Abstract
The GTPase elongation factor (EF)-G is responsible for promoting the translocation of the messenger RNA–transfer RNA complex on the ribosome, thus opening up the A site for the next aminoacyl-tRNA. Chemical modification and cryo-EM studies have indicated that tRNAs can bind the ribosome in an alternative ‘hybrid’ state after peptidyl transfer and before translocation, though the relevance of this state during translation elongation has been a subject of debate. Here, using pre–steady-state kinetic approaches and mutant analysis, we show that translocation by EF-G is most efficient when tRNAs are bound in a hybrid state, supporting the argument that this state is an authentic intermediate during translation.
Translation elongation is the multistep process performed by the ribosome to sequentially add mRNA-encoded amino acids to the growing polypeptide chain. There are three iterated steps performed by the ribosome during the elongation cycle: (i) a tRNA-selection step in which the ribosome and elongation factor (EF)-Tu select the next aminoacyl-tRNA to enter the cycle; (ii) peptide-bond formation catalyzed in the active site of the large ribosomal subunit; and (iii) translocation of the mRNA–tRNA complex through the subunit interface region, facilitated by EF-G, with associated GTP hydrolysis. The tRNA substrates are at the heart of each of these steps, where they have key functional roles1–3.
Early studies on the ribosome identified binding sites for two tRNA substrates, the A site for the aminoacyl-tRNA and the P site for the peptidyl-tRNA. Further biochemistry eventually identified a third site, the E or exit site, where deacylated tRNAs bind after loss of the peptidyl moiety and before release into solution4. An understanding of the order and timing with which tRNA substrates occupy these three sites (on both the large and small subunits) during the elongation cycle is central to a detailed molecular view of the process of translation.
The hybrid state of tRNA binding on the ribosome was first proposed in the 1960s as an elegant way to understand how controlled tRNA movement through the ribosome might be accomplished5. The basic feature of such a hybrid state of binding is that tRNAs would move independently with respect to the two subunits of the ribosome during the steps of the translation cycle. Fluorescence studies provided early biochemical clues that such a state of tRNA binding might be populated during translation6, and subsequent chemical-modification analysis provided clear and detailed experimental support for the hybrid state7. These studies have suggested that after peptide-bond formation, the A- and P-site tRNAs occupy an intermediate state on the ribosome, where the acceptor ends of the tRNAs have moved with respect to the large ribosomal subunit, but the anticodon ends remain in their original positions relative to the small ribosomal subunit (a hybrid state; Fig. 1a). Thus, ‘hybrid-bound’ tRNAs are described as binding in P/E and A/P states (where the first letter represents the binding site on the small subunit and the second letter the site on the large subunit). Complete translocation of the mRNA–tRNA complex is then catalyzed by EF-G with associated GTP hydrolysis.
Figure 1.
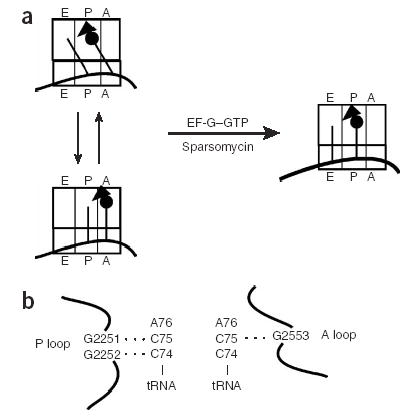
Schematic drawing of relationship between pre- and post-translocation state ribosomes and of known rRNA–tRNA pairing interactions. (a) Model of tRNA sampling between hybrid and classic state of tRNA binding before EF-G– or sparsomycin-mediated translocation. (b) Scheme of Watson-Crick base-pairing interactions between the P and A loops of the 23S rRNA and the CCA ends of tRNAs bound in the P and A site.
High-resolution crystal structures of the 70S ribosome have provided detailed molecular information on three tRNAs bound in classic (that is, nonhybrid) states8. Cryo-EM studies have provided suggestive views of hybrid-bound tRNAs on the ribosome9,10. Kinetic studies have also argued for the relevance of such a hybrid intermediate during elongation. For example, studies examining the role of the terminal adenosine nucleotide of a deacylated P-site tRNA have concluded that its interactions with the large-subunit E site (that is, binding of the deacylated tRNA to the P/E state) are crucial for translocation efficiency11. Semenkov et al.12 have compared the behavior of two distinct tRNA substrates, formylmethionyl (fMet)-Phe-tRNAPhe and Phe-tRNAPhe, when bound to the A site of pre-translocation-state ribosomes (the P site was filled with deacylated tRNA). The premise of the studies by Semenkov et al. hinges on the argument that the dipeptidyl-tRNA species favors the hybrid state of tRNA binding (A/P), whereas the aminoacyl-tRNA favors the classical state (A/A); our independent subsequent studies have supported these assignments13. The conclusion of these studies was that the formation of the hybrid state makes important energetic contributions to translocation12. Nevertheless, key issues still remain. For example, the aminoacyl-tRNA species evaluated in the original study is not a physiologically relevant one: translocation follows peptidyl transfer, so there is minimally a dipeptidyl-tRNA in a pretranslocation complex. How can we compare the behavior of two such different substrates? Moreover, what is the magnitude of the contribution to translocation catalysis of the hybrid binding state? Here we address these specific concerns by manipulating different tRNA–ribosomal RNA base-pairing interactions to form well-defined hybrid and classical pretranslocation complexes (using the well-studied Escherichia coli translation system), and we study the kinetic properties of these complexes in several different translocation assays.
RESULTS
General strategy for binding tRNAs to the ribosome
The goal of this study was to analyze the rate and efficiency of translocation of tRNA–ribosome complexes in different, well-defined states. Biochemical and structural studies have previously identified specific Watson-Crick pairing interactions between the universally conserved elements of the 23S large-ribosomal-subunit rRNA, termed the P and A loops, and the CCA ends of tRNAs bound in the P and A sites, respectively (Fig. 1b). These well-defined rRNA-tRNA interactions specify whether tRNA is bound to the A or P site on the 50S subunit14–16. In vitro genetic studies have further established that canonical interactions between conserved guanosines in the rRNA and cytidines in the tRNA can be substituted with other Watson-Crick pairing partners with some success15,16. Here we take advantage of these functional substitutions in the tRNA and rRNA to drive binding of tRNA substrates to defined states on the ribosome. ‘Classically bound’ tRNAs bind in the P/P and A/A states, whereas in the alternative configuration, ‘hybrid-bound’ tRNAs bind in the P/E and A/P states. As ribosome complexes carrying a deacylated tRNA in the P site and essentially any form of tRNA in the A site (peptidyl, aminoacyl or deacylated) are recognized and translocated by EF-G, the studies herein seek to define whether EF-G–promoted translocation is more efficient on ribosomes carrying classically bound or hybrid-bound tRNAs.
Effects on sparsomycin-dependent translocation
As an initial approach, we constructed a set of rRNA and tRNA variants and evaluated their translocation competency in a simplified assay. Translocation, generally catalyzed by the GTPase EF-G, is also promoted by the peptidyl transfer–specific antibiotic sparsomycin17. Atomic-resolution structures of sparsomycin and a peptidyl-tRNA analog bound to the 50S subunit indicate that sparsomycin binds in the A site region and makes extensive contacts with the peptidyl-tRNA18. This view of sparsomycin readily explains how it stabilizes P-site tRNA interactions with the ribosome19 and further suggests that binding of sparsomycin stimulates translocation by occluding the A site and restricting the backward movement of the 3′ end of the peptidyl-tRNA20. According to such a model, for sparsomycin to promote translocation, the A site must be accessible for sparsomycin to bind (that is, the tRNAs should be bound in hybrid configurations). It follows that classically bound tRNAs would not allow sparsomycin to bind, and thus such complexes should be refractory to sparsomycin-catalyzed translocation. For our purposes, the assay provides a convenient approach for defining hybrid-bound ribosome–tRNA complexes using an easily accessible assay for translocation.
For the initial experiments, ribosome complexes were constructed using wild-type (WT) ribosomes (MRE600) bound to a defined mRNA species (m301) carrying tyrosine and phenylalanine codons in the P and A sites, respectively. Sequential binding of deacylated tRNATyr and N-acetylphenylalanyl (AcPhe)-tRNAPhe established a pretranslocation state on ribosomes that was recognized by EF-G to promote three-nucleotide movement of the mRNA–tRNA complex (Fig. 2a, lanes 1–5), as characterized by primer-extension analysis21. Pretranslocation complexes carrying two tRNA substrates showed a characteristic doublet pattern in the primer-extension analysis, as previously noted22. Incubation of these same ribosome complexes with sparsomycin promoted three-nucleotide movement equivalent to that previously reported17. Notably, the EF-G–promoted event predominantly yielded an unexpected upstream toeprint (Fig. 2a, asterisk), whereas the sparsomycin-promoted product was seen solely as the expected three-nucleotide movement downstream on the mRNA. Although this phenomenon has previously been referred to as inaccurate translocation23, it is more consistent with explanations invoking low complex stability and tRNA rebinding events that occur without the initial bias of ordered tRNA binding (discussed in ref. 17). Thus, in the sparsomycin-catalyzed reaction, peptidyl-tRNA binding is stabilized by sparsomycin19, whereas in the absence of sparsomycin, peptidyl-tRNA falls off the ribosome after EF-G–dependent translocation and rebinds in a preferred position on the mRNA.
Figure 2.
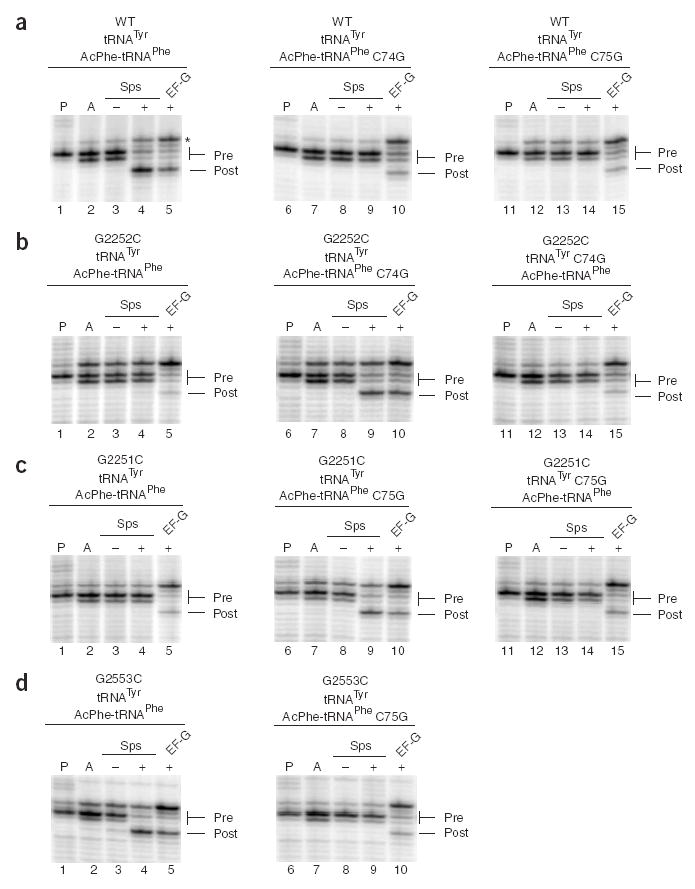
Toeprinting analysis of sparsomycin- and EF-G–mediated translocation on m301 messenger RNA. (a–d) Pretranslocation complexes were assembled by binding deacylated tRNATyr, tRNATyr C74G or tRNATyr C75G to the P site (P lanes), then binding AcPhe-tRNAPhe, AcPhe-tRNAPhe C74G or AcPhe-tRNAPhe C75G to the A site (A lanes). Complexes were incubated with sparsomycin (+Sps), 2.5% (v/v) DMSO (−Sps) or EF-G–GTP (+EF-G). Ribosomes used were either WT (a), G2252C (b), G2251C (c) or G2553C (d). Asterisk in a indicates the alternative, preferred binding site for AcPhe-tRNAPhe on ribosomes programmed with m301 mRNA.
To test the role of the tRNA binding state in translocation, we next looked at sparsomycin-catalyzed translocation in similar ribosome complexes with a series of tRNA and rRNA variants. tRNAPhe and tRNATyr CCA-end variants were prepared by in vitro transcription, aminoacylated and N-acetylated and HPLC-purified as previously described15, and variant ribosomes were prepared and purified using a recently described affinity-tagging procedure24. Substitutions in AcPhe-tRNAPhe such as C74G and C75G should disrupt pairing with the P loop of WT ribosomes and thus occupation of the hybrid state by these A-site tRNAs (Fig. 2a, lanes 6–15). Similarly, changes in the P loop of the ribosome (G2252C and G2251C) should prevent WT A-site tRNAs from occupying a hybrid state of binding. For both of these examples, the data showed that translocation by sparsomycin was inhibited (Fig. 2b,c, lanes 1–5). Mutations in the A loop are predicted to favor a hybrid state of tRNA binding (by disfavoring binding of the A-site tRNA to the A loop), and indeed, these changes had no impact on sparsomycin-mediated translocation (Fig. 2d, lanes 1–5). Similarly, changes in the P-site tRNA (deacylated tRNATyr) predicted to disfavor binding to the P loop (and thus to favor P/E binding) had no apparent affect on sparsomycin-dependent translocation (Supplementary Fig. 1 online). These data are consistent with a view where stable binding of A-site AcPhe-tRNAPhe to the P loop (that is, formation of the hybrid state) is essential for efficient sparsomycin-dependent translocation.
We next combined the rRNA and tRNA variants to examine whether compensatory changes that stabilize various pairing interactions affect translocation in a predictable fashion. As anticipated, when interactions between the P loop and the A-site tRNA, AcPhe-tRNAPhe, were restored by combining either G2252C ribosomes with C74G AcPhe-tRNAPhe or G2251C ribosomes with C75G AcPhe-tRNAPhe, sparsomycin-dependent translocation activity was restored (Fig. 2b,c, lanes 6–10). By contrast, complexes that stabilized favorable interactions between the P loop and the P-site deacylated tRNATyr (G2252C and C74G tRNATyr in Fig. 2b, lanes 11–15, or G2251 and C75G tRNATyr in Fig. 2c, lanes 11–15) or between the A loop and the A-site AcPhe-tRNAPhe (G2553C and C75G AcPhe-tRNAPhe in Fig. 2d, lanes 6–10) were not competent for sparsomycin-dependent translocation. Although complete data are shown only for certain rRNA and tRNA mutations, we were similarly able to rescue other nucleotide substitutions at position G2252 with compensatory mutant tRNAs in the A site (though not other G2251 substitutions, as previously noted25). These data are consistent with the view that sparsomycin-mediated translocation depends on the accessibility of the A site of the large ribosomal subunit and therefore on whether tRNAs bind the ribosome predominantly in a hybrid or classic configuration.
Sparsomycin-dependent translocation of tRNAMet species
Although sparsomycin was able to promote efficient translocation on the m301 mRNA described above, sparsomycin was unable to promote translocation on our standard gene32 mRNA loaded with deacylated tRNAfMet and AcPhe-tRNAPhe in the P and A sites, respectively (Fig. 3a, lanes 1–4). Previous hints in the literature led us to reason that unique sequence elements in tRNAfMet might favor binding of this specialized initiator tRNA in the classic (P/P) rather than the hybrid (P/E) state26, thus preventing sparsomycin-mediated translocation. Indeed, substitution of initiator tRNAfMet with elongator tRNAMet on the gene32 mRNA substantially increased the efficiency of sparsomycin-dependent translocation (Fig. 3a, lanes 5–8). In these experiments, both the elongator and initiator tRNAs were generated by in vitro transcription reactions27; in the case of the initiator transcript, we introduced a C1G substitution to increase the yield of the transcription reaction. In control experiments, the behaviors of the native and in vitro–transcribed initiator tRNAs were indistinguishable in the sparsomycin-dependent translocation assay (Supplementary Fig. 2 online).
Figure 3.
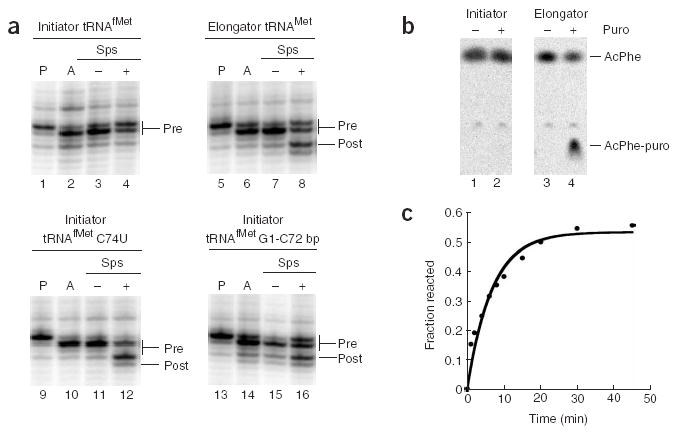
Analysis of sparsomycin-mediated translocation and puromycin reactivity of pretranslocation ribosome complexes carrying initiator and elongator tRNAMet. (a) Sparsomycin-and EF-G–mediated translocation on MRE600 ribosomes and gene32 mRNA, monitored by toeprinting analysis. Pretranslocation complexes were assembled by binding deacylated tRNAfMet, tRNAMet, tRNAfMet C74U or tRNAfMet containing the C1G-A72C base pair (bp) to the P site (P lanes), then binding AcPhe-tRNAPhe to the A site (A lanes). Complexes were incubated with sparsomycin (+Sps) or with 2.5% DMSO as a control (−Sps). (b) Hybrid reactivity of [14C]AcPhe-tRNAPhe with puromycin, analyzed by electrophoretic TLC. Pretranslocation complexes prepared with deacylated initiator tRNAfMet (lanes 1 and 2) or elongator tRNAMet (lanes 3 and 4) in the P site and [14C]AcPhe-tRNAPhe in the A site were incubated with (+) or without (−) puromycin. (c) Time course of reactivity of [14C]AcPhe-tRNAPhe with puromycin in a pretranslocation complex with elongator tRNAMet.
Further support for the importance of the identity of the deacylated tRNA species and its interactions with the ribosome in specifying sparsomycin-dependent translocation comes from two independent experiments. First, the incorporation of a C74U mutation into the initiator tRNAfMet body, known to disfavor binding of this tRNA to the P loop (and thus probably favoring a P/E binding state), stimulated sparsomycin-dependent translocation (Fig. 3a, lanes 9–12). Second, the incorporation of a Watson-Crick base-pairing interaction (C1G-A72C) at the top of the acceptor stem in the initiator tRNAfMet promotes sparsomycin-dependent translocation (Fig. 3a, lanes 13–16). Previous studies have shown that the absence of a Watson-Crick base pair at the top of the acceptor stem is a unique structural feature of E. coli initiator tRNAs that is an important determinant for the formylation enzymes and for the overall function of initiator tRNA28,29. These results are consistent with the mutational analysis reported above and support the argument that hybrid ribosomal complexes are good targets for sparsomycin-mediated translocation.
We next measured the reactivity of peptidyl-tRNA moieties bound in the pretranslocation state with puromycin, as an independent means to evaluate the state of tRNA binding on the ribosome. Previous studies have established that pretranslocation ribosome complexes carrying AcPhe-tRNAPhe in the A site do not react with puromycin, whereas those carrying a dipeptidyl tRNA species (such as fMet-Phe-tRNAPhe) react to completion (albeit at reduced rates compared to classically bound peptidyl-tRNA)13. These data are most simply explained by a model where tRNA derivatized with a single amino acid (AcPhe) does not occupy the hybrid state, whereas a dipeptidyl moiety (fMet-Phe) favors such a binding mode. For this study, the puromycin reactivity of pretranslocation-state ribosome complexes with AcPhe-tRNAPhe in complexes carrying either an initiator tRNAfMet or an elongator tRNAMet in the P site (Fig. 3b) was compared. Consistent with the sparsomycin results reported above, the complex carrying the initiator tRNAfMet was essentially unreactive to puromycin, whereas the complex carrying the elongator tRNAMet was fully reactive (rate of 0.12 min−1 in the presence of 1 mM puromycin at 37 °C) (Fig. 3c). Equivalent rates were obtained with ribosome complexes programmed with a different mRNA, m301, loaded with tRNATyr and AcPhe-tRNAPhe (Supplementary Fig. 2). The observed rate of hybrid puromycin reactivity is consistent with those previously reported for pretranslocation-bound dipeptidyl-tRNA13,30.
EF-G–dependent translocation of rRNA and tRNA variants
Although sparsomycin-mediated translocation is in some way related to authentic EF-G–mediated translocation on the ribosome, comparisons between the processes should be made cautiously. For example, the simplest explanation for the general results presented above is that sparsomycin can only promote translocation on ribosome complexes to which it can bind (that is, where the A site on the large subunit is not occupied by the peptidyl-tRNA species). On the basis of the consistent results from our assays of sparsomycin-mediated translocation and puromycin reactivity, we next asked what the effects of the same tRNA and rRNA mutations might be on the rates of EF-G–mediated translocation. For this analysis, we used a pre–steady-state fluorescence-based translocation assay26,31 to directly evaluate whether the state of tRNA binding on the ribosome affects the observed rates of EF-G–mediated translocation. Pretranslocation complexes were formed by the sequential addition of [35S]fMet-tRNAfMet (and initiation factors and GTP) and Phe-tRNAPhe (in a ternary complex with EF-Tu–GTP) to generate pyrene-labeled ribosome complexes essentially as described in refs. 26,31. Analysis of dipeptide formation established that pretranslocation complex formation was efficient enough to allow for subsequent analysis by stopped-flow fluorescence of the bulk ribosome population (>70% for wild-type complexes and as low as 60% with mutant tRNAs). The state of tRNA binding (hybrid versus classic) in the various pretranslocation complexes was established using puromycin-reactivity assays (Supplementary Table 1 online)13,30.
Control experiments first evaluated the rate of translocation of wild-type dipeptidyl-tRNA pretranslocation complexes with both MRE600 and wild-type aptamer-tagged24 ribosomes. The kinetic profiles of these reactions fit readily to single exponential curves and yielded observed rate constants of 3.8 ± 0.4 s−1 and 3.7 ± 0.4 s−1, respectively (for representative traces, see Fig. 4). We next substituted C74G tRNA both in the P site (deacylated C74G tRNAfMet) and in the A site (C74G fMet-Phe-tRNAPhe) and determined the effect of the substitution on the rate of EF-G–mediated translocation. Whereas the C74G tRNA mutant had no effect on the rate of translocation when substituted for the P-site tRNA (4.6 ± 0.2 s−1), the rate was reduced by 2.5-fold when the mutant was substituted for the A-site tRNA (1.5 ± 0.2 s−1) (Table 1). Thus, when pairing interactions between the A-site tRNA and the P loop of the 23S rRNA are disrupted, the ribosomal complex is a less robust substrate for EF-G. As predicted from a hybrid-state model (and the sparsomycin results above), when potential pairing between the A-site tRNA and the P loop was restored (with C74G tRNA in the A site and G2252C ribosomes), translocation rates were restored to wild-type levels (5.0 ± 0.4 s−1). Similar overall results were observed when the other pairing interaction (G2251 and C75) between the A-site tRNA and the P loop was perturbed and then restored to a non–wild-type pairing interaction (G2251C and C75G) (Table 1). In this case, substitution of C75G tRNA in the A site in wild-type ribosomes decreased the rate of translocation by 2.7-fold (from 3.8 ± 0.4 s−1 to 1.4 ± 0.1 s−1), and this activity was restored (to 9.3 ± 0.4 s−1) when G2551C ribosomes were instead used to form the pretranslocation complexes. It seems to be generally true that isolated substitutions in the P loop or in the A-site tRNA decrease the rate of translocation and that these deficiencies are suppressed when compensatory changes in the other component (tRNA or rRNA) are made.
Figure 4.
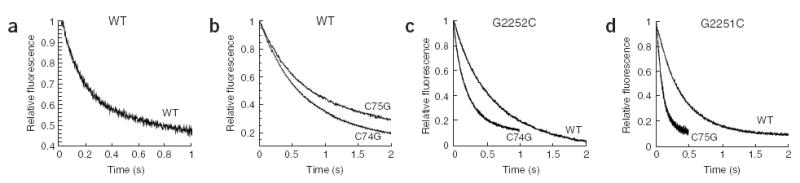
Pre–steady-state kinetic analysis of EF-G–mediated translocation. (a–d) Pretranslocation complexes with tRNAfMet in the P site and dipeptidyl-tRNA in the A site were rapidly mixed with EF-G–GTP in the stopped-flow apparatus, and fluorescence was measured as a function of time. The A-site tRNAs used are indicated next to the trace (WT, C75G or C74G). The ribosomes used are indicated at top of panel (WT in a and b, G2252 in c and G2251 in d).
Table 1.
EF-G translocation of WT, G2252C and G2251C mutant ribosomes
| Translocation rate (s−1)
|
Translocation rate (s−1)
|
||||||
|---|---|---|---|---|---|---|---|
| P-site tRNA | A-site tRNA | WT | G2252C | P-site tRNA | A-site tRNA | WT | G2251C |
| WT | WT | 3.8 | 1.5 | WT | WT | 3.8 | 2.1a |
| WT | C74G | 1.5 | 5.0 | WT | C75G | 1.4 | 9.3 |
Rates are averages of two to four independent experiments. Standard deviations were below 13%, except where indicated.
s.d. = 22%.
As a final test, a distinct pairing interaction between C75 of the tRNA substrate and G2553 in the A loop of the 23S rRNA was used to probe the requirements for optimal translocation on the ribosome. Substitution of G2553 of the A loop with C, U or A increased the overall rate of translocation (to 10.7 ± 1.1, 12.2 ± 0.6 or 12.4 ± 0.6 s−1, respectively) relative to wild-type (3.8 s−1) (Table 2). When the G2553C ribosomes are loaded with C75G tRNAs (thus restoring the potential A-loop pairing), the rate of translocation is diminished (1.1 ± 0.1 s−1), consistent with models in which translocation is optimal when tRNAs are able to bind in a hybrid state on the ribosome. In these complexes, unlike the fully wild-type complexes, the C75G tRNA mutation will also disrupt pairing with the P loop, thus reinforcing a classical state of tRNA binding.
Table 2.
EF-G translocation with WT and G2553 mutant ribosomes
| Translocation rate (s−1)
|
|||||
|---|---|---|---|---|---|
| P-site tRNA | A-site tRNA | WT | G2553C | G2553U | G2553A |
| WT | WT | 3.8 | 10.7 | 12.2a | 12.4a |
| WT | C75G | 1.4 | 1.1 | ND | ND |
All rates except those indicated are averages of at least two independent experiments.
Standard deviations were below 11%. ND, not determined.
In light of previous studies32,33 and their cautionary notes regarding nucleotide purity, we have carefully examined the nucleotide dependence of our translocation experiments. First, although our GTP stock does contain GDP (3.7%) (Supplementary Fig. 3 online), our translocation rate is dependent on added GTP and is unaffected by the additional of equimolar amounts of GDP (with GTP) (Supplementary Table 2 online). In addition, the incubation of an energy-regenerating system with our GTP stock, which effectively eliminates the GDP contamination, had no effect on the observed rate constants for translocation (Supplementary Fig. 3 and Supplementary Table 3 online). These data strongly support the argument that the reported translocation rates reflect a step in translocation catalysis downstream of GDP exchange by EF-G.
EF-G–dependent translocation of tRNAMet species
Similar experiments comparing the rates of translocation with initiator and elongator methionyl-tRNAs in the P site and either dipeptidyl-tRNAPhe or AcPhe-tRNAPhe in the A site were next performed. Rate constants for translocation were two- to four-fold higher when elongator rather than initiator tRNAMet was loaded in the pretranslocation P site (16.6 ± 0.4 s−1 versus 3.8 ± 0.4 s−1 for dipeptidyl-tRNAPhe; 4.0 ± 0.2 s−1 versus 1.8 ± 0.3 s−1 for AcPhe-tRNAPhe) (Table 3). Similar differences in rate constants have been reported in comparisons of the initiator and elongator tRNAs in pretranslocation complexes containing only deacylated tRNAs26. Whereas in earlier studies natural and in vitro–transcribed tRNAs were directly compared, we here make comparisons only between equivalently derived tRNAs. These data are consistent with both sparsomycin-dependent translocation and puromycin-reactivity data presented above and support a model where ribosome complexes carrying tRNAs bound in the hybrid state are optimal substrates for EF-G–mediated translocation.
Table 3.
Comparison of P-site initiator versus elongator tRNAMet in single-turnover EF-G translocation
| Translocation rate (s−1)
|
||
|---|---|---|
| A-site tRNA | Initiator tRNAfMet | Elongator tRNAMet |
| AcPhe-tRNAPhe | 1.8 | 4.0 |
| dipeptidyl-tRNAPhe | 3.8a | 16.6b |
Rates are averages of at least two independent experiments.
Standard deviations were below 16%.
fMet-Phe-tRNAPhe in A site.
AcMet-Phe-tRNAPhe in A site.
DISCUSSION
The quantitative data presented above provide evidence that the hybrid state of tRNA binding on the ribosome is a kinetically relevant intermediate during the translation elongation cycle. We have taken advantage of known Watson-Crick pairing interactions between the CCA ends of the P- and A-site tRNA substrates14–16 to form defined classically bound and hybrid-bound pretranslocation-state ribosome complexes. Two distinct assays yielded consistent results indicating that optimal translocation occurs in complexes where tRNAs are bound in a hybrid state. Although the overall effects of disfavoring hybrid-state binding on EF-G–mediated translocation are not dramatic (from two- to four-fold, or <1 kcal of energy), they provide evidence that a hybrid ribosomal complex is the preferred substrate for EF-G and thus is likely to be a biologically relevant elongation intermediate. In an iterated process such as the translation elongation cycle, such small effects could have substantial in vivo consequences. The results complement earlier observations from cryo-EM studies that trapped EF-G binds ribosomes with a single, apparently hybrid tRNA10, but never ribosomes with two tRNAs bound (that is, the physiologically relevant ribosome complex).
These data also extend earlier work comparing translocation rates with a dipeptidyl-tRNA versus an aminoacyl-tRNA in the A site12. These studies reported substantial rate differences for translocation with the two tRNA substrates (~130-fold), though conclusions about the contribution of the hybrid state to these differences in rate are limited. Although the dipeptidyl- and aminoacyl-tRNA probably do bind predominantly in the hybrid (A/P) and classic (A/A) states, respectively13,34, contributions to translocation kinetics are probably made by the different lengths of the polypeptide (aminoacyl versus peptidyl) or by the loss of the positive charge on the amino moiety. A simple aminoacyl-tRNA is not a physiologically relevant pretranslocation A-site substrate for the ribosome: by definition, a pretranslocation substrate must have previously undergone at least one round of peptidyl transfer. Furthermore, differences in the identity or composition of the peptidyl moiety have been shown previously to exert substantial effects on ribosome binding and translocation efficiency23.
To avoid these potential ambiguities, we have compared ribosome–tRNA complexes that differ only in the identity of single targeted nucleotides in the P or A loop of the ribosome and/or in the CCA end of the tRNA. The premise of this approach is that both the ribosome and tRNA components in this system tolerate the single-nucleotide substitutions and that restoration of binding occurs when the compensatory change (in the other component) is made. Although full restoration of function is not necessarily anticipated with compensatory changes15,16, the levels of activity observed in the restoration experiments were substantial (compare 3.8 to 5.0 s−1 in Table 1).
The base-pair-swap experiments were complemented by comparison of the impact on translocation of initiator and elongator tRNAs in the P site. In two distinct ribosome complexes, one carrying N-acetylated Phe-tRNAPhe (thought to bind predominantly in a classic (A/A) configuration) and another carrying dipeptidyl-tRNA (thought to bind in a hybrid (A/P) configuration) in the A site, the initiator tRNA in the P site negatively affects the rate of translocation by EF-G. Perhaps the structural properties of initiator tRNA that make it uniquely suited for binding in the P site29 similarly limit occupation of the E site by this tRNA species. The importance of E-site interactions for efficient translocation has been established in previous tRNA-truncation experiments11. Our data extend these earlier results by establishing that both tRNAs should bind in a hybrid configuration to promote optimal translocation.
Although our data indicate that the hybrid state of tRNA binding is a kinetically relevant intermediate yielding maximal rates of translocation, the question remains as to which specific step in translocation is affected by these different states of the ribosome. Several previous studies have indicated that the binding affinity between the ribosome and EF-G is largely unaffected by numerous factors, including the identity of the bound nucleotide (GTP versus GDP)35 and the presence of the L7/L12 stalk protein36. On the basis of these observations and our own determination of the K1/2 of EF-G for the pre–steady-state ribosome complexes (Supplementary Fig. 4 online), we would argue that EF-G binding is unaffected by the state of tRNAs bound to the ribosome. Another possibility is that the composition of the ribosome complex affects the rate of GTP hydrolysis by EF-G, though an earlier pre–steady-state analysis of GTP hydrolysis argues that the rates of this reaction are not affected by the state of tRNA binding37. Our nucleotide-dependence studies further indicate that GDP exchange on EF-G is not likely to be rate limiting in our system, though recent studies have pointed to a role for the ribosome as the guanine nucleotide exchange factor for this process33.
Another recent study has identified a rate-limiting step for translocation by EF-G that follows GTP hydrolysis and precedes and limits both translocation and inorganic phosphate (Pi) release31. It seems likely that the configuration of tRNAs bound to the ribosome affects this rate-limiting ‘unlocking’ step in translocation. tRNAs occupying hybrid states on the ribosome are bound less tightly than those bound classically12 and are thus better poised for movement stimulated by EF-G and the rearrangements that take place as a consequence of GTP hydrolysis. Molecular insight into how unlocking of the ribosome might be specifically affected by the configuration of bound tRNAs will come from further structural and biochemical approaches.
METHODS
Ribosomes, tRNAs, mRNAs and translation factors
Ribosomes were prepared from E. coli MRE600 in mid-log phase. Crude ribosomes were isolated and then were either further purified over 10%–40% (w/v) sucrose gradients or subjected to an additional pelleting through a sucrose cushion as described in ref. 38. Aptamer-tagged wild-type and variant ribosomes were prepared as described from plasmid p278MS2A626G22,38. Mutations at positions G2251 and G2252 were introduced using site-directed mutagenesis (XL-QuikChange, Stratagene). Mutations at position G2553 were described in ref. 12.
gene32 mRNA24 and m301 mRNA17 were prepared by run-off T7 transcription27 and purified as described in ref. 24. Pyrene-labeled mRNA 95.2 was prepared by synthesis of an RNA oligonucleotide (5′-AAGGAGGUAAAAAUG UUUGCU-3′-NH2) from Dharmacon and labeling at the 3′end with 1-pyrenebutanoic acid, succinimidylester (P-130, Molecular Probes) according to the manufacturer’s protocol.
tRNATyr, tRNAMet and tRNAfMet were PCR-amplified from genomic DNA from MRE600 with primers (listed in Supplementary Methods online) containing the T7 promoter and were cloned into pCR2.1-TOPO (Invitrogen), and their sequences were confirmed. tRNAPhe was amplified from plasmid p67CF10 (ref. 39). tRNAs were transcribed from PCR templates prepared from these plasmids essentially as described in ref. 15. Mutations in the 5′and 3′ends of the tRNA were introduced with primers containing the respective mutations. tRNAs were aminoacylated and formylated using S100 extracts and [35S]methionine or [14C]phenylalanine essentially as described in refs. 25,40. N-acetylation of aminoacyl-tRNAs was performed using acetic anhydride as described in ref. 41. N-acetylated tRNAs were purified over an RP-HPLC C4 column (Vydac, 10 × 250 mm, 10–15 μm) using a linear gradient of 0%–25% (v/v) ethanol in 50 mM ammonium acetate and 50 mM MgCl2 (pH 5.5) over 60 min.
Initiation factors IF1, IF2 and IF3 were purified as described in ref. 42. His6- tagged EF-Tu and His6-tagged EF-G were purified over Ni-NTA resin and the His6 tags were removed by tobacco etch virus (TEV) protease cleavage essentially as described in ref. 34.
Sparsomycin (NSC number 59729; obtained from Drug Synthesis & Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, US National Cancer Institute) was resuspended in DMSO and diluted to 5 mM in 25% (v/v) DMSO. GTP was purchased from Amersham Bioscience and resuspended in H2O, and the pH was adjusted with 10 M KOH to 7.0. The GDP content was determined on a ResourceQ column (6 ml, Amersham Biosciences), with a 0–350 mM NaCl gradient in 20 mM Tris-HCl (pH 7.5) over 30 min at 5 ml min−1. The GTP stock used for all experiments contained 3.7% contaminating GDP.
Toeprint assay
Primer-extension analysis of translocation promoted either by sparsomycin or EF-G–GTP was performed essentially as described in ref. 17. 32P-labeled primer (0.6 μM of 5′-TTTATCTTCAGAAGAAAAACC-3′) complementary to the 3′end of m301 mRNA was annealed to mRNA template (8 μM) at 65 °C, incubated for 3 min and snap-cooled on ice. 70S ribosomes (either wild-type MRE600 (1 μM) or aptamer-tagged wild type or variants (2 μM)), tRNATyr (6 μM) and m301 mRNA (0.7 μM, preannealed to the labeled primer) were incubated in buffer A (50 mM Tris-HCl (pH 7.5), 100 mM NH4Cl, 20 mM MgCl2 and 6 mM β-mercaptoethanol) for 10 min at 37 °C and 1 μl of the reaction was removed for primer extension (P lane). The A site was then filled with AcPhe-tRNAPhe or one of its mutant variants (C74G or C75G) at 3 μM for 5 min at 37 °C, and again a 1-μl aliquot was removed for primer extension (A lane). For sparsomycin-mediated translocation, 4.5 μl pretranslocation complex was mixed with 0.5 μl 5 mM sparsomycin (+Sps lanes) or 25% (v/v) DMSO (−Sps lanes) and incubated for 30 min at 37 °C. For EF-G–mediated translocation, 4 μl pretranslocation complex was mixed with 1 μl EF-G–GTP in 1× buffer A to final concentrations of 0.05 μM EF-G and 300 μM GTP (+EF-G lanes) and incubated for 1 min at room temperature. From each reaction, 1 μl was removed and added to 9 μl primer-extension mix (10 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 60 mM NH4Cl, 6 mM β-mercaptoethanol, 375 μM each dNTP, 1 mM viomycin, 3 units AMV reverse transcriptase (Seikagaku Amerika)) and reverse-transcribed for 10 min at 37 °C. Reactions were precipitated and separated on 6% (w/v) denaturing PAGE. Dried gels were exposed to GP Phosphor-Screens (Amersham Pharmacia) and scanned in a Typhoon 9410 (Amersham Pharmacia). Data were analyzed using ImageQuant 5.2. Toeprinting assays with gene32 mRNA (Fig. 3) were performed as above except that gene32 mRNA was annealed to a 3′ gene32 primer and the indicated tRNAMet variant was used as a P-site substrate (tRNAfMet, tRNAMet, tRNAfMetC1G-A72C or tRNAfMetC74U). Puromycin reactivity of these pretranslocation complexes was tested by incubation with 1 mM puromycin at 37 °C for 45 min (Fig. 3b) or the time indicated (Fig. 3c). Products were hydrolyzed and separated by electrophoretic TLC.
Pretranslocation complexes for stopped-flow experiments
Initiation complex was prepared by incubation of 1 μM MRE600 ribosomes or 2 μM purified tagged ribosomes (supplemented with 1 μM gradient-purified 30S subunits) with 8 μM pyrene-labeled mRNA 95.2, 3 μM each of IF1, IF2 and IF3, 2 mM GTP and 3 μM [35S]fMet-tRNAfMet in buffer B (50 mM Tris-HCl (pH 7.5), 7 mM MgCl2, 70 mM NH4Cl, 30 mM KCl). Ternary complex was formed by incubation of EF-Tu (20 μM), GTP (2 mM) and [14C]Phe-tRNAPhe (10 μM) in 1× buffer B for 15 min at 37 °C. EF-Tu (100 μM) and GTP (2 mM) were preincubated together at 37 °C for 15 min to allow for exchange of GDP bound to EF-Tu. Equal volumes of initiation complex and ternary complex were mixed and allowed to form pretranslocation complex for 5 min at 37 °C. The extent of peptide bond formation was determined using electrophoretic thin-layer chromatography (TLC) as described in ref. 24. Pretranslocation complexes formed with [35S]AcMet-tRNAMet were assembled as above except that IFs were omitted and the MgCl2 concentration was raised to 15 mM in buffer B. For pretranslocation complexes with [14C]AcPhe-tRNAPhe in the A site, 1 μM MRE600 ribosomes and 8 μM pyrene-labeled mRNA 95.2 were incubated with 5 μM tRNAfMet or tRNAMet for 20 min at 37 °C, and [14C]AcPhe-tRNAPhe was then added to 5 μM and the mixture incubated for 10 min at 37 °C.
The MgCl2 concentration of pretranslocation complexes was raised to 20 mM and then pelleted through a 0.9-ml 1.1 M sucrose cushion in buffer B containing 20 mM MgCl2 (MLA130 rotor, 75,000 r.p.m., 2 h, 4 °C). After pelleting and resuspension, tRNA was quantitated by scintillation counting or electrophoretic TLC. Pellets containing pretranslocation complexes were resus-pended in buffer B to a final concentration of 0.2 μM and rapidly mixed with 6 μM EF-G and 2 mM GTP in buffer B at 25 °C. Fluorescence was monitored as a function of time using λex = 343 nm and a 375-nm cut-off filter (Schott APFF375) on the emission side. Kinetic experiments were performed on an SX.180MV-R stopped-flow fluorometer (Applied Photophysics). Time courses were evaluated by single-exponential fitting (Kaleidagraph).
Supplementary Material
Acknowledgments
We thank C. Merryman and other members of the lab for discussions, J. Lorsch (Johns Hopkins University) for critical reading of the manuscript and support with our kinetic studies on the ribosome, K. Fredrick (The Ohio State University) for the m301 plasmid, S. Blanchard (Cornell University) for the EF-G construct and O. Uhlenbeck (Northwestern University) for plasmid p67CF10. The work was supported by an Erwin Schrödinger fellowship (J2172) from the Austrian Science Foundation to S.D., by the US National Institutes of Health and by salary support from the Howard Hughes Medical Institute.
Footnotes
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
Published online at http://www.nature.com/nsmb/
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html
References
- 1.Cochella L, Green R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–1180. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinger JS, Parnell KM, Dorner S, Green R, Strobel SA. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nat Struct Mol Biol. 2004;11:1101–1106. doi: 10.1038/nsmb841. [DOI] [PubMed] [Google Scholar]
- 3.Piepenburg O, et al. Intact aminoacyl-tRNA is required to trigger GTP hydrolysis by elongation factor Tu on the ribosome. Biochemistry. 2000;39:1734–1738. doi: 10.1021/bi992331y. [DOI] [PubMed] [Google Scholar]
- 4.Rheinberger HJ, Sternbach H, Nierhaus KH. Three tRNA binding sites on Escherichia coli ribosomes. Proc Natl Acad Sci USA. 1981;78:5310–5314. doi: 10.1073/pnas.78.9.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bretscher MS. Translocation in protein synthesis: a hybrid structure model. Nature. 1968;218:675–677. doi: 10.1038/218675a0. [DOI] [PubMed] [Google Scholar]
- 6.Hardesty B, Odom OW, Deng H-Y. In: The Structure, Function and Genetics of Ribosomes. Hardesty B, Kramer G, editors. Springer-Verlag; New York, USA: 1986. pp. 495–508. [Google Scholar]
- 7.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 8.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 9.Agrawal RK, et al. Effect of buffer conditions on the position of tRNA on the 70 S ribosome as visualized by cryoelectron microscopy. J Biol Chem. 1999;274:8723–8729. doi: 10.1074/jbc.274.13.8723. [DOI] [PubMed] [Google Scholar]
- 10.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 11.Lill R, Robertson JM, Wintermeyer W. Binding of the 3′terminus of tRNA to 23S rRNA in the ribosomal exit site actively promotes translocation. EMBO J. 1989;8:3933–3938. doi: 10.1002/j.1460-2075.1989.tb08574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Semenkov YP, Rodnina MV, Wintermeyer W. Energetic contribution of tRNA hybrid state formation to translocation catalysis on the ribosome. Nat Struct Biol. 2000;7:1027–1031. doi: 10.1038/80938. [DOI] [PubMed] [Google Scholar]
- 13.Sharma D, Southworth DR, Green R. EF-G-independent reactivity of a pre-translocation state ribosome complex with the aminoacyl tRNA substrate puromycin supports an intermediate (hybrid) state of tRNA binding. RNA. 2004;10:102–113. doi: 10.1261/rna.5148704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 15.Samaha RR, Green R, Noller HF. A base pair between tRNA and 23S rRNA in the peptidyl transferase centre of the ribosome. Nature. 1995;377:309–314. doi: 10.1038/377309a0. [DOI] [PubMed] [Google Scholar]
- 16.Kim DF, Green R. Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol Cell. 1999;4:859–864. doi: 10.1016/s1097-2765(00)80395-0. [DOI] [PubMed] [Google Scholar]
- 17.Fredrick K, Noller HF. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300:1159–1162. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
- 18.Hansen JL, Schmeing TM, Moore PB, Steitz TA. Structural insights into peptide bond formation. Proc Natl Acad Sci USA. 2002;99:11670–11675. doi: 10.1073/pnas.172404099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monro RE, Celma ML, Vazquez D. Action of sparsomycin on ribosome-catalysed peptidyl transfer. Nature. 1969;222:356–358. doi: 10.1038/222356a0. [DOI] [PubMed] [Google Scholar]
- 20.Southworth DR, Green R. Ribosomal translocation: sparsomycin pushes the button. Curr Biol. 2003;13:R652–R654. doi: 10.1016/s0960-9822(03)00574-8. [DOI] [PubMed] [Google Scholar]
- 21.Hartz D, McPheeters DS, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- 22.Jerinic O, Joseph S. Conformational changes in the ribosome induced by translational miscoding agents. J Mol Biol. 2000;304:707–713. doi: 10.1006/jmbi.2000.4269. [DOI] [PubMed] [Google Scholar]
- 23.Fredrick K, Noller HF. Accurate translocation of mRNA by the ribosome requires a peptidyl group or its analog on the tRNA moving into the 30S P site. Mol Cell. 2002;9:1125–1131. doi: 10.1016/s1097-2765(02)00523-3. [DOI] [PubMed] [Google Scholar]
- 24.Youngman EM, Brunelle JL, Kochaniak AB, Green R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- 25.Green R, Samaha RR, Noller HF. Mutations at nucleotides G2251 and U2585 of 23 S rRNA perturb the peptidyl transferase center of the ribosome. J Mol Biol. 1997;266:40–50. doi: 10.1006/jmbi.1996.0780. [DOI] [PubMed] [Google Scholar]
- 26.Studer SM, Feinberg JS, Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J Mol Biol. 2003;327:369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- 27.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laursen BS, Sorensen HP, Mortensen KK, Sperling-Petersen HU. Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev. 2005;69:101–123. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.RajBhandary UL. Initiator transfer RNAs. J Bacteriol. 1994;176:547–552. doi: 10.1128/jb.176.3.547-552.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J Mol Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 31.Savelsbergh A, et al. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 32.Czworkowski J, Moore PB. The conformational properties of elongation factor G and the mechanism of translocation. Biochemistry. 1997;36:10327–10334. doi: 10.1021/bi970610k. [DOI] [PubMed] [Google Scholar]
- 33.Zavialov AV, Hauryliuk VV, Ehrenberg M. Guanine-nucleotide exchange on ribosome-bound elongation factor G initiates the translocation of tRNAs. J Biol. 2005;4:9. doi: 10.1186/jbiol24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katunin VI, Savelsbergh A, Rodnina MV, Wintermeyer W. Coupling of GTP hydrolysis by elongation factor G to translocation and factor recycling on the ribosome. Biochemistry. 2002;41:12806–12812. doi: 10.1021/bi0264871. [DOI] [PubMed] [Google Scholar]
- 36.Mohr D, Wintermeyer W, Rodnina MV. GTPase activation of elongation factors Tu and G on the ribosome. Biochemistry. 2002;41:12520–12528. doi: 10.1021/bi026301y. [DOI] [PubMed] [Google Scholar]
- 37.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 38.Youngman EM, Green R. Affinity purification of in vivo-assembled ribosomes for in vitro biochemical analysis. Methods. 2005;36:305–312. doi: 10.1016/j.ymeth.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 39.Sampson J, DiRenzo A, Behlen L, Uhlenbeck O. Nucleotides in yeast tRNAPhe required for the specific recognition by its cognate synthetase. Science. 1989;243:1363–1366. doi: 10.1126/science.2646717. [DOI] [PubMed] [Google Scholar]
- 40.Dubnoff JS, Mairtra U. Isolation and properties of protein factors involved in polypeptide chain initiation in Escherichia coli. Methods Enzymol. 1971;20:248–261. [Google Scholar]
- 41.Moazed D, Noller HF. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989;57:585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- 42.Brunelle JL, Youngman EM, Sharma D, Green R. The interaction between C75 of tRNA and the A loop of the ribosome stimulates peptidyl transferase activity. RNA. 2006;12:33–39. doi: 10.1261/rna.2256706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


