Abstract
We have developed a new primer design strategy for PCR amplification of distantly related gene sequences based on consensus-degenerate hybrid oligonucleotide primers (CODEHOPs). An interactive program has been written to design CODEHOP PCR primers from conserved blocks of amino acids within multiply-aligned protein sequences. Each CODEHOP consists of a pool of related primers containing all possible nucleotide sequences encoding 3–4 highly conserved amino acids within a 3′ degenerate core. A longer 5′ non-degenerate clamp region contains the most probable nucleotide predicted for each flanking codon. CODEHOPs are used in PCR amplification to isolate distantly related sequences encoding the conserved amino acid sequence. The primer design software and the CODEHOP PCR strategy have been utilized for the identification and characterization of new gene orthologs and paralogs in different plant, animal and bacterial species. In addition, this approach has been successful in identifying new pathogen species. The CODEHOP designer (http://blocks.fhcrc.org/codehop.html) is linked to BlockMaker and the Multiple Alignment Processor within the Blocks Database World Wide Web (http://blocks.fhcrc.org).
INTRODUCTION
Sequence comparison of multiple members of protein families has proven to be an important approach in the analysis of protein structure and function. The identification of blocks of conserved amino acid sequences has revealed the presence of protein motifs and domains that play important roles in protein function. However, such sequence analysis is limited by the number of individual protein sequences available for comparison. Even with the recent advances in whole genome sequencing, the acquisition of new members of a targeted protein family is limited. Previous methods to isolate unknown family members by PCR have relied on either degenerate primers consisting of a pool of primers containing most or all of the possible nucleotide sequences encoding a conserved amino acid motif or consensus primers consisting of a single primer containing the most common nucleotide at each codon position within the motif. Although these strategies have been successful in isolating closely related sequences, they have generally failed when sequences were more distantly related or were in low copy number. We have developed the COnsensus-DEgenerate Hybrid Oligonucleotide Primer (CODEHOP) PCR strategy for the identification of new members of protein families, which overcomes problems inherent in both degenerate and consensus methods for primer design (1).
CODEHOP STRATEGY
Short regions of proteins with high levels of conservation can be represented as ungapped blocks of multiply aligned protein sequences (Fig. 1) (2). CODEHOPs are derived from these conserved sequence blocks (http://blocks.fhcrc.org/codehop.html), and are used in PCR to amplify the region between them. A CODEHOP PCR primer consists of a pool of primers each containing a different sequence in the 3′ degenerate core region where each primer provides one of the possible codon combinations encoding a targeted 3–4 conserved amino acid motif within the sequence block (Fig. 2). In addition, each primer in the pool has an identical 5′ consensus clamp region derived from the most probable nucleotide at each position encoding the conserved amino acids flanking the targeted motif. Amplification initiates by annealing and extension of primers in the pool with the most similarity in the 3′ degenerate core to the target template (Fig. 3). Annealing is stabilized by the 5′ consensus clamp which partially matches the target template. Once the primer is incorporated, it becomes the template for subsequent amplification cycles. Because all primers are identical in the 5′ consensus clamp region, they all will anneal at high stringency during subsequent rounds of amplification. This increases the efficiency of the PCR amplification and differentiates the CODEHOP technique from the less efficient consensus PCR or degenerate PCR techniques. The CODEHOP technique has been validated by the successful amplification of new members of protein families that have proven challenging using conventional methods (1).
Figure 1.
Block representation of a highly conserved region within the family of DNA methyltransferases. The ungapped multiple sequence alignment of Block D of the DNA methyltransferases was obtained from a comparison of eight DNA methyltransferase sequences: MTCH_ARATH, AAC02665 (CMT1); MTCH_CARAR, AAB95486 (CMT1); MTDM_ARATH AAF14882 (MET2); MTDM_MOUSE AAC40061 (MET1); MTDM_HUMAN AAF23609 (MET1); MTDM_XENLA JC5145 (MET1); MTDM_CHICK Q92072 (MET1); MTDM_PARLI CAD43079 (MET1) using BlockMaker. Information regarding the size of the block and its position within the original sequences is indicated. In addition, a relative sequence weight has been assigned to each aligned sequence to reduce redundancy and emphasize diversity in the multiple sequence alignment (5). As shown, the sequence weights for chromomethylase sequences, MTCH_CARAR and MTCH_ARATH, were increased four times to bias CODEHOP design towards chromomethylases. The block output of BlockMaker is hypertext linked to the CODEHOP designer.
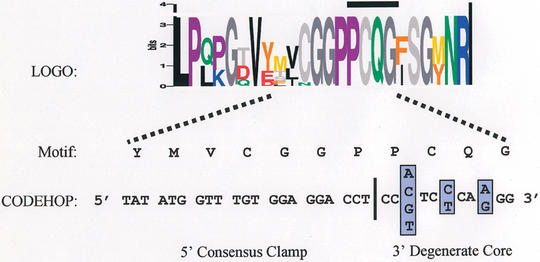
Figure 2.
Anatomy of a CODEHOP PCR primer. A CODEHOP PCR primer is targeted to an amino acid sequence motif within a block of amino acids conserved between different members of a protein family. In this example, the sequence logo representation (9) shows the amino acid conservation within the DNA methyltransferase Block D (Fig. 1), which was successfully used to isolate Arabidopsis thaliana CMT2 and CMT3 proteins (1). The height of each amino acid is proportional to its degree of conservation. The CODEHOP PCR primer is targeted to the highly conserved ‘PCQG’ motif and consists of a pool of 16 related primers in which the 3′ degenerate core contains all of the possible nucleotide sequences encoding the ‘PCQG’ motif. The 5′ consensus clamp, immediately upstream, contains the most probable nucleotide at each position flanking the ‘PCQG’ motif.
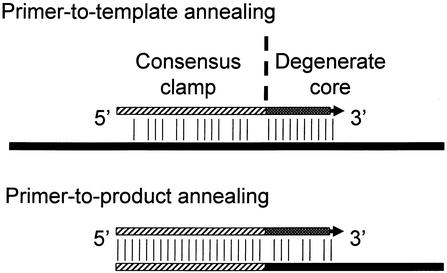
Figure 3.
The CODEHOP PCR strategy. CODEHOP PCR primers target conserved motifs of 3–4 amino acids using a short 3′ degenerate core containing all codon possibilities for the motif. Annealing of the degenerate core of the primer to the starting template is stabilized by the 5′ consensus clamp. During subsequent rounds of amplification, annealing of primer to PCR product is driven by the identical match with the 5′ consensus clamp between the incorporated primer and primers remaining in the pool. This strategy permits amplification of distantly related sequences with limited homology to known family members.
CODEHOP PROGRAM
The CODEHOP program consists of the following nine steps.
In order to identify target amino acid motifs which are conserved between members of a gene family, a set of related sequences are aligned and conserved regions are determined. This can be done conveniently using the BlockMaker program (http://blocks.fhcrc.org/blockmkr/make_blocks.html) or multiple alignment processor (http://blocks.fhcrc.org/process_blocks.html) at the Blocks Database website. The resulting conserved blocks of amino acid sequences can be used directly as input for the CODEHOP program through a hyperlink (see Fig. 1). Additionally, blocks derived from annotated protein families can be obtained directly from the Blocks Database (http://blocks.fhcrc.org).
A position-specific scoring matrix (PSSM) is computed for each block using the odds-ratio method (3).
For each position of the block, a consensus amino acid is chosen as the highest scoring amino acid in the matrix.
The most common codon for each consensus amino acid in the block is determined using the selected codon usage table. This selection is used for the default 5′ consensus clamp determination in step 8.
A DNA PSSM is calculated from the amino acid matrix in step 2 using the selected codon usage table. The score for each amino acid within a position in the matrix is divided among its codons in proportion to their relative frequencies from the codon usage table and the scores for each of the four different nucleotides are combined in each DNA matrix position.
The degeneracy is determined by the number of bases at each position of the DNA matrix.
Degenerate core regions are evaluated by scanning the DNA matrix in the 3′–5′ direction to determine the overall codon degeneracy within 11 or 12 positions ending on a codon boundary. The degeneracy of a core region is the product of the number of possible nucleotides in each position. A core region must start on an invariant 3′ nucleotide position and have a maximum degeneracy of 128 (current default), which can be changed.
A 5′ consensus clamp region from steps 4 or 5 is added to candidate degenerate core regions. The length of the clamp region is controlled by an annealing temperature calculation (4). The current default is 60°C, which usually provides a clamp of ∼20 nucleotides.
For primers corresponding to the opposite strand of DNA, steps 7 and 8 are repeated on the reverse complement of the DNA matrix from step 5.
Examples of CODEHOP PCR primers designed from Blocks D and E of the cytosine DNA methyltransferases are provided in Figure 4.
Figure 4.
CODEHOP designer output. The biased Block D of the DNA methyltransferases (Fig. 1) was used to design CODEHOP PCR primers using the default 60°C (A) or 65°C (B) annealing temperature parameter. The consensus amino acid residues and predicted CODEHOP PCR primers (5′–3′) are shown. The preferred CODEHOP has 16-fold degeneracy. The change in the 5′ clamp length obtained with the higher annealing temperature setting in (B) is evident. The anti-sense CODEHOP PCR primer designed from the complementary strand of the DNA encoding Block F of the DNA methyltransferase sequences is shown (C). The primers shown here are very similar but not identical to those utilized in our previous study to identify new DNA methyltransferases (1), since the codon usage table for A.thaliana has changed since 1998. The degeneracy and length for each core and the length and annealing temperature of each clamp are indicated.
USING THE CODEHOP PROGRAM
The CODEHOP designer can be biased towards selected sequences within a block of multiply aligned sequences. This is useful for targeting specific orthologous or paralogous members of a protein family. The set of blocks returned by BlockMaker for CODEHOP input may be analyzed phylogenetically to permit convenient selection of a subset of related blocks using the ProWeb TreeViewer link from the BlockMaker output. To emphasize or de-emphasize a particular sequence, the weight provided for each sequence segment in the BlockMaker output (5) can be manually altered, thus changing its contribution to the primer design (Fig. 1). Low abundance nucleotides within the 3′ degenerate codon positions can be excluded by increasing the degeneracy ‘strictness’ value in the range from 0 to 1. Also, members of a protein family from a specific organism or genome can be targeted by selecting the applicable codon usage table for that genome. Finally, the most common codon encoding the consensus amino acid determined for the 5′ consensus clamp region can replace the most favored nucleotide chosen from DNA PSSM.
Other user-defined parameters are available to alter the primer design and output. First, the specificity and function of a CODEHOP PCR primer may be altered by changing the length of the 5′ consensus clamp region through an annealing temperature parameter (default=60°C). The presence of nucleotide runs within the 5′ consensus clamp region can be limited through a polynucleotide parameter (default=5) and the core/clamp boundary may be restricted to a codonboundary. Finally, the program output can show all possible primers or only the single most degenerate primer in each region.
Selection of the optimal CODEHOP PCR primer is primarily based on minimal degeneracy across the 3′ degenerate core and secondarily on the clamp score, which indicates the quality of the match between the 5′ non-degenerate clamp and the sequence block given a codon usage table. If no primer is identified from an input sequence block, the block may be biased using the methods described above to remove or de-emphasize the most distantly related sequences. Conversely, the default degeneracy limit of 128 may be increased to 256 or higher, which may identify highly conserved motifs that contain amino acids with higher codon degeneracies.
CONCLUSIONS
Since publication of the original CODEHOP manuscript (1) and implementation of the CODEHOP designer program on the WWW in 1998 more than 70 studies have been published in which CODEHOP PCR primers have been designed and successfully utilized to amplify distantly related sequences from organisms as diverse as fish, frog, protozoa, plants, viruses and bacteria (http://courses.washington.edu/bioinfo/CODEHOP/Codehop%20Genes.html). The methodology has been further expanded in the exploration of conservation and diversity of gene families in higher plants (6) and the characterization of viral genomes (7,8). Useful features of the CODEHOP designer include reweighting sequences in order to bias the design of CODEHOPs towards targeted protein families and the ProWeb TreeView selection of input sequences based on their phylogenetic relationship. The CODEHOP web site provides information for ‘Getting Started’, a detailed ‘Help’ page, and a description of the ‘CODEHOP Algorithm’. We are currently working on enhancements to the CODEHOP PCR strategy and the output of the designer program.
REFERENCES
- 1.Rose T.M., Schultz,E.R., Henikoff,J.G., Pietrokovski,S., McCallum,C.M. and Henikoff,S. (1998) Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res., 26, 1628–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henikoff J.G., Pietrokovski,S., McCallum,C.M. and Henikoff,S. (2000) Blocks-based methods for detecting protein homology. Electrophoresis, 21, 1700–1706. [DOI] [PubMed] [Google Scholar]
- 3.Henikoff J.G. and Henikoff,S. (1996) Using substitution probabilities to improve position-specific scoring matrices. Comput. Appl. Biosci., 12, 135–143. [DOI] [PubMed] [Google Scholar]
- 4.Rychlik W., Spencer,W.J. and Rhoads,R.E. (1990) Optimization of the annealing temperature for DNA amplification in vitro. Nucleic Acids Res., 18, 6409–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henikoff S. and Henikoff,J.G. (1994) Position-based sequence weights. J. Mol. Biol., 243, 574–578. [DOI] [PubMed] [Google Scholar]
- 6.Morant M., Hehn,A. and Werck-Reichhart,D. (2002) Conservation and diversity of gene families explored using the CODEHOP strategy in higher plants. BMC Plant Biol., 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schultz E.R., Rankin,G.W.,Jr., Blanc,M.P., Raden,B.W., Tsai,C.C. and Rose,T.M. (2000) Characterization of two divergent lineages of macaque rhadinoviruses related to Kaposi's sarcoma-associated herpesvirus. J. Virol., 74, 4919–4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rose T.M., Ryan,J.T., Schultz,E.R., Raden,B.W., Tsai,C.-C. (2003) Analysis of 4.3 Kb of the divergent locus-B of macaque retroperitoneal fibromatosis-associated herpesvirus (RFHV) reveals close similarity to Kaposi's sarcoma-associated herpesvirus (KSHV) in gene sequence and genome organization. J. Virol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneider T.D. and Stephens,R.M. (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res., 18, 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]