Abstract
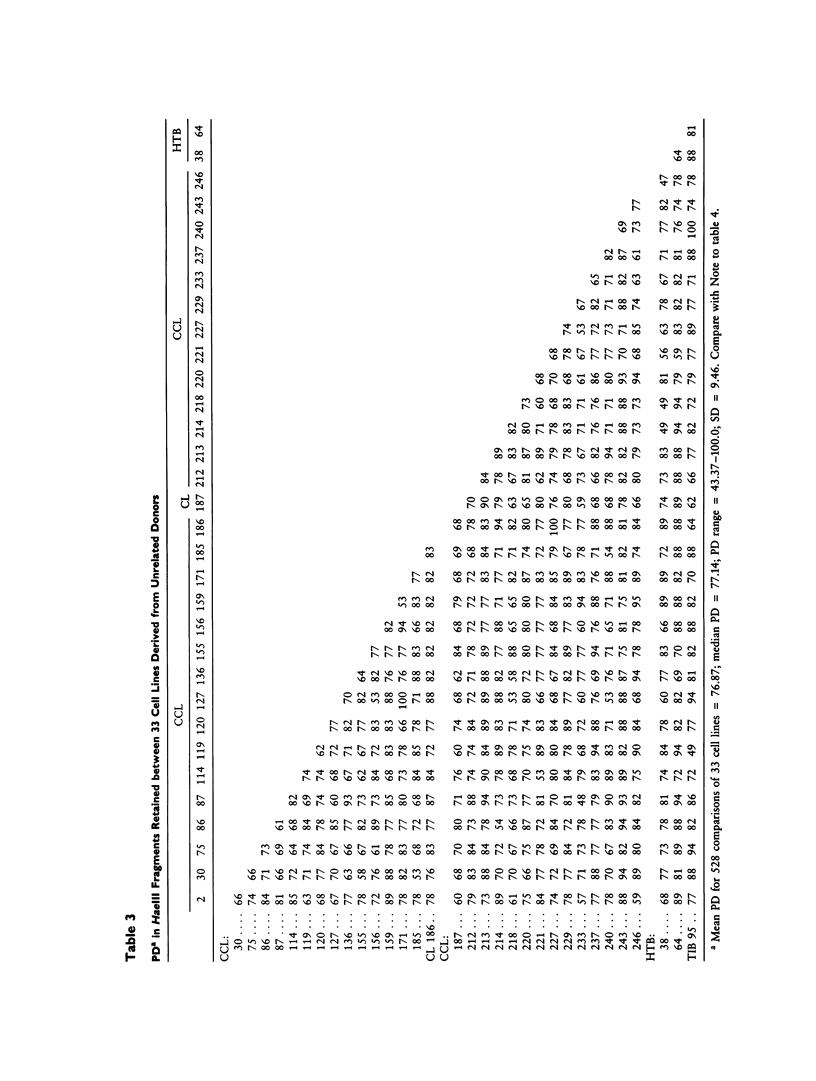
DNA fingerprints of 46 human cell lines were derived using minisatellite probes for hypervariable genetic loci. The incidence of 121 HaeIII DNA fragments among 33 cell lines derived from unrelated individuals was used to estimate allelic and genotypic frequencies for each fragment and for composite individual DNA fingerprints. We present a quantitative estimate of the extent of genetic difference between individuals, an estimate based on the percentage of restriction fragments at which they differ. The average percent difference (APD) among pairwise combinations from the population of 33 unrelated cell lines was 76.9%, compared with the APD in band sharing among cell lines derived from the same individual (less than or equal to 1.2%). Included in this survey were nine additional cell lines previously implicated as HeLa cell derivatives, and these lines were clearly confirmed as such by DNA fingerprints (APD less than or equal to 0.6%). On the basis of fragment frequencies in the tested cell line population, a simple genetic model was developed to estimate the frequencies of each DNA fingerprint in the population. The median incidence was 2.9 X 10(-17), and the range was 2.4 X 10(-21) to 6.6 X 10(-15). This value approximates the probability that a second cell line selected at random from unrelated individuals will match a given DNA fingerprint. Related calculations address the chance that any two DNA fingerprints would be identical among a large group of cell lines. This estimate is still very slight; for example, the chance of two or more common DNA fingerprints among 1 million distinct individuals is less than .001. The procedure provides a straightforward, easily interpreted, and statistically robust method for identification and individualization of human cells.
Full text
PDF














Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell G. I., Selby M. J., Rutter W. J. The highly polymorphic region near the human insulin gene is composed of simple tandemly repeating sequences. Nature. 1982 Jan 7;295(5844):31–35. doi: 10.1038/295031a0. [DOI] [PubMed] [Google Scholar]
- Burke T., Bruford M. W. DNA fingerprinting in birds. Nature. 1987 May 14;327(6118):149–152. doi: 10.1038/327149a0. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins T., Lapierre L. A., Fiers W., Strominger J. L., Pober J. S. Recombinant human tumor necrosis factor increases mRNA levels and surface expression of HLA-A,B antigens in vascular endothelial cells and dermal fibroblasts in vitro. Proc Natl Acad Sci U S A. 1986 Jan;83(2):446–450. doi: 10.1073/pnas.83.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrone S., Pellegrino M. A., Reisfeld R. A. A rapid method for direct HL-A typing of cultured lymphoid cells. J Immunol. 1971 Aug;107(2):613–615. [PubMed] [Google Scholar]
- GEY G. O. Some aspects of the constitution and behavior of normal and malignant cells maintained in continuous culture. Harvey Lect. 1954;50:154–229. [PubMed] [Google Scholar]
- Gartler S. M. Genetic markers as tracers in cell culture. Natl Cancer Inst Monogr. 1967 Sep;26:167–195. [PubMed] [Google Scholar]
- Gill P., Jeffreys A. J., Werrett D. J. Forensic application of DNA 'fingerprints'. Nature. 1985 Dec 12;318(6046):577–579. doi: 10.1038/318577a0. [DOI] [PubMed] [Google Scholar]
- Hsu S. H., Schacter B. Z., Delaney N. L., Miller T. B., McKusick V. A., Kennett R. H., Bodmer J. G., Young D., Bodmer W. F. Genetic characteristics of the HeLa cell. Science. 1976 Jan 30;191(4225):392–394. doi: 10.1126/science.1246620. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Brookfield J. F., Semeonoff R. Positive identification of an immigration test-case using human DNA fingerprints. 1985 Oct 31-Nov 6Nature. 317(6040):818–819. doi: 10.1038/317818a0. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Morton D. B. DNA fingerprints of dogs and cats. Anim Genet. 1987;18(1):1–15. doi: 10.1111/j.1365-2052.1987.tb00739.x. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Wilson V., Thein S. L. Hypervariable 'minisatellite' regions in human DNA. Nature. 1985 Mar 7;314(6006):67–73. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- Jones H. W., Jr, McKusick V. A., Harper P. S., Wuu K. D. George Otto Gey. (1899-1970). The HeLa cell and a reappraisal of its origin. Obstet Gynecol. 1971 Dec;38(6):945–949. [PubMed] [Google Scholar]
- Lander E. S. DNA fingerprinting on trial. Nature. 1989 Jun 15;339(6225):501–505. doi: 10.1038/339501a0. [DOI] [PubMed] [Google Scholar]
- Lavappa K. S., Macy M. L., Shannon J. E. Examination of ATCC stocks for HeLa marker chromosomes in human cell lines. Nature. 1976 Jan 22;259(5540):211–213. doi: 10.1038/259211a0. [DOI] [PubMed] [Google Scholar]
- Lynch M. Estimation of relatedness by DNA fingerprinting. Mol Biol Evol. 1988 Sep;5(5):584–599. doi: 10.1093/oxfordjournals.molbev.a040518. [DOI] [PubMed] [Google Scholar]
- Mann D. L., O'Brien S. J., Gilbert D. A., Reid Y., Popovic M., Read-Connole E., Gallo R. C., Gazdar A. F. Origin of the HIV-susceptible human CD4+ cell line H9. AIDS Res Hum Retroviruses. 1989 Jun;5(3):253–255. doi: 10.1089/aid.1989.5.253. [DOI] [PubMed] [Google Scholar]
- Mann D. L., Popovic M., Sarin P., Murray C., Reitz M. S., Strong D. M., Haynes B. F., Gallo R. C., Blattner W. A. Cell lines producing human T-cell lymphoma virus show altered HLA expression. Nature. 1983 Sep 1;305(5929):58–60. doi: 10.1038/305058a0. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Leppert M., O'Connell P., Wolff R., Holm T., Culver M., Martin C., Fujimoto E., Hoff M., Kumlin E. Variable number of tandem repeat (VNTR) markers for human gene mapping. Science. 1987 Mar 27;235(4796):1616–1622. doi: 10.1126/science.3029872. [DOI] [PubMed] [Google Scholar]
- Nelson-Rees W. A., Daniels D. W., Flandermeyer R. R. Cross-contamination of cells in culture. Science. 1981 Apr 24;212(4493):446–452. doi: 10.1126/science.6451928. [DOI] [PubMed] [Google Scholar]
- Nelson-Rees W. A., Flandermeyer R. R., Hawthorne P. K. Banded marker chromosomes as indicators of intraspecies cellular contamination. Science. 1974 Jun 7;184(4141):1093–1096. doi: 10.1126/science.184.4141.1093. [DOI] [PubMed] [Google Scholar]
- Nelson-Rees W. A., Flandermeyer R. R. HeLa cultures defined. Science. 1976 Jan 9;191(4222):96–98. doi: 10.1126/science.1246601. [DOI] [PubMed] [Google Scholar]
- Nelson-Rees W. A., Hunter L., Darlington G. J., O'Brien S. J. Characteristics of HeLa strains: permanent vs. variable features. Cytogenet Cell Genet. 1980;27(4):216–231. doi: 10.1159/000131490. [DOI] [PubMed] [Google Scholar]
- O'Brien S. J., Shannon J. E., Gail M. H. A molecular approach to the identification and individualization of human and animal cells in culture: isozyme and allozyme genetic signatures. In Vitro. 1980 Feb;16(2):119–135. doi: 10.1007/BF02831503. [DOI] [PubMed] [Google Scholar]
- O'Brien S. U., Kleiner G., Olson R., Shannon J. E. Enzyme polymorphisms as genetic signatures in human cell cultures. Science. 1977 Mar 25;195(4284):1345–1348. doi: 10.1126/science.841332. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Gil A., Maniatis T. The structure of the human zeta-globin gene and a closely linked, nearly identical pseudogene. Cell. 1982 Dec;31(3 Pt 2):553–563. doi: 10.1016/0092-8674(82)90311-7. [DOI] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeders S. T., Breuning M. H., Davies K. E., Nicholls R. D., Jarman A. P., Higgs D. R., Pearson P. L., Weatherall D. J. A highly polymorphic DNA marker linked to adult polycystic kidney disease on chromosome 16. Nature. 1985 Oct 10;317(6037):542–544. doi: 10.1038/317542a0. [DOI] [PubMed] [Google Scholar]
- Stoker N. G., Cheah K. S., Griffin J. R., Pope F. M., Solomon E. A highly polymorphic region 3' to the human type II collagen gene. Nucleic Acids Res. 1985 Jul 11;13(13):4613–4622. doi: 10.1093/nar/13.13.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker J., Webb M. B., Debenham P. G. Fingerprinting cell lines: use of human hypervariable DNA probes to characterize mammalian cell cultures. Somat Cell Mol Genet. 1988 Nov;14(6):519–525. doi: 10.1007/BF01535307. [DOI] [PubMed] [Google Scholar]
- Wetton J. H., Carter R. E., Parkin D. T., Walters D. Demographic study of a wild house sparrow population by DNA fingerprinting. Nature. 1987 May 14;327(6118):147–149. doi: 10.1038/327147a0. [DOI] [PubMed] [Google Scholar]
- Wong Z., Wilson V., Jeffreys A. J., Thein S. L. Cloning a selected fragment from a human DNA 'fingerprint': isolation of an extremely polymorphic minisatellite. Nucleic Acids Res. 1986 Jun 11;14(11):4605–4616. doi: 10.1093/nar/14.11.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman A. R., White R. A highly polymorphic locus in human DNA. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6754–6758. doi: 10.1073/pnas.77.11.6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden P. D., Wiid I. J., Albrecht C. F., Theron E., Thornley A. L., Hoal-van Helden E. G. Cross-contamination of human esophageal squamous carcinoma cell lines detected by DNA fingerprint analysis. Cancer Res. 1988 Oct 15;48(20):5660–5662. [PubMed] [Google Scholar]



