Abstract
NEBcutter, version 1.0, is a program available via a web server (http://tools.neb.com/NEBcutter) that will accept an input DNA sequence and produce a comprehensive report of the restriction enzymes that will cleave the sequence. It produces a variety of outputs including restriction enzyme maps, theoretical digests and links into the restriction enzyme database, REBASE (http://www.neb.com/rebase). Importantly, its table of recognition sites is updated daily from REBASE and it marks all sites that are potentially affected by DNA methylation (Dam, Dcm, etc.). Many options exist to choose the enzymes used for digestion, including all known specificities, subsets of those that are commercially available or sets of enzymes that produce compatible termini.
INTRODUCTION
The Type II restriction enzymes are among the most valuable tools available to researchers in molecular biology. These enzymes recognize short DNA sequences (4–8 nucleotides) and cleave at, or close to, their recognition sites (1,2). A comprehensive database (REBASE) contains information about these enzymes including their recognition specificities and their sensitivity to DNA methylation (3). In the 1970s they were used extensively to provide physical maps of small DNAs and, in the 1980s, were used to map large DNAs. As methods for the determination of DNA sequence improved, it became convenient to search those sequences for potential restriction enzyme cleavage sites since that would permit the facile further manipulation of specific fragments. In fact, until the advent of the polymerase chain reaction (PCR) (4), they provided the most convenient way to manipulate individual genes and move them from one vector to another. For a while, it seemed that the ability of PCR to permit precise amplification of individual stretches of DNA might render the use of restriction enzymes obsolete. However, they merely found new utility by then serving as diagnostic reagents to show that DNA constructs had been made correctly. They still provide one of the cheapest and most convenient ways to characterize DNA constructs. They have also found use in analyzing the genomes of higher organisms using restriction fragment length polymorphisms (RFLPs) as physical markers (5) or by directly detecting the presence of single nucleotide polymorphisms (SNPs) (6).
Since most DNA constructs are now quickly sequenced, tools to locate restriction enzyme sites within these constructs are especially valuable. Every commercial software package to manipulate DNA sequences always includes one or more modules to detect restriction enzyme recognition sites. However, most rely on data files of recognition sites that are out-of-date, do not have links into REBASE or are cumbersome to use. One other web server, Webcutter (http://www.firstmarket.com/cutter/cut2.html), which cuts DNA with restriction enzymes, is also available, although its maintenance status is unclear. In this paper we describe a new tool, NEBcutter, that is freely available on the web and which analyzes DNA sequences for the presence of restriction enzyme sites in a convenient and easy to use manner.
METHODS
NEBcutter consists of a set of cooperating program modules. The module that finds recognition sites implements a brute force algorithm in C (gcc version 2.96, http://www.gnu.org/software/gcc). Most calculations (digests, filterings, etc.) as well as queue management tasks are also implemented in C. PHP scripts (version 4.0, http://www.php.net/) generate the dynamic HTML pages of the user interface. Graphics are created by C programs calling GD library functions (GD version 1.3, http://www.boutell.com/gd). The program package runs on a 440 MHz, single processor Sun NteraT1 server, in a SunOS 5.9 environment, running the Apache web server (version 1.3, http://httpd.apache.org).
The algorithm that detects open reading frames (ORFs) defines them as maximal length segments of DNA from start codons to stop codons in the same reading frame, assuming bacterial sequences and codons. Semi non-overlapping tiling of the input sequence with such ORFs is done according to the rules of a heuristic algorithm. This algorithm prefers longer ORFs to shorter ones and progressively discards short ORFs which significantly overlap with some longer ORF(s). This is intended to give a cursory view of where genes might be found in the input sequence.
Using NEBcutter
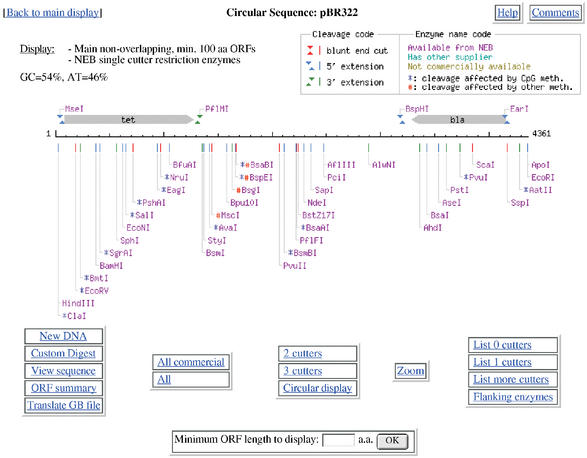
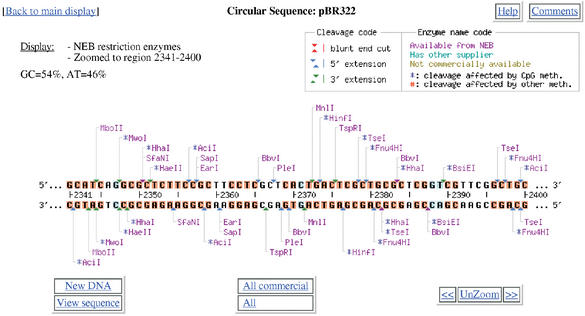
NEBcutter accepts an input sequence, which can be pasted in, picked up from a local file or retrieved from NCBI as a GenBank file via its accession number. Various options are available to select the size of ORFs to be displayed and the set of restriction enzymes to be used. The program calculates the positions of all restriction enzyme sites noting those that might potentially be blocked by overlapping methylation and finds the ORFs in the sequence. It then displays a schematic diagram of the sequence, the long ORFs, based on the rules described in the Methods and all restriction enzymes that cut it just once. The initial display also shows the enzymes that can be used in a complete digest to excise each ORF that is displayed. Figure 1 shows a typical digest, in this case a linear view of the plasmid pBR322. If the original DNA sequence is circular, then both linear and circular displays are offered. From the initial display there are many options to go further, including custom digestion with enzymes of your choice and various displays of the digest. It is also possible, from linear displays, to zoom in on a selected region of the sequence for a more detailed view. One option allows a particular region to be selected and those enzymes that cut immediately adjacent to the region to be displayed. This is useful to find enzymes able to excise the desired region. If zooming leads to a region of <60 nucleotides then the actual sequence is displayed (Fig. 2), together with any translation that is appropriate. On this display all enzymes that cut the sequence are shown, all bases that form parts of a restriction enzyme recognition sequence are highlighted and moving the mouse over an enzyme name will produce a box with the recognition sequence noted and the sequence itself becomes underlined in the display.
Figure 1.
The linear display of a digest of the plasmid pBR322. Note that the two main ORFs are flanked by sites (MseI and PflMI; BspHI and EarI) that could be used to excise them from the complete plasmid sequence. In this display only enzymes that cleave the sequence once are shown. Options for further digestion and display are available through the buttons at the bottom of the display.
Figure 2.
Zoomed display of a short region from pBR322. Note that bases highlighted in red and blue form parts of restriction enzyme recognition sites. The red ones are unambiguous, whereas the blue ones are degenerate bases. Thus BsiEI recognizes the sequence CGRYCG. The outer CG residues are invariant, while the inner bases are degenerate and, in the specific sequence shown, are GT. Bases shown in black do not form part of any restriction enzyme recognition site. This display can be useful to find restriction enzymes able to distinguish SNP alleles.
On any main display, moving the mouse over a restriction enzyme name will show its recognition sequence and the precise base number at which it cleaves. If more than one site is present for the enzyme then all other sites are underlined. Clicking on the enzyme name will produce a page with a summary of information about the enzyme including its methylation sensitivity, the kinds of ends produced, isoschizomers that are available and a list of other enzymes that can produce compatible ends. For NEB enzymes, information about digestion conditions and a link to the NEB web site is provided.
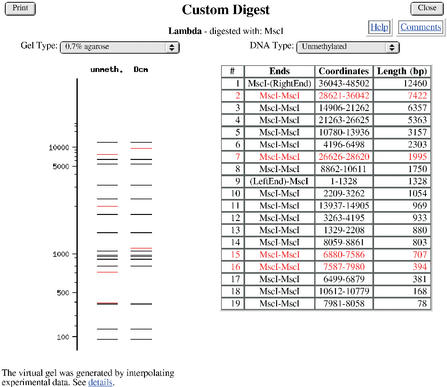
A key feature of NEBcutter is that it incorporates everything that is recorded in REBASE about the methylation sensitivity of any of the enzymes displayed when they overlap a Dam site (GATC), a Dcm site (CCWGG), a CpG site, an EcoKI site (AACN6GTGC) or an EcoBI (TGAN8TGCT) site. When theoretical digests are performed, the results include a comparison of the effects of methylation highlighting the bands that shift (Fig. 3).
Figure 3.
A theoretical digest showing the effects of overlapping Dcm methylation on the MscI sites (recognition sequence: TGGCCA) present in the sequence if the bacteriophage lambda had been grown in an E.coli strain expressing the Dcm methyltransferase (recognition sequence: CCWGG). Fragments that are affected by Dcm methylation are shown in red. The virtual gel is based on interpolation of a cubic spline curve generated from experimental data produced with fragments of known size.
On every page hot links are provided that lead back to the main page, provide help in interpreting the display or allow users to send comments back to the authors. The interface has been designed to be as intuitive as possible and to provide easy access to the main functions useful to researchers attempting to cleave their DNA. The current version of NEBcutter has processed more than 400 000 sequences from users. We are currently preparing a number of enhancements to NEBcutter and version 2.0 is scheduled for release by July 1st.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dana Macelis for help in providing REBASE data and Max Heiman for generously raising no objections to our using the name NEBcutter, which is very close to Webcutter, his program that also provides restriction enzyme cleavage information.
REFERENCES
- 1.Pingoud A. and Jeltsch,A. (2001) Structure and function of type II restriction endonucleases. Nucleic Acids Res., 29, 3705–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts R.J. and Halford,S.E. (1993) Type II restriction enzymes. In Linn,S.M., Lloyd,R.S. and Roberts,R.J. (eds). Nucleases. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp. 35–88. [Google Scholar]
- 3.Roberts R.J., Vincze,T., Posfai,J. and Macelis,D. (2003) REBASE—restriction enzymes and methyltransferases. Nucleic Acids Res., 31, 418–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saiki R.K., Gelfand,D.H., Stoffel,S., Scharf,S.J., Higuchi,R., Horn,G.T., Mullis,K.B. and Erlich,H.A. (1988) Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science, 239, 487–491. [DOI] [PubMed] [Google Scholar]
- 5.Lander E.S. and Botstein,D. (1986) Strategies for studying heterogeneous genetic traits in humans by using a linkage map of restriction fragment length polymorphisms. Proc. Natl Acad. Sci. USA, 83, 7353–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hao K., Niu,T., Sangokoya,C., Li,J. and Xu,X. (2002) SNPkit: an efficient approach to systematic evaluation of candidate single nucleotide polymorphisms in public databases. Biotechniques, 33, 822–828. [DOI] [PubMed] [Google Scholar]