Abstract
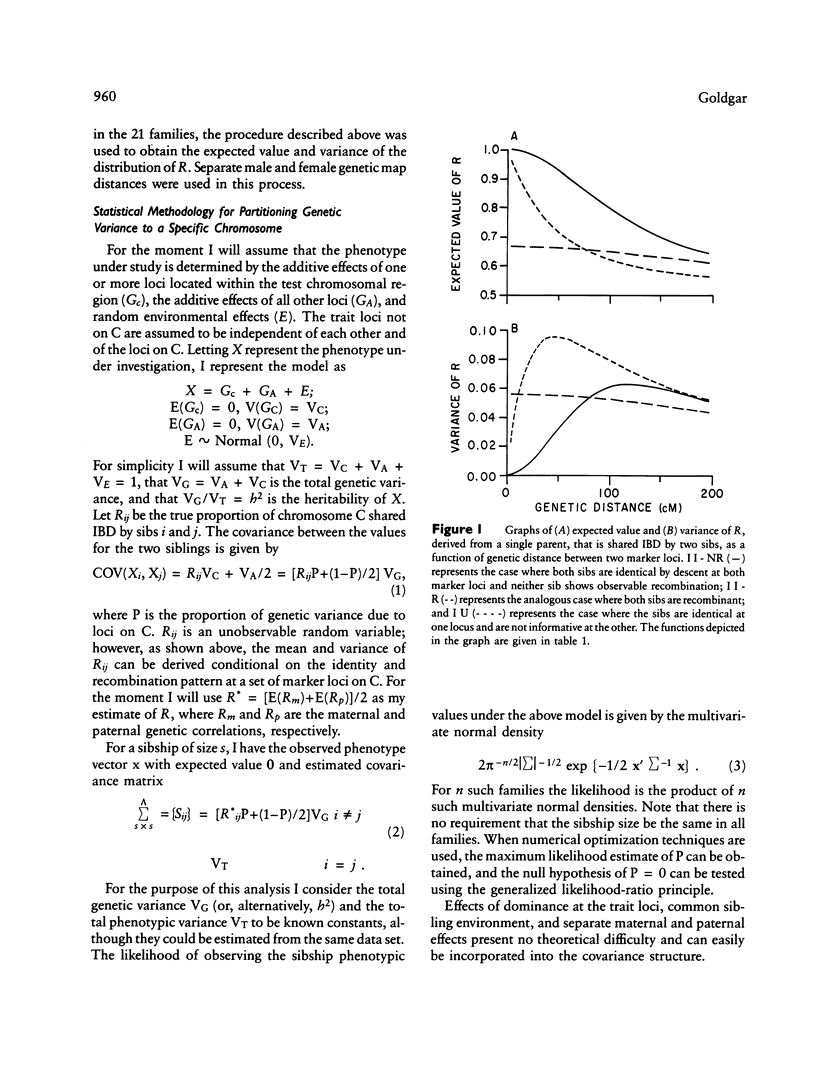
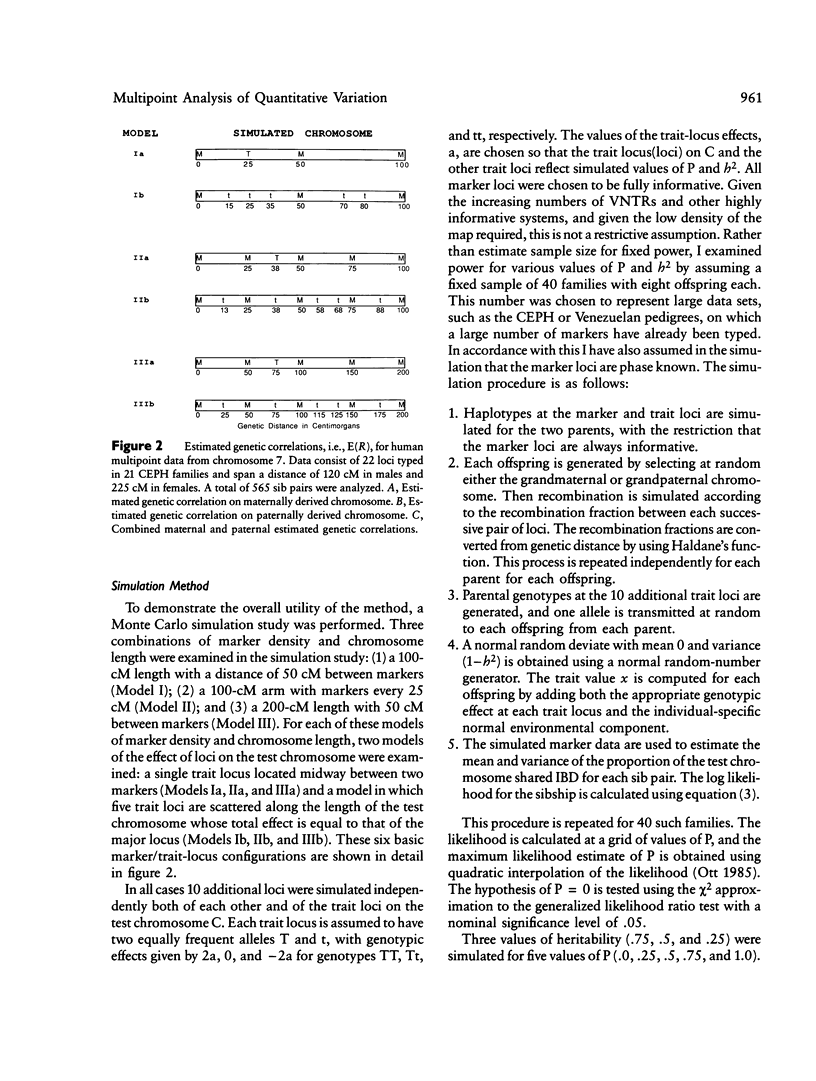
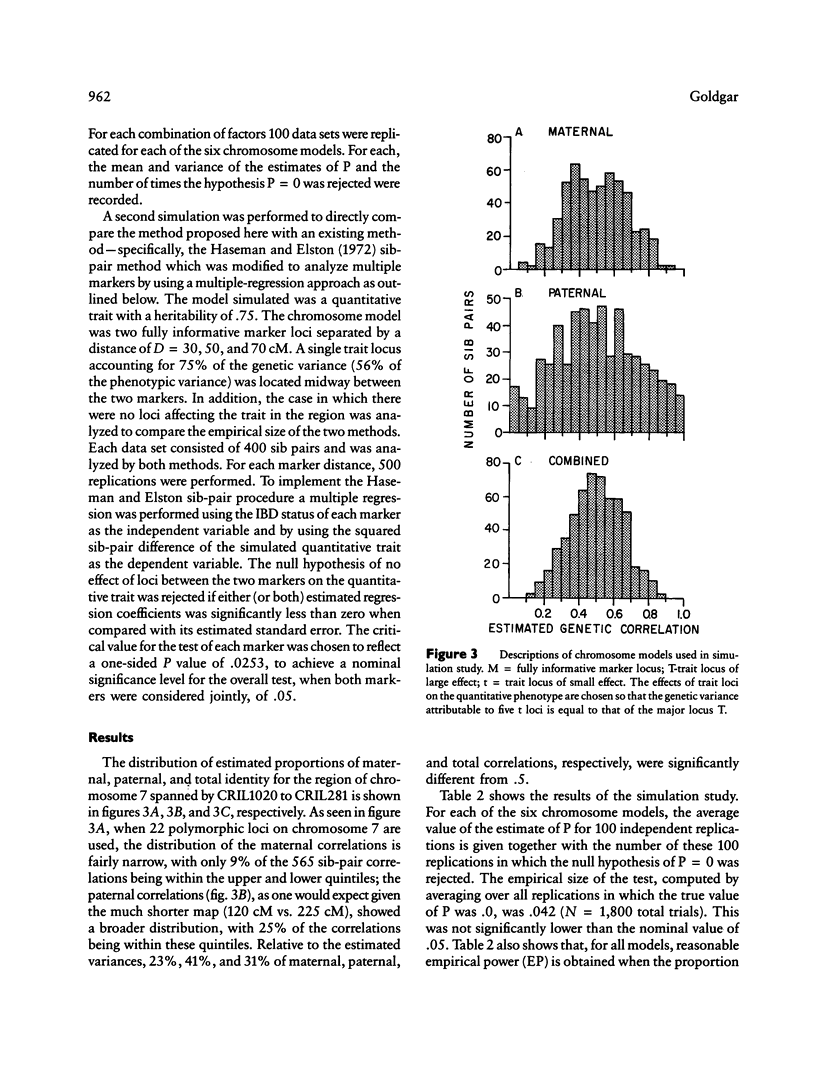
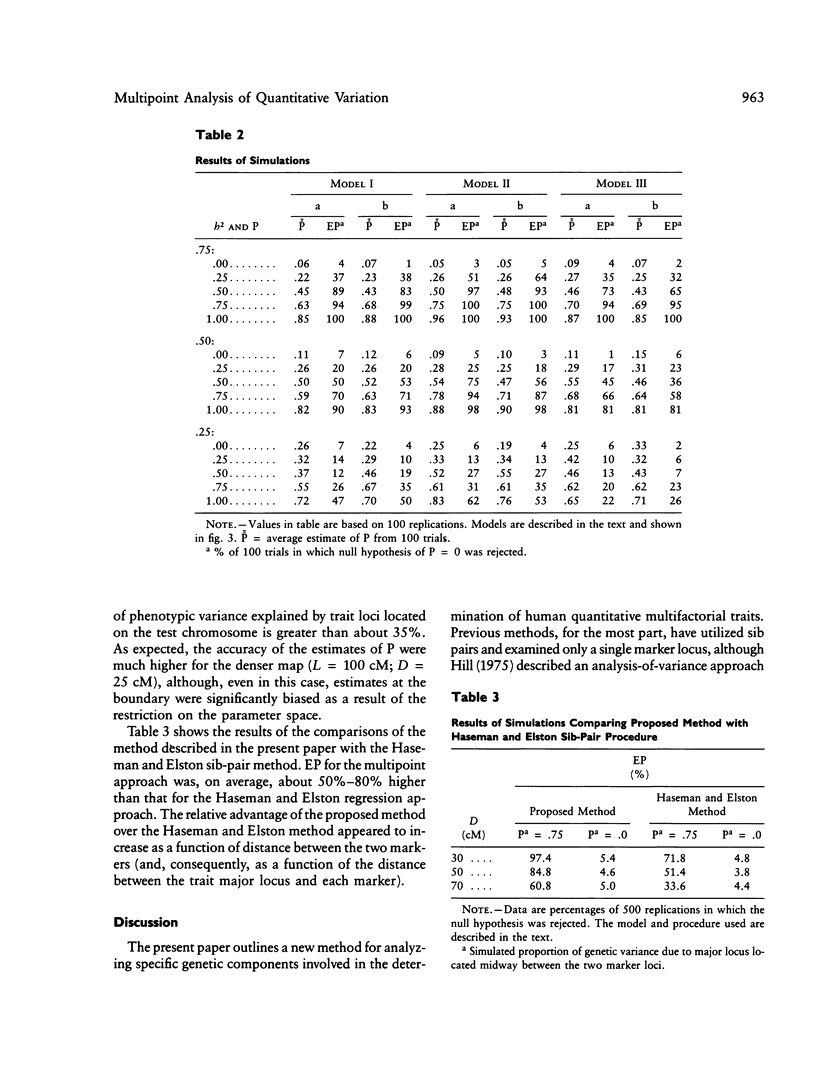
A unique method of partitioning human quantitative genetic variation into effects due to specific chromosomal regions is presented. This method is based on estimating the proportion of genetic material, R, shared identical by descent (IBD) by sibling pairs in a specified chromosomal region, on the basis of their marker genotypes at a set of marker loci spanning the region. The mean and variance of the distribution of R conditional on IBD status and recombination pattern between two marker loci are derived as a function of the distance between the two loci. The distribution of the estimates of R is exemplified using data on 22 loci on chromosome 7. A method of using the estimated R values and observed values of a quantitative trait in a set of sibships to estimate the proportion of total genetic variance explained by loci in the region of interest is presented. Monte Carlo simulation techniques are used to show that this method is more powerful than existing methods of quantitative linkage analysis based on sib pairs. It is also shown through simulation studies that the proposed method is sensitive to genetic variation arising from both a single locus of large effect as well as from several loosely linked loci of moderate phenotypic effect.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amos C. I., Elston R. C., Wilson A. F., Bailey-Wilson J. E. A more powerful robust sib-pair test of linkage for quantitative traits. Genet Epidemiol. 1989;6(3):435–449. doi: 10.1002/gepi.1370060306. [DOI] [PubMed] [Google Scholar]
- Barker D., Green P., Knowlton R., Schumm J., Lander E., Oliphant A., Willard H., Akots G., Brown V., Gravius T. Genetic linkage map of human chromosome 7 with 63 DNA markers. Proc Natl Acad Sci U S A. 1987 Nov;84(22):8006–8010. doi: 10.1073/pnas.84.22.8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H., Green P., Helms C., Cartinhour S., Weiffenbach B., Stephens K., Keith T. P., Bowden D. W., Smith D. R., Lander E. S. A genetic linkage map of the human genome. Cell. 1987 Oct 23;51(2):319–337. doi: 10.1016/0092-8674(87)90158-9. [DOI] [PubMed] [Google Scholar]
- GIBSON J. B., THODAY J. M. Effects of disruptive selection. VI. A second chromosome polymorphism. Heredity (Edinb) 1962 Feb;17:1–26. doi: 10.1038/hdy.1962.1. [DOI] [PubMed] [Google Scholar]
- HARRISON B. J., MATHER K. Polygenic variability in chromosomes of Drosophila melanogaster obtained from the wild. Heredity (Edinb) 1950 Dec;4(3):295–312. doi: 10.1038/hdy.1950.23. [DOI] [PubMed] [Google Scholar]
- Haseman J. K., Elston R. C. The investigation of linkage between a quantitative trait and a marker locus. Behav Genet. 1972 Mar;2(1):3–19. doi: 10.1007/BF01066731. [DOI] [PubMed] [Google Scholar]
- Hill A. P. Quantitative linkage: a statistical procedure for its detection and estimation. Ann Hum Genet. 1975 May;38(4):439–449. doi: 10.1111/j.1469-1809.1975.tb00633.x. [DOI] [PubMed] [Google Scholar]
- Lander E. S., Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989 Jan;121(1):185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N., Lange K. Application of a recombination model in calculating the variance of sib pair genetic identity. Ann Hum Genet. 1979 Oct;43(2):177–186. doi: 10.1111/j.1469-1809.1979.tb02010.x. [DOI] [PubMed] [Google Scholar]
- Suarez B. K., Reich T., Fishman P. M. Variability in sib pair genetic identity. Hum Hered. 1979;29(1):37–41. doi: 10.1159/000153013. [DOI] [PubMed] [Google Scholar]


