Abstract
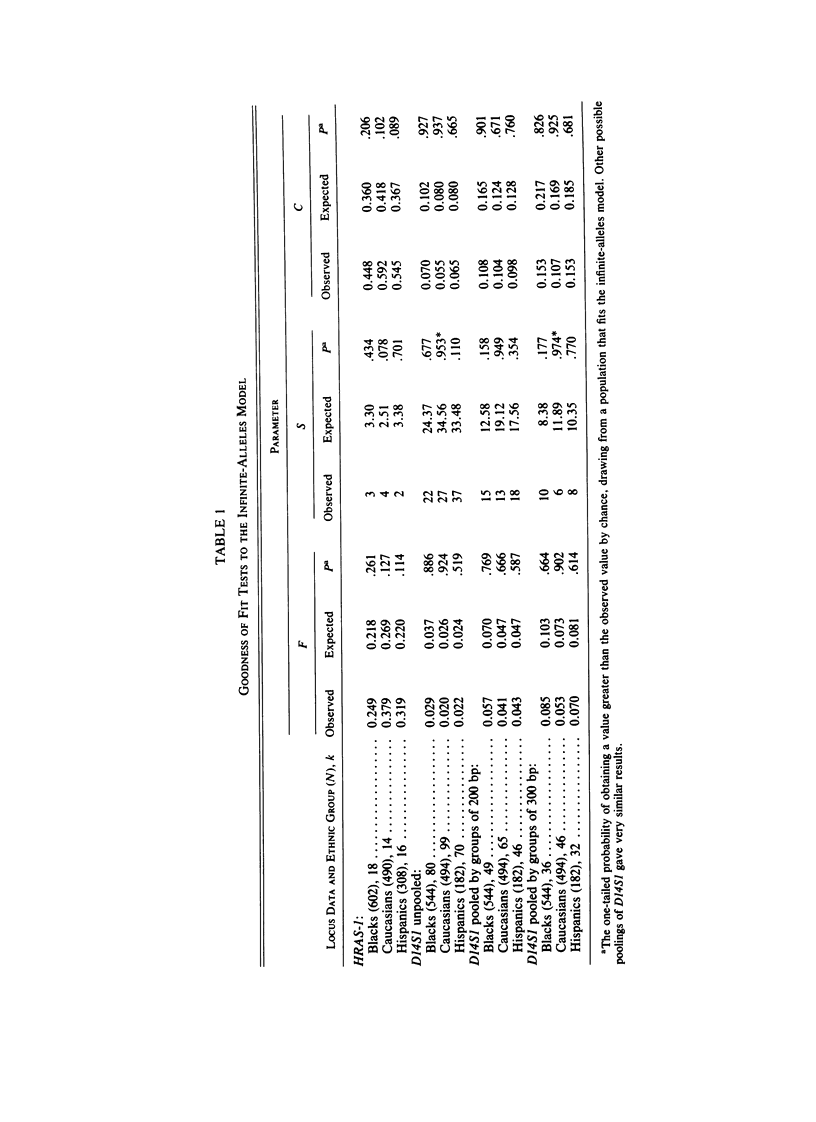
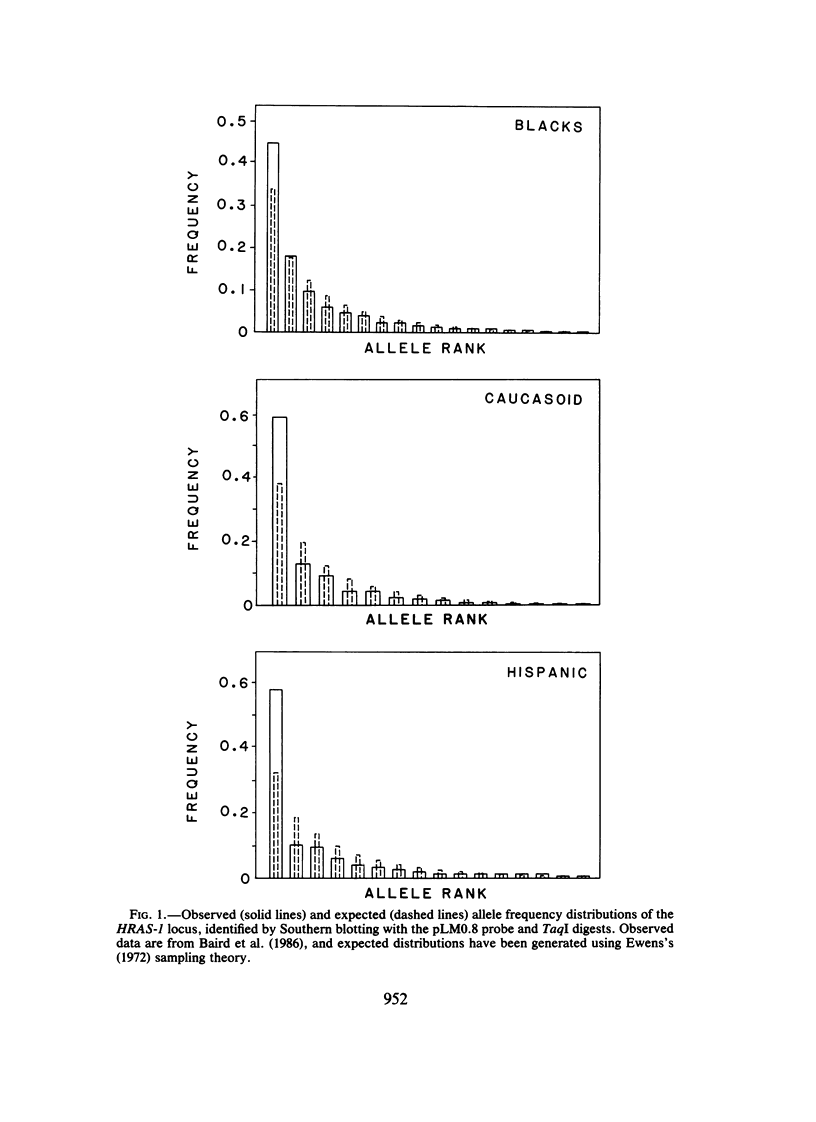
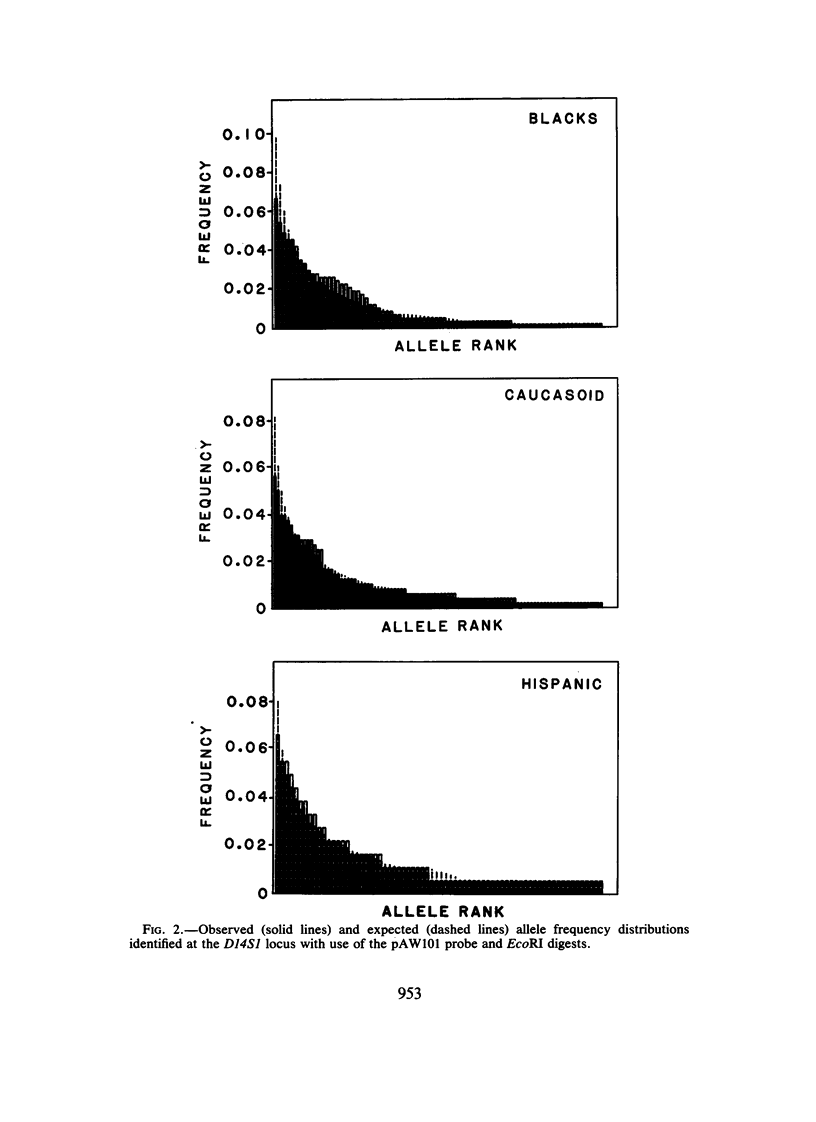
The allele frequency data of Baird et al. were tested using Ewens-Watterson sampling theory for goodness of fit to the infinite-alleles model of neutral evolution. Although probes of both the HRAS-1 and D14S1 loci identify highly diverse restriction-fragment-length polymorphisms, the observed values of gene identity (F) and the common allele frequency (C) are not significantly different from the neutral expectation. Allele frequency distributions show a tendency toward a deficit in diversity for HRAS-1 and a slight excess diversity for D14S1. The direction of these departures is consistent with potential selective effects of the Harvey-ras oncogene and hitchhiking of the D14S1 locus to closely linked immunoglobulin genes. Direct chi 2-tests of goodness of fit of the observed and expected allele frequency distributions reveal significant departures in the caucasoid and Hispanic HRAS-1 distributions but not in any of the other tests.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baird M., Balazs I., Giusti A., Miyazaki L., Nicholas L., Wexler K., Kanter E., Glassberg J., Allen F., Rubinstein P. Allele frequency distribution of two highly polymorphic DNA sequences in three ethnic groups and its application to the determination of paternity. Am J Hum Genet. 1986 Oct;39(4):489–501. [PMC free article] [PubMed] [Google Scholar]
- Balazs I., Purrello M., Rubinstein P., Alhadeff B., Siniscalco M. Highly polymorphic DNA site D14S1 maps to the region of Burkitt lymphoma translocation and is closely linked to the heavy chain gamma 1 immunoglobulin locus. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7395–7399. doi: 10.1073/pnas.79.23.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capon D. J., Chen E. Y., Levinson A. D., Seeburg P. H., Goeddel D. V. Complete nucleotide sequences of the T24 human bladder carcinoma oncogene and its normal homologue. Nature. 1983 Mar 3;302(5903):33–37. doi: 10.1038/302033a0. [DOI] [PubMed] [Google Scholar]
- Ewens W. J. The sampling theory of selectively neutral alleles. Theor Popul Biol. 1972 Mar;3(1):87–112. doi: 10.1016/0040-5809(72)90035-4. [DOI] [PubMed] [Google Scholar]
- Fuerst P. A., Chakraborty R., Nei M. Statistical studies on protein polymorphism in natural populations. I. Distribution of single locus heterozygosity. Genetics. 1977 Jun;86(2 Pt 1):455–483. [PMC free article] [PubMed] [Google Scholar]
- Gerald P. S., Grzeschik K. H. Report of the Committee on the Genetic Constitution of Chromosomes 10, 11, and 12. Cytogenet Cell Genet. 1984;37(1-4):103–126. doi: 10.1159/000132006. [DOI] [PubMed] [Google Scholar]
- Hedrick P. W., Thomson G. Evidence for balancing selection at HLA. Genetics. 1983 Jul;104(3):449–456. doi: 10.1093/genetics/104.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith T. P., Brooks L. D., Lewontin R. C., Martinez-Cruzado J. C., Rigby D. L. Nearly identical allelic distributions of xanthine dehydrogenase in two populations of Drosophila pseudoobscura. Mol Biol Evol. 1985 May;2(3):206–216. doi: 10.1093/oxfordjournals.molbev.a040348. [DOI] [PubMed] [Google Scholar]
- Keith T. P. Frequency Distribution of Esterase-5 Alleles in Two Populations of DROSOPHILA PSEUDOOBSCURA. Genetics. 1983 Sep;105(1):135–155. doi: 10.1093/genetics/105.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klitz W., Thomson G., Baur M. P. Contrasting evolutionary histories among tightly linked HLA loci. Am J Hum Genet. 1986 Sep;39(3):340–349. [PMC free article] [PubMed] [Google Scholar]
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978 Jul;89(3):583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson G. The effect of a selected locus on linked neutral loci. Genetics. 1977 Apr;85(4):753–788. doi: 10.1093/genetics/85.4.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G. A. An analysis of multi-allelic data. Genetics. 1978 Jan;88(1):171–179. doi: 10.1093/genetics/88.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittam T. S., Clark A. G., Stoneking M., Cann R. L., Wilson A. C. Allelic variation in human mitochondrial genes based on patterns of restriction site polymorphism. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9611–9615. doi: 10.1073/pnas.83.24.9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman A. R., White R. A highly polymorphic locus in human DNA. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6754–6758. doi: 10.1073/pnas.77.11.6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Martinville B., Giacalone J., Shih C., Weinberg R. A., Francke U. Oncogene from human EJ bladder carcinoma is located on the short arm of chromosome 11. Science. 1983 Feb 4;219(4584):498–501. doi: 10.1126/science.6297001. [DOI] [PubMed] [Google Scholar]


