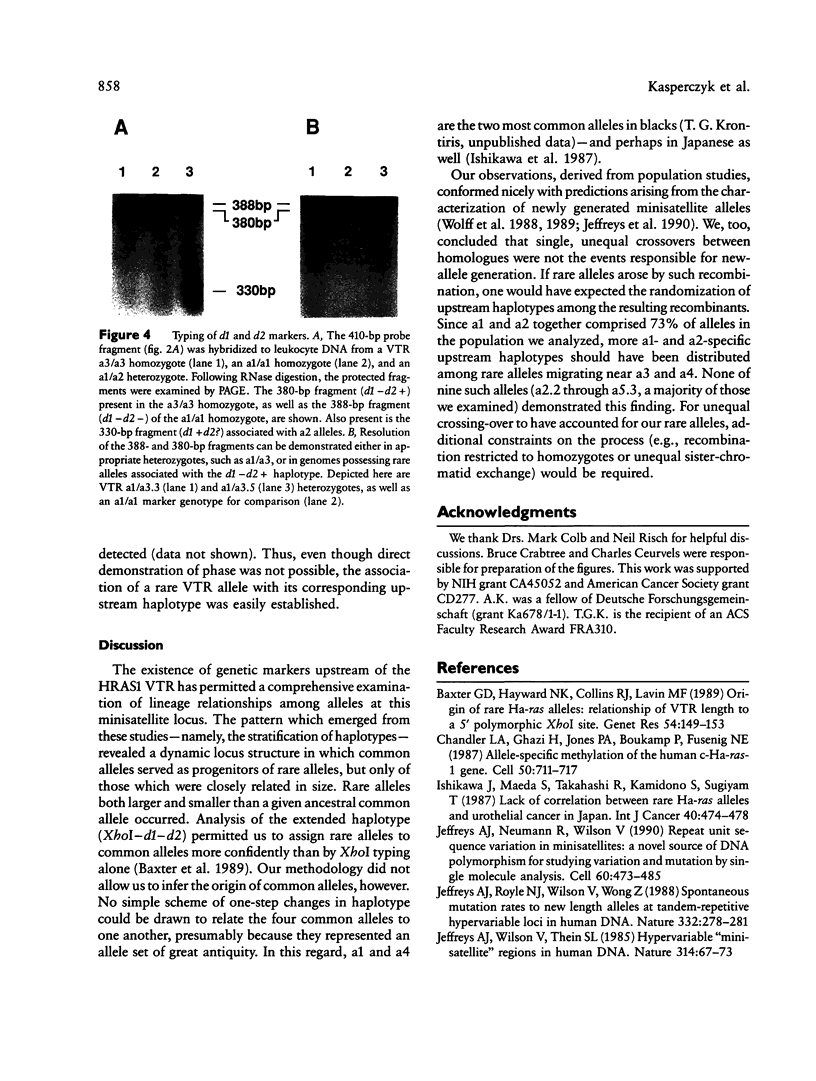
Three genetic markers within the promoter-exon 1 region of the HRAS1 locus have been employed to investigate lineage relationships among alleles of the highly polymorphic variable tandem repeat (VTR) immediately downstream of the HRAS1 gene. These markers were in absolute linkage disequilibrium with the HRAS1 VTR, allowing the assignment of unique upstream haplotypes to each of the four common VTR alleles. Analysis of 17 rare alleles revealed a stratification of allele fragment size and upstream haplotype in which each rare VTR allele possessed the markers characteristic of the common allele nearest in size. Therefore, hyperallelism emanated from the four common alleles in a defined fashion, the size of a rare allele specifying its origin. As discussed below, this result implies that unequal crossing-over between homologues is unlikely to be the predominant mechanism for generating new VTR alleles at this minisatellite locus.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baxter G. D., Hayward N. K., Collins R. J., Lavin M. F. Origin of rare Ha-ras alleles: relationship of VTR length to a 5' polymorphic Xho I site. Genet Res. 1989 Oct;54(2):149–153. doi: 10.1017/s0016672300028524. [DOI] [PubMed] [Google Scholar]
- Chandler L. A., Ghazi H., Jones P. A., Boukamp P., Fusenig N. E. Allele-specific methylation of the human c-Ha-ras-1 gene. Cell. 1987 Aug 28;50(5):711–717. doi: 10.1016/0092-8674(87)90329-1. [DOI] [PubMed] [Google Scholar]
- Ishikawa J., Maeda S., Takahashi R., Kamidono S., Sugiyama T. Lack of correlation between rare Ha-ras alleles and urothelial cancer in Japan. Int J Cancer. 1987 Oct 15;40(4):474–478. doi: 10.1002/ijc.2910400407. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Neumann R., Wilson V. Repeat unit sequence variation in minisatellites: a novel source of DNA polymorphism for studying variation and mutation by single molecule analysis. Cell. 1990 Feb 9;60(3):473–485. doi: 10.1016/0092-8674(90)90598-9. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Royle N. J., Wilson V., Wong Z. Spontaneous mutation rates to new length alleles at tandem-repetitive hypervariable loci in human DNA. Nature. 1988 Mar 17;332(6161):278–281. doi: 10.1038/332278a0. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Wilson V., Thein S. L. Hypervariable 'minisatellite' regions in human DNA. Nature. 1985 Mar 7;314(6006):67–73. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- Kasperczyk A., Mermer B. A., Parkinson D. R., Lonergan J. A., Krontiris T. G. Allele-specific deletion in exon I of the HRAS1 gene. Am J Hum Genet. 1989 Nov;45(5):689–696. [PMC free article] [PubMed] [Google Scholar]
- Krontiris T. G., DiMartino N. A., Colb M., Parkinson D. R. Unique allelic restriction fragments of the human Ha-ras locus in leukocyte and tumour DNAs of cancer patients. 1985 Jan 31-Feb 6Nature. 313(6001):369–374. doi: 10.1038/313369a0. [DOI] [PubMed] [Google Scholar]
- Krontiris T. G., DiMartino N. A., Mitcheson H. D., Lonergan J. A., Begg C., Parkinson D. R. Human hypervariable sequences in risk assessment: rare Ha-ras alleles in cancer patients. Environ Health Perspect. 1987 Dec;76:147–153. doi: 10.1289/ehp.8776147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R. M., Larin Z., Maniatis T. Detection of single base substitutions by ribonuclease cleavage at mismatches in RNA:DNA duplexes. Science. 1985 Dec 13;230(4731):1242–1246. doi: 10.1126/science.4071043. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Leppert M., O'Connell P., Wolff R., Holm T., Culver M., Martin C., Fujimoto E., Hoff M., Kumlin E. Variable number of tandem repeat (VNTR) markers for human gene mapping. Science. 1987 Mar 27;235(4796):1616–1622. doi: 10.1126/science.3029872. [DOI] [PubMed] [Google Scholar]
- Shih C., Weinberg R. A. Isolation of a transforming sequence from a human bladder carcinoma cell line. Cell. 1982 May;29(1):161–169. doi: 10.1016/0092-8674(82)90100-3. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Wolff R. K., Nakamura Y., White R. Molecular characterization of a spontaneously generated new allele at a VNTR locus: no exchange of flanking DNA sequence. Genomics. 1988 Nov;3(4):347–351. doi: 10.1016/0888-7543(88)90126-7. [DOI] [PubMed] [Google Scholar]
- Wolff R. K., Plaetke R., Jeffreys A. J., White R. Unequal crossingover between homologous chromosomes is not the major mechanism involved in the generation of new alleles at VNTR loci. Genomics. 1989 Aug;5(2):382–384. doi: 10.1016/0888-7543(89)90076-1. [DOI] [PubMed] [Google Scholar]