Abstract
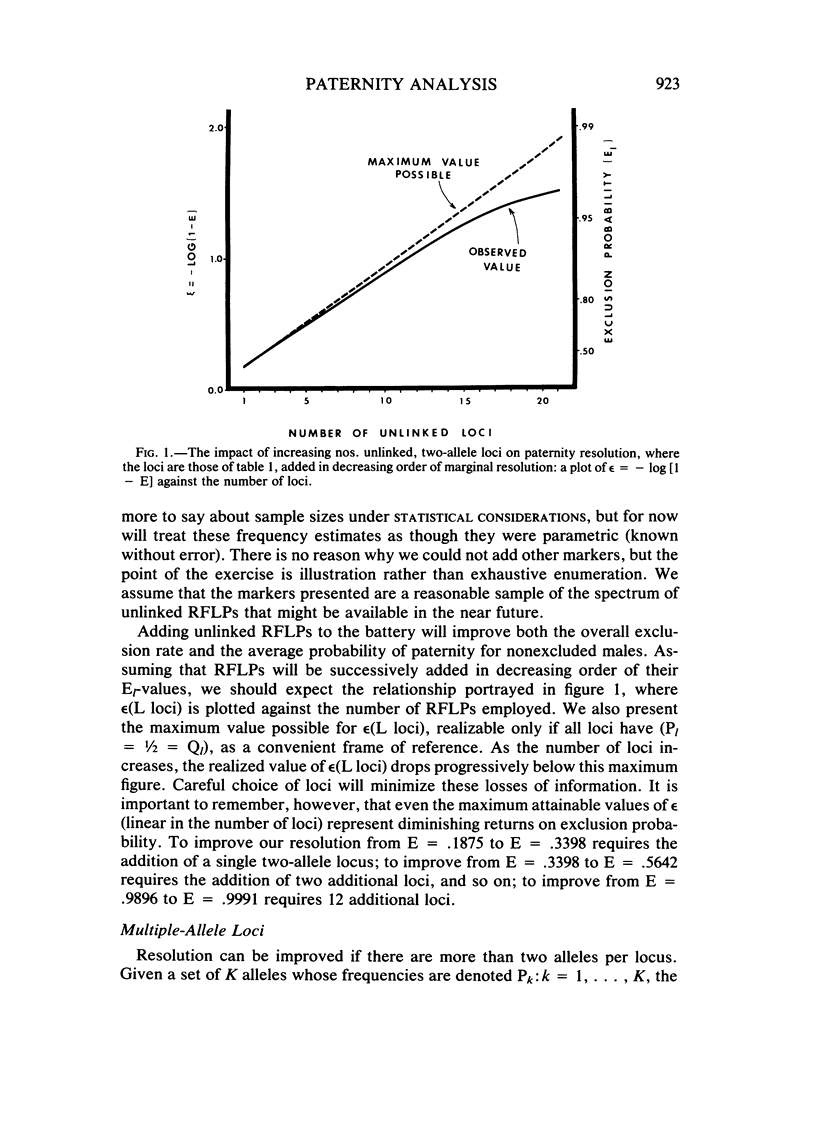
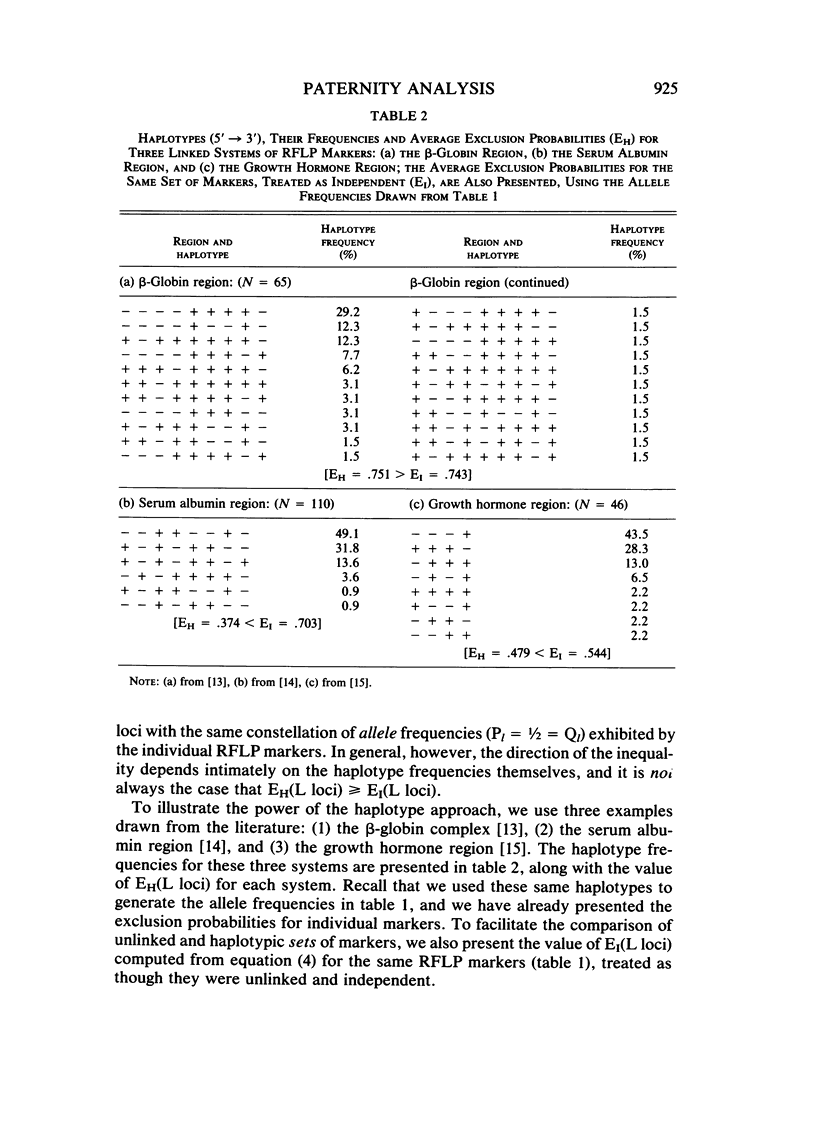
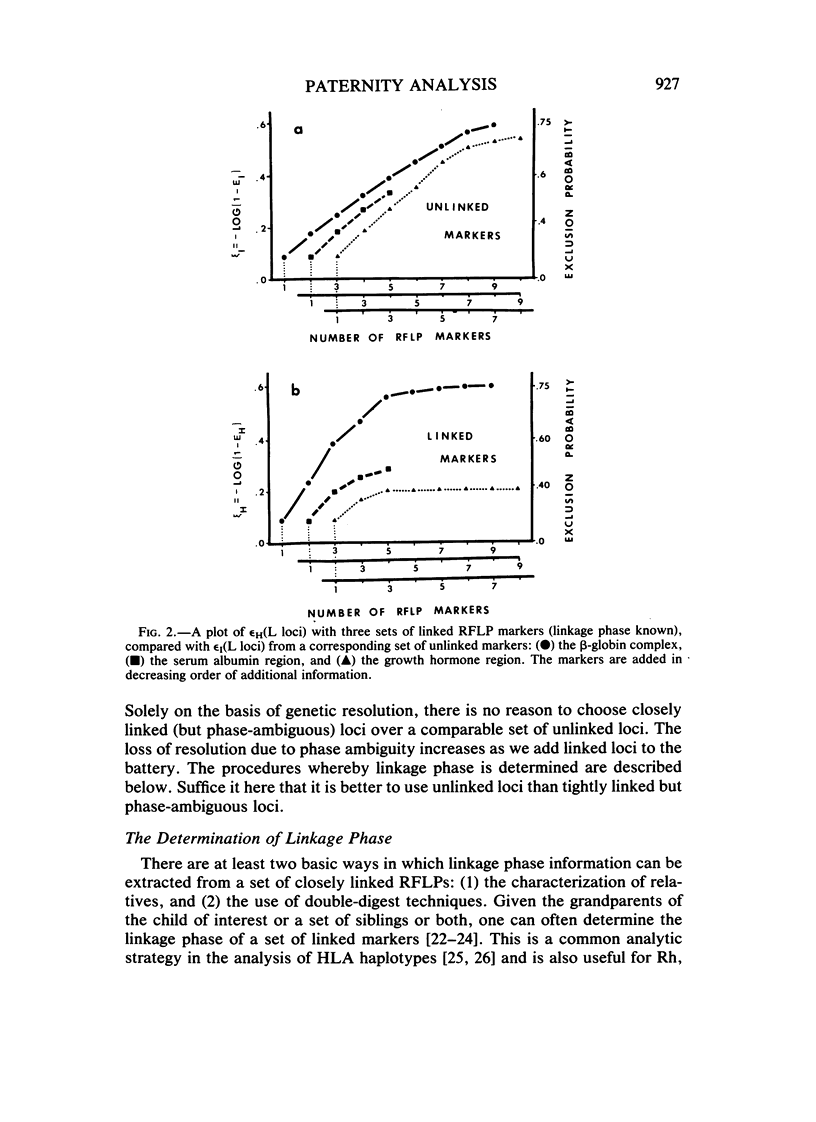
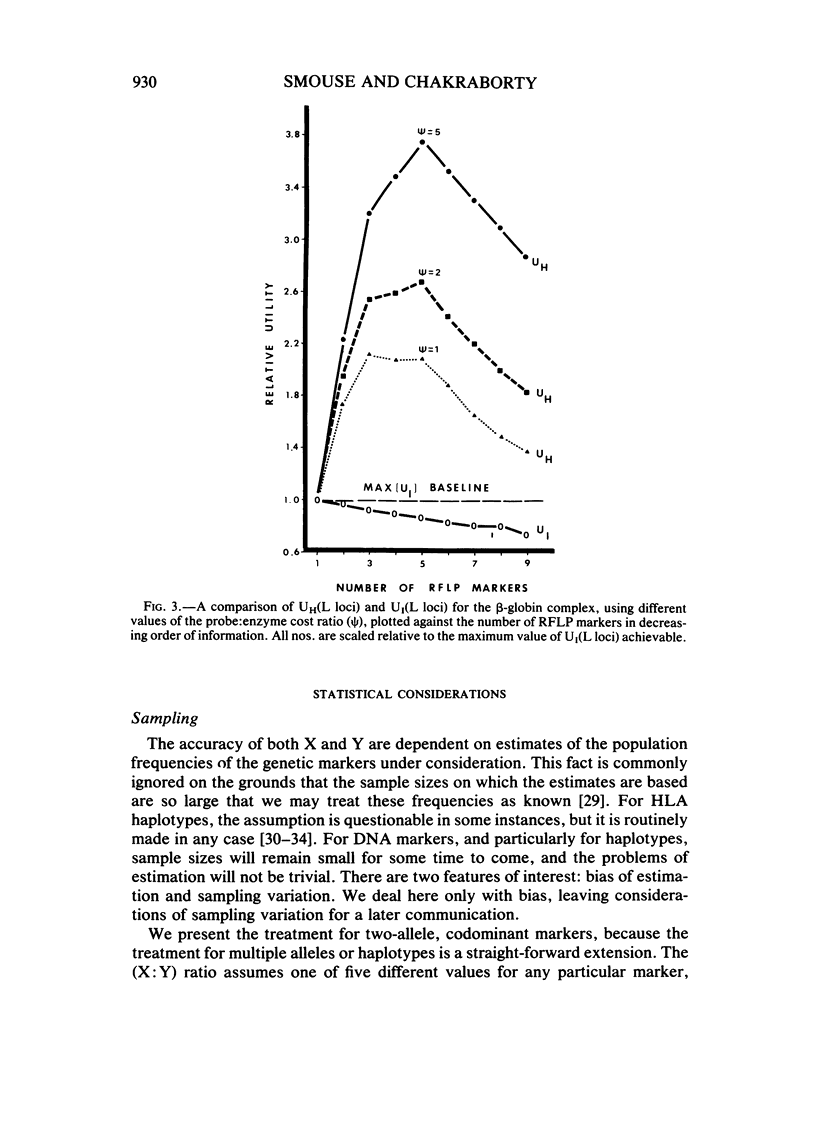
This paper examines the utility of restriction fragment length polymorphisms (RFLPs) for paternity analysis. While, on the average, 99% of falsely accused males can be excluded with the standard battery of blood group antigens, red cell enzymes, serum proteins, and HLA antigens, there are still mother-child pairs for whom the exclusion probability is not high. It has been suggested that additional resolution would be available with RFLPs. We have examined the strategic aspects of using RFLPs for paternity analysis, comparing the efficacy and cost of a multimarker haplotypic set with those of a comparable set of unlinked RFLPs, using published frequencies for the beta-globin complex, the serum albumin region, and the growth hormone region. There are four major findings. (1) Greater resolution is obtained with a carefully chosen set of tightly linked RFLPs producing chromosomal haplotypes than with a comparable set (same allele frequencies) of unlinked markers, but only if it is possible to establish linkage phase unambiguously. (2) Assay of linked sets is cheaper than is the assay of unlinked markers, but the cost advantage is optimized with sets of no more than two or three linked markers. (3) Also, with more than two or three tightly linked markers, the haplotypic frequencies are too poorly estimated to provide a reliable measure of the probability of paternity for unexcluded males, given the sample sizes likely to be available in the near future. (4) Optimal resolution, minimal cost, and acceptable accuracy are obtained with several independent sets of no more than two or three tightly linked RFLP markers each. With current technology, RFLP analysis is more expensive for the same level of genetic resolution than is the standard battery, but gradual replacement of the latter can be anticipated as economies of scale reduce the cost of the DNA technology.
Full text
PDF
















Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baur M. P., Mayr W. R., Rittner C. Algorithm for the computation of plausibilities of paternity in the HLA system. Z Immunitatsforsch Immunobiol. 1976 Nov;152(3):209–219. [PubMed] [Google Scholar]
- Bech-Hansen N. T., Linsley P. S., Cox D. W. Restriction fragment length polymorphisms associated with immunoglobulin C gamma genes reveal linkage disequilibrium and genomic organization. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6952–6956. doi: 10.1073/pnas.80.22.6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. I., Karam J. H., Rutter W. J. Polymorphic DNA region adjacent to the 5' end of the human insulin gene. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5759–5763. doi: 10.1073/pnas.78.9.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R., Ferrell R. E. Correlation of paternity index with probability of exclusion and efficiency criteria of genetic markers for paternity testing. Forensic Sci Int. 1982 Mar-Apr;19(2):113–124. doi: 10.1016/0379-0738(82)90038-x. [DOI] [PubMed] [Google Scholar]
- Chakraborty R., Hedrick P. W. Paternity exclusion and the paternity index for two linked loci. Hum Hered. 1983;33(1):13–23. doi: 10.1159/000153341. [DOI] [PubMed] [Google Scholar]
- Chakraborty R., Ryman N. Use of odds of paternity computations in determining the reliability of single exclusions in paternity testing. Hum Hered. 1981;31(6):363–369. doi: 10.1159/000153239. [DOI] [PubMed] [Google Scholar]
- Chakraborty R., Shaw M., Schull W. J. Exclusion of paternity: the current state of the art. Am J Hum Genet. 1974 Jul;26(4):477–488. [PMC free article] [PubMed] [Google Scholar]
- Chakravarti A., Buetow K. H. A strategy for using multiple linked markers for genetic counseling. Am J Hum Genet. 1985 Sep;37(5):984–997. [PMC free article] [PubMed] [Google Scholar]
- Chakravarti A., Buetow K. H., Antonarakis S. E., Waber P. G., Boehm C. D., Kazazian H. H. Nonuniform recombination within the human beta-globin gene cluster. Am J Hum Genet. 1984 Nov;36(6):1239–1258. [PMC free article] [PubMed] [Google Scholar]
- Chakravarti A., Nei M. Utility and efficiency of linked marker genes for genetic counseling. II. Identification of linkage phase by offspring phenotypes. Am J Hum Genet. 1982 Jul;34(4):531–551. [PMC free article] [PubMed] [Google Scholar]
- Chakravarti A., Phillips J. A., 3rd, Mellits K. H., Buetow K. H., Seeburg P. H. Patterns of polymorphism and linkage disequilibrium suggest independent origins of the human growth hormone gene cluster. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6085–6089. doi: 10.1073/pnas.81.19.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper D. N., Schmidtke J. DNA restriction fragment length polymorphisms and heterozygosity in the human genome. Hum Genet. 1984;66(1):1–16. doi: 10.1007/BF00275182. [DOI] [PubMed] [Google Scholar]
- Goodbourn S. E., Higgs D. R., Clegg J. B., Weatherall D. J. Molecular basis of length polymorphism in the human zeta-globin gene complex. Proc Natl Acad Sci U S A. 1983 Aug;80(16):5022–5026. doi: 10.1073/pnas.80.16.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A. J., Wilson V., Thein S. L. Hypervariable 'minisatellite' regions in human DNA. Nature. 1985 Mar 7;314(6006):67–73. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- Mayr W. R. Grundlagen zur Berechnung der Vaterschaftswahrscheinlichkeit im HL-A-System. Z Immunitatsforsch Exp Klin Immunol. 1972 Jul;144(1):18–27. [PubMed] [Google Scholar]
- Mayr W. R., Pausch V. Die Berechnung der Vaterschaftsausschlusschance im HL-A-System. Z Immunitatsforsch Exp Klin Immunol. 1975 Nov;150(5):447–457. [PubMed] [Google Scholar]
- Migone N., Feder J., Cann H., van West B., Hwang J., Takahashi N., Honjo T., Piazza A., Cavalli-Sforza L. L. Multiple DNA fragment polymorphisms associated with immunoglobulin mu chain switch-like regions in man. Proc Natl Acad Sci U S A. 1983 Jan;80(2):467–471. doi: 10.1073/pnas.80.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrenweiser H. W. Enzyme-deficiency variants: frequency and potential significance in human populations. Isozymes Curr Top Biol Med Res. 1983;10:51–68. [PubMed] [Google Scholar]
- Morgan K., Holmes T. M., Schlaut J., Marchuk L., Kovithavongs T., Pazderka F., Dossetor J. B. Genetic variability of HLA in the Dariusleut Hutterites. A comparative genetic analysis of the Hutterities, the Amish, and other selected Caucasian populations. Am J Hum Genet. 1980 Mar;32(2):246–257. [PMC free article] [PubMed] [Google Scholar]
- Murray J. C., Mills K. A., Demopulos C. M., Hornung S., Motulsky A. G. Linkage disequilibrium and evolutionary relationships of DNA variants (restriction enzyme fragment length polymorphisms) at the serum albumin locus. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3486–3490. doi: 10.1073/pnas.81.11.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel J. V. The detection of increased mutation rates in human populations. Perspect Biol Med. 1971;14(4):522–537. doi: 10.1353/pbm.1971.0043. [DOI] [PubMed] [Google Scholar]
- Neel J. V., Ueda N., Satoh C., Ferrell R. E., Tanis R. J., Hamilton H. B. The frequency in Japanese of genetic variants of 22 proteins. V. Summary and comparison with data on Caucasians from the British Isles. Ann Hum Genet. 1978 May;41(4):429–441. doi: 10.1111/j.1469-1809.1978.tb00913.x. [DOI] [PubMed] [Google Scholar]
- Nei M. Proportion of informative families for genetic counseling with linked marker genes. Jinrui Idengaku Zasshi. 1979 Sep;24(3):131–142. doi: 10.1007/BF01888684. [DOI] [PubMed] [Google Scholar]
- Rothman E. D., Neel J. V., Hoppe F. M. Assigning a probability for paternity in apparent cases of mutation. Am J Hum Genet. 1981 Jul;33(4):617–628. [PMC free article] [PubMed] [Google Scholar]
- Salmon D., Salmon C. Blood groups and genetic markers polymorphism and probability of paternity. Transfusion. 1980 Nov-Dec;20(6):684–694. doi: 10.1046/j.1537-2995.1980.20681057158.x. [DOI] [PubMed] [Google Scholar]
- Selvin S. Probability of nonpaternity determined by multiple allele codominant systems. Am J Hum Genet. 1980 Mar;32(2):276–278. [PMC free article] [PubMed] [Google Scholar]
- Wyman A. R., White R. A highly polymorphic locus in human DNA. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6754–6758. doi: 10.1073/pnas.77.11.6754. [DOI] [PMC free article] [PubMed] [Google Scholar]


