Abstract
Since a large number of computationally predicted exons are not supported by existing sequence (e.g. ESTs) or experimental (e.g. expression analysis) data they need to be validated by other methods. ETOPE is designed to test computational predictions by using signals that have not been included in any current computational prediction method. The test is based on the ratio of non-synonymous to synonymous substitution rates between sequences from different genomes. It has been previously shown, by empirical data and computer simulation, to be a powerful criterion for identifying protein-coding regions. The ETOPE is available at http://nekrut.uchicago.edu/etope/.
INTRODUCTION
Considerable progress has been made in fine-tuning the algorithms for computational gene prediction (1). Most of the popular gene prediction programs available today, such as Genscan (2), use sophisticated pattern recognition approaches to find protein-coding regions within genomic DNA and to assemble them into genes. However, such patterns can occur by chance, leading to a high false-positive rate in most current gene prediction algorithms (3,4). This is why computationally predicted genes are routinely compared to experimental data or known sequences (most often to ESTs) to test their validity. This use of ESTs for validation of computational predictions has three limitations. Firstly, although huge numbers of EST sequences have been generated over the past 10 years, they do not represent all genes in a genome because they are dominated by highly expressed transcripts (1). Secondly, the majority of sequenced ESTs are human- or mouse-derived. For example, a current release of the UniGene database contains 3 850 573 human EST sequences but only 334 543 rat ESTs. This poses a challenge for the reliable annotation of the rat genome, which is almost completely sequenced. Thirdly, ESTs can be low quality sequences that contain considerable amounts of contamination. Thus, a match between a computationally predicted gene and an EST sequence does not guarantee the validity of the prediction.
We have developed a tool for validation of computational predictions that may overcome these limitations. The key feature of our approach is that it does not rely on homology information per se, but tests whether the homologous sequences are truly protein-coding by using evolutionary signals within these sequences. It is a comparative genomics approach and involves two steps. Firstly, a computational prediction from one species (e.g. human) is compared against sequences from another suitably diverged species (e.g. mouse) to find a homologous reading frame (orthologous or paralogous). Secondly, if there is a match, we examine the nucleotide substitutions within the resulting alignment to detect the pattern of functional conservation characteristic of the vast majority of protein-coding regions. Specifically, we calculate two values: KA, the number of non-synonymous substitutions per non-synonymous site and KS, the number of synonymous substitutions per synonymous site. These values are calculated using the approximate method of Yang and Neilsen (5), which takes into account the transition/transversion and codon usage biases. Next, we test whether the non-synonymous rate KA is lower than the synonymous rate KS by Fisher's exact test. If the ratio KA/KS is significantly smaller than 1 (P<0.05), then the computational prediction is most likely a true coding exon because a mutation that changes an amino acid is less likely to survive than a mutation that does not (6). This test has been shown to have a false-positive rate of 3% at the 5% significance level and only 0.7% at the 1% significance level (7). In addition, Guigo et al. (4) demonstrated empirically that the majority of computational predictions with the KA/KS<1 between human and mouse are confirmed by RT–PCR experiments and can be considered true genes.
THE ETOPE PROGRAM
The ETOPE test engine is implemented in Perl. It has been tested to work reliably with an Apache web server (http://www.apache.org) under Linux (Intel-based) and Solaris (Sparc-based) operating systems. It is currently installed on a Pentium III 450 MHz machine (nekrut.uchicago.edu) with 771 Mb of RAM running Red Hat Linux 7.0. To run ETOPE it requires NCBI BLAST (8), ClustalW 1.8 (9) and PAML (10) to be installed on the host machine. The PAML package is modified to run as the Apache user. Below we outline the test algorithm.
Finding matches. The nucleotide sequence of a potential protein-coding region, say a computationally predicted exon, is provided by the user. It is then compared against a user-specified database containing nucleotide sequences from a suitably diverged species (10–30% coding sequence divergence). For example, the user may compare a human computational prediction against a mouse sequence database. Presently available databases are listed in Table 1. The comparison is carried out using MEGABLAST (11) with the following parameters: -W 16 -t 11 -q -2. We use the discontinuous word approach (12) to achieve a high sensitivity and to maximize the system performance. After the search is completed, the program chooses a single database hit that has no stop codons in the alignable region and has the maximum alignment score. This step outputs an alignment between the reading frame of the computational prediction and the putative reading frame of the database hit.
Processing translations. The computational prediction and the database portions of the alignment from the previous step are translated. The resultant translations are realigned using ClustalW (9). After the global alignment is constructed, terminal gaps (if any) are removed so that both sequences have the same length. Corresponding nucleotide sequences are also trimmed so that they correspond exactly to the translations (nt length=aa length×3). The alignment of translations generated by ClustalW is now used as a guide to realign corresponding nucleotide sequences. This ensures that nucleotide sequences are aligned ‘codon-to-codon’ and that all gaps (if any) are placed between codons.
Estimation of KA and KS and the KA/KS test. Using the alignments from the previous step, the program estimates the KA and KS using the approximate method of Yang and Nielsen (5). The method reports the number of synonymous and non-synonymous sites (LS and LN) and the synonymous and non-synonymous rates KS (KS=SS/LS; SS=number of synonymous changes between the two sequences in the alignment) and KA (KA=SN/LN; SN=number of non-synonymous changes). To test whether KA/KS<1 (P<0.05) the program does the following: (i) it creates a two-way contingency table with rows containing numbers of synonymous and non-synonymous sites and columns containing numbers of changed and unchanged sites; (ii) it then tests independence between the numbers of changed synonymous and non-synonymous sites using Fisher's exact test (implemented in a separate Perl module) following the procedure from Sokal and Rohlf (13). If the test is significant at the 5% level, it provides further evidence that the region is protein-coding.
Table 1. Installed ETOPE databasesa.
| Name | Description | Version of the source database | No. of sequences | Species to useb |
|---|---|---|---|---|
| gs_hs_10_30 | Human Genscan predictions | Human Ensembl 10.30 | 73 128 | R, M, D, C, P |
| gs_mm_10_3 | Mouse Genscan predictions | Mouse Ensembl 10.3 | 110 379 | H, D, C, P |
| known_hs_10_30 | Known human genes | Human Ensembl 10.30 | 22 439 | R, M, D, C, P |
| known_mm_10_3 | Known mouse genes | Mouse Ensembl 10.3 | 16 000 | H, D, C, P |
| novel_hs_10_30 | Novel human genes identified by Ensembl | Human Ensembl 10.30 | 5 189 | R, M, D, C, P |
| novel_mm_10_3 | Novel mouse genes identified by Ensembl | Mouse Ensembl 10.3 | 12 097 | H, D, C, P |
| ug_hs_158 | Human Unigene cluster representatives (Hs.seq.uniq) | Human Unigene build 158 | 128 826 | R, M, D, C, P |
| ug_mm_119 | Mouse Unigene cluster representatives (Mm.seq.uniq) | Mouse Unigene build 119 | 94 528 | H, D, C, P |
| RIKEN_mm | RIKEN full length cDNAs | Fantom2 | 37 086 | H, D, C, P |
aDatabase versions as of publication submission.
bComputational predictions from these species can be compared against the corresponding database. R, rat; M, mouse; D, dog; C, cow; P, pig; H, human.
ETOPE INTERFACE
ETOPE accepts two types of input: (i) a single sequence (http://nekrut.uchicago.edu/etope/single.html) and (ii) a Genscan output (http://nekrut.uchicago.edu/etope/gs.html). The single sequence input page is designed to test the entire sequence of a computational prediction from any gene finding program. The Genscan input page allows the user to test individual exons of a Genscan gene model. The Genscan input page is useful for validation of suboptimal exons that do not have a high P-value associated with them. Both pages have a simple intuitive design. Presently, we only support the Genscan input page because it is the most widely used gene prediction tool. In the future we may add pages that accept output of other gene prediction programs.
Single sequence input page
The single sequence input page contains a text box for pasting the sequence to be analyzed, a pull-down menu for database selection and a pull-down menu for selecting the reading frame. First, the user provides a FASTA formatted nucleotide sequence to be tested. The sequence should represent a protein-coding region without untranslated regions. Next, the user chooses the database against which the sequence will be compared. Because some computational predictions are truncated, their first nucleotide may not correspond to the first codon position. ETOPE allows the user to specify whether the reading frame starts with the first (zero, default value), second (one), or third (two) position within the sequence using the reading frame menu. Pressing the ‘Run ETOPE’ button causes ETOPE to compare the input sequence against the selected database and to report the results of the KA/KS test.
Genscan page
The Genscan page contains a text box for pasting Genscan results and a pull-down menu for database selection. The user provides the output of Genscan and selects the database against which the Genscan prediction should be tested. Once the ‘Run ETOPE’ button is pressed, the ETOPE will process Genscan results and will display a table listing all Genscan exons. Suboptimal exons (P<75%) will be highlighted with yellow. The user can now press the ‘Analyze’ button next to exons of interest to perform the KA/KS test.
Output
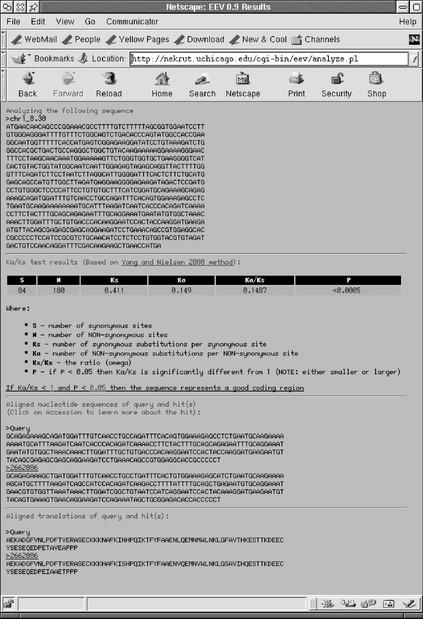
Both the single sequence and Genscan output versions of ETOPE generate the same type of output. An output contains four parts: (i) the original query sequence; (ii) the results of the KA/KS test; (iii) the portion of the query sequence that matches the database and the corresponding database hit; and (iv) the translations of the query sequence and the database hit. Figure 1 shows the result of testing a rat Genscan prediction chr1_8.30 from the University of California at Santa Cruz rat genome database (http://genome.ucsc.edu) against human Unigene sequences (database ug_hs_158; Table 1). The KA/KS=0.1487 and the test is highly significant (P<0.0005). The rat Genscan prediction used in this example does not match any annotated rat sequences such as known genes or ESTs. The matching sequence has a gi # 2662086 displayed as a hyperlink. Clicking on the hyperlink causes the web browser to display an NCBI web page containing the description of the sequence.
Figure 1.
ETOPE output page.
WHEN TO USE ETOPE
ETOPE compares the coding region of a computationally predicted gene against sequences from other species to find and to statistically test patterns of functional conservation. For ETOPE to work efficiently, the average divergence of coding sequences between the two species (a species from which computational prediction was taken, and the species it is to be compared against) should be in the range ∼10–30% (7). For each database in Table 1, we list appropriately divergent species that can be compared against that database. How can it be useful? For example, a complex trait mapping experiment identified a 150 000 kb candidate locus on rat chromosome 1 between positions 38 050 000 and 38 200 000. The next step is to identify genes within this interval. This can be done by using the University of California Santa Cruz genome browser (14). According to the UCSC browser this region contains 15 gene predictions generated by five different gene finders (Twinscan, SLAM, SGP, FgenesH++ and Genscan). None of the predictions overlap with any rat genes or EST sequences. To identify predictions that are likely to be real genes they can be tested by ETOPE against the human sequence database to further examine its coding potential. Among the 15 predictions, eight were confirmed by ETOPE. Other uses of ETOPE may include testing of genes that are reported as ‘putative’ or ‘pseudogenes’ in genome annotation databases and testing EST sequences for protein-coding capacity.
FUTURE ETOPE UPGRADES
ETOPE provides a simple and reliable way to test the coding potential of computationally predicted genes that are not supported by existing sequence data. ETOPE upgrades will involve algorithm improvements and hardware upgrades. Specifically, we will modify the algorithm to find and test not only the best hit from the database but a user specified number of hits. Work is underway to parallelize the ETOPE test engine. Currently ETOPE is limited to the databases listed in Table 1 due to hardware limitations. Planned hardware upgrades will allow users to test computational predictions against genomic sequences. We will add the following databases: human genomic sequences, mouse genomic sequences, rat genomic sequences, all human ESTs, all mouse ESTs and all rat ESTs.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Henrik Kaessmann for suggestions, Rick Blocker for UNIX system administration assistance and Rick Stevens for granting access to the Argonne National Laboratory Linux cluster. This study was supported by NIH grants GM30998, GM65499 and HD38287 and by Academia Sinica, Taiwan.
REFERENCES
- 1.Zhang M.Q. (2002) Computational prediction of eukaryotic protein-coding genes. Nature Rev. Genet., 3, 698–710. [DOI] [PubMed] [Google Scholar]
- 2.Burge C. and Karlin,S. (1997) Prediction of complete gene structures in human genomic DNA. J. Mol. Biol., 268, 78–94. [DOI] [PubMed] [Google Scholar]
- 3.Rogic S., Macworth,A.K. and Ouellette,F.B. (2001) Evaluation of gene-finding programs on mammalian sequences. Genome Res., 11, 817–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guigo R., Dermitzakis,E.T., Agarwal,P., Ponting,C.P., Parra,G., Reymond,A., Abril,J.F., Keibler,E., Lyle,R., Ucla,C. et al. (2003) Comparison of mouse and human genomes followed by experimental verification yields an estimated 1019 additional genes. Proc. Natl Acad. Sci. USA, 100, 1140–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Z. and Nielsen,R. (2000) Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol., 17, 32–43. [DOI] [PubMed] [Google Scholar]
- 6.Li W.-H. (1997) Molecular Evolution. Sinauer, Sunderland, MA. [Google Scholar]
- 7.Nekrutenko A., Makova,K.D. and Li,W.-H. (2002) The K(A)/K(S) ratio test for assessing the protein-coding potential of genomic regions: an empirical and simulation study. Genome Res., 12, 198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 9.Thompson J.D., Higgins,D.G. and Gibbson,T.J. (1994) ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Z. (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci., 13, 555–556. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z., Schwartz,S., Wagner,L. and Miller,W. (2000) A greedy algorithm for aligning DNA sequences. J. Comput. Biol., 7, 203–214. [DOI] [PubMed] [Google Scholar]
- 12.Ma B., Tromp,J. and Li,M. (2002) PatternHunter: faster and more sensitive homology search. Bioinformatics, 18, 440–445. [DOI] [PubMed] [Google Scholar]
- 13.Sokal R.R. and Rohlf,F.J. (2000) Biometry. W.H. Freeman and Co., New York, NY. [Google Scholar]
- 14.Kent J., Sugnet,C.W., Furey,T.S., Roskin,K.M., Pringle,T.H., Zahler,A.M. and Haussler,D. (2002) The human genome browser at UCSC. Genome Res., 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]