Abstract
Ribosomal frameshifting signals are found in mobile genetic elements, viruses and cellular genes of prokaryotes and eukaryotes. Typically they comprise a slippery sequence, X XXY YYZ, where the frameshift occurs, and a stimulatory mRNA element. Here we studied the influence of host translational environment and the identity of slippery sequence-decoding tRNAs on the frameshift mechanism. By expressing candidate signals in Escherichia coli, and in wheatgerm extracts depleted of endogenous tRNAs and supplemented with prokaryotic or eukaryotic tRNA populations, we show that when decoding AAG in the ribosomal A-site, E.coli tRNALys promotes a highly unusual single-tRNA slippage event in both prokaryotic and eukaryotic ribosomes. This event does not appear to require slippage of the adjacent P-site tRNA, although its identity is influential. Conversely, asparaginyl-tRNA promoted a dual slippage event in either system. Thus, the tRNAs themselves are the main determinants in the selection of single- or dual-tRNA slippage mechanisms. We also show for the first time that prokaryotic tRNAAsn is not inherently ‘unslippery’ and induces efficient frameshifting when in the context of a eukaryotic translation system.
Keywords: frameshifting/pseudoknot/ribosome/tRNA/translation
Introduction
The elongation phase of protein synthesis is a precise process and mechanisms exist to promote translational fidelity (reviewed in Czworkowski and Moore, 1996). However, a growing number of examples have been described of highly efficient ‘programmed’ frameshift sites (see Farabaugh, 1996, 2000 for reviews). Such frameshifts, which can occur at frequencies approaching 100%, are not errors in the classical sense in that they generate authentic proteins and are stimulated specifically by elements encoded in the mRNA. For this reason, they are considered more as extensions of the genetic code (recoding sites) (Gesteland and Atkins, 1996) rather than ‘natural’ errors, although there may be mechanistic similarities between the two (Farabaugh and Bjork, 1999). There is considerable interest in how programmed frameshifting occurs, as this may provide insights into normal frame maintenance, tRNA movement and the unwinding of mRNA secondary structures by ribosomes. Programmed –1 ribosomal frameshift signals are found most commonly in eukaryotic RNA viruses and Escherichia coli insertional elements (IS), where they facilitate expression of replicases and transposases, respectively (see Chandler and Fayet, 1993; Brierley, 1995; Futterer and Hohn, 1996; Farabaugh, 2000 for reviews). Examples have also been described in conventional cellular genes of E.coli (dnaX) (Blinkowa and Walker, 1990; Flower and McHenry, 1990; Tsuchihashi and Kornberg, 1990), Bacillus subtilis (cdd) (Mejlhede et al., 1999) and mammalian cells (Edr) (Shigemoto et al., 2001). The mRNA signals that cause frameshifting typically comprise two essential elements: a heptanucleotide ‘slippery’ sequence, where the ribosome changes reading frame, and an adjacent stimulatory signal. This can be an RNA secondary structure (often an RNA pseudoknot) located a few nucleotides downstream (Jacks et al., 1988; Brierley et al., 1989; ten Dam et al., 1990), a Shine–Dalgarno-like (SD-like) sequence upstream (Mejlhede et al., 1999) or a combination of both (Larsen et al., 1994; Rettberg et al., 1999). The slippery sequence consists of two homopolymeric triplets, conforming in the vast majority of cases to the motif X-XXY-YYZ. Frameshifting at this sequence is thought to occur by simultaneous (also referred to as ‘dual’ or ‘tandem’) slippage of the peptidyl and aminoacyl tRNAs, which are translocated from the zero (X-XXY-YYN) to the –1 phase (XXX-YYY) (Jacks et al., 1988). Following the slip, the tRNAs remain base-paired to the mRNA in at least two out of three anticodon positions. In prokaryotic systems, the generality of the simultaneous slippage model of frameshifting is not fully established. At most naturally occuring E.coli frameshift signals, including dnaX (slippery sequence A-AAA-AAG) (Tsuchihashi and Brown, 1992), IS911 (A-AAA-AAG) (Chandler and Fayet, 1993) and the G-T ORF of bacteriophage λ (G-GGA-AAG) (Levin et al., 1993), a simultaneous slippage mechanism is employed. However, in IS1, frameshifting is thought to occur by –1 slippage of a single lysyl-tRNA at the sequence A-AAA (from the underlined codon onto the overlapping AAA codon), despite the fact that the AAAA stretch is embedded within two potential and conventional slippery sequences (U-UUA-AAA-AAC) (Sekine and Ohtsubo, 1992). There are other examples of frameshift signals where the occurrence of a dual slippage mechanism is questionable on the grounds that re-pairing of the frameshifted P-site tRNA with the mRNA does not allow the formation of stable base-pairs at the non-wobble positions. These include IS3 (C CAA AAG) (Sekine et al., 1994), the bacteriophage T7 gene 10 signal (G-GUU-UUC) (Condron et al., 1991) and that of equine arteritis virus 1a/1b (G-UUA-AAC) (den Boon et al., 1991; Brierley et al., 1992).
In mechanistic terms, the position of the tRNAs during the slippage event is a key issue. Evidence supports the view that in the vast majority of cases, the slippery sequence-decoding tRNAs are present in the P- and A-sites of the ribosome (see Atkins et al., 2001 for a review). However, there are a few exceptions. In an E.coli expression system, frameshifting at the human immunodeficiency virus type 1 (HIV-1) gag/pro frameshift signal (U-UUU-UUA) is thought to occur when the tRNAs are in the ribosomal E- and P-sites (Horsfield et al., 1995). This hypothesis was proposed following the discovery that the presence of a termination codon immediately downstream of the U-UUU-UUA stretch reduced frameshifting some 5- to 10-fold. There are also two examples of –1 frameshift signals where a single tRNA slippage event is proposed. At the CP/12K signal of potato virus M (A-AAA-UGA), a P-site slip is proposed, with lysyl-tRNA slipping back by 1 nucleotide (nt) from the underlined codon when the A-site is unoccupied (Gramstat et al., 1994). The stimulation of such movements of peptidyl-tRNA by termination codons is not without precedent in other classes of frameshift signal (see Atkins et al., 2001). However, a –1 frameshift signal in the B.subtilis cytidine deaminase gene (cdd; CGA-AAG) (Mejlhede et al., 1999) is particularly unusual. Frameshifting at this site appears to occur without P-site slippage, with the A-site tRNALys repairing from AAG to AAA. How this can occur without simultaneous P-site tRNA slippage is not known, although displacement of the wobble pair (A:I) of the P-site tRNA by the first base of the re-pairing A-site tRNA is a possibility (Mejlhede et al., 1999). An event that bears similarity to the cdd frameshift event has been described for a viral frameshift system (Brierley et al., 1997). An investigation into the functionality of the frameshift signal of the coronavirus infectious bronchitis virus (IBV; U-UUA-AAC) in E.coli revealed that a variant with slippery sequence U-CUA-AAG was highly active (40%) (Brierley et al., 1997), despite possessing a slippery sequence which should allow only a single-tRNA slip in the A-site (from AAG back to AAA). This was unexpected, since viral frameshifting was generally believed to occur by simultaneous slippage and, as with cdd, no obvious mechanism exists to account for movement of just the A-site tRNA.
At present, we do not know whether such unusual A-site single-tRNA slippage events are restricted to prokaryotic ribosomes. There is a need to investigate more thoroughly the prevalence of such events and the features that can influence the selection of single- or dual-tRNA slippage mechanisms of frameshifting. Here, we employed the IBV frameshift signal as a model system to investigate whether the mechanism employed is determined by the host translational environment (prokaryotic versus eukaryotic) and/or the nature of the tRNAs decoding the slippery sequence in the ribosomal A- and/or P-sites, focusing on tRNALys and tRNAAsn. To achieve this, we exploited a recently developed methodology for the preparation of tRNA-dependent in vitro translation systems (Jackson et al., 2001). By expressing candidate frameshifting signals both in E.coli and in wheatgerm extracts (WG) depleted of endogenous tRNAs and supplemented with purified prokaryotic or eukaryotic tRNA populations, we show that, when present in the A-site, E.coli tRNALys can promote an unusual single-tRNA slippage event in both prokaryotic and eukaryotic ribosomes. We also demonstrate that prokaryotic tRNAAsn is not inherently ‘unslippery’ and is perfectly capable of inducing efficient frameshifting when placed in the environment of a eukaryotic translation system. However, with this tRNA, a dual slippage event is promoted. These findings provide strong evidence that it is the tRNAs themselves that are the main determinants in the selection of a single- or dual-tRNA slippage mechanism. Models for the disposition of tRNAs on the ribosome during the unusual single-tRNA slip are discussed.
Results
Single- and dual-tRNA slippage events during ribosomal frameshifting in E.coli
In E.coli, the high frameshift efficiencies elicited by ribosomal frameshift signals containing slippery sequences of the general organization X-XXA-AAG (Weiss et al., 1989) have been ascribed to the presence in this organism of a single tRNALys isoacceptor with anticodon 3′UUU*5′. The modification (*) on the wobble base of this tRNA (5-methylaminomethyl-2-thiouridine; mnm5s2U) has been proposed to weaken the mRNA– tRNA interaction, permitting efficient frameshifting (Tsuchihashi, 1991). Although mammalian cells harbour a lysyl-tRNA closely related to the E.coli molecule [with anticodon 3′UUU**5′, where U** is 5-methylcarbonylymethyl-2-thiouridine (mcm5s2U)], the U-UUA-AAG signal does not stimulate efficient frameshifting in mammalian systems, probably because two additional tRNALys isoacceptors are present with anticodon 3′UUC5′ which outcompete the mcm5s2U-containing isoacceptor (Tsuchihashi and Brown, 1992). With slippery sequences of the order X XXA AAC, decoded by a single tRNAAsn isoacceptor with anticodon 3′UUQ5′ (Q is queuosine), the situation is reversed in that in mammalian systems such signals are usually highly efficient, but in E.coli promote only low levels of frameshifting. At present, we have no explanation for the different functionality of tRNAAsn in the two systems.
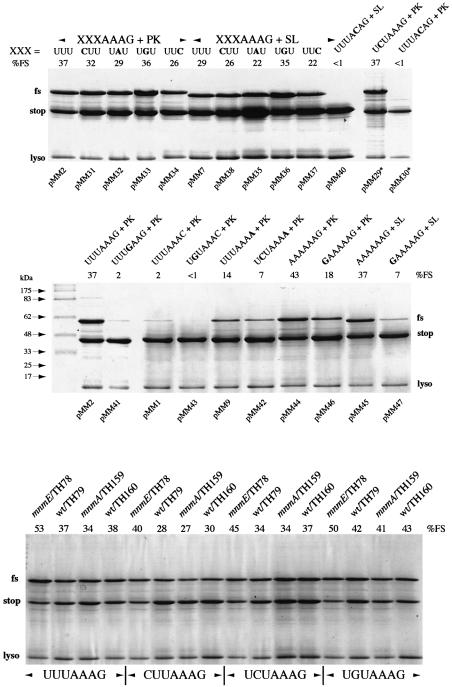
It is clear that the tRNA species decoding the slippery sequence in the ribosomal A-site can have a major impact on the magnitude of frameshifting. It is also possible that the actual mechanism of frameshifting may change depending upon the identity of the tRNA. In a study of frameshifting at the IBV signal, we made the unexpected observation that a variant with slippery sequence U-CUA-AAG (wild type is U-UUA-AAC) showed highly efficient frameshifting in E.coli (40%) but poor frameshifting (<1%) in the rabbit reticulocyte lysate in vitro translation system (RRL) (Brierley et al., 1997). In E.coli, the P-site codon in the mutant, U-CUA-AAG, is probably decoded by a minor tRNALeu isoacceptor with anticodon 3′GAU5′ (Inokuchi and Yamao, 1995) and if it were to slip into the –1 reading frame, in accordance with the simultaneous slippage model, it could form only a single G-U base-pair with the –1 frame codon (see Table I). This level of re-pairing seemed insufficient to account for the level of frameshifting seen and we speculated that when the highly frameshift-prone tRNALys is present in the ribosomal A-site, it may induce frameshifting by a different mechanism, one where the requirements for re-pairing of the adjacent P-site tRNA were less stringent. However, we could not rule out the possibility that the tRNALeu isoacceptor was itself unusually slippery and in some way tolerant of restricted post-slippage pairing. For this reason, we examined here a broader range of IBV slippery sequence mutants, decoded by a variety of tRNAs, and measured frameshifting in E.coli BL21 cells (see Materials and methods). In the pMM plasmid series, the various slippery sequences were present in combination with the IBV pseudoknot or a related hairpin–loop structure (Brierley et al., 1991) (see Figure 1) and specify the synthesis of a 33 kDa non-frameshift product and a 50 kDa –1 frameshift product. As can be seen in Figure 2, variants of U-UUA-AAG with individual point mutations that were predicted to disrupt slippage of the P-site tRNA retained high levels of frameshifting (Figure 2). This was the case even when the expected number of P-site post-slippage tRNA anticodon:mRNA codon contacts formed was only one or less (taking into account pairing specificities of tRNAs, including anticodon modifications; see Table I). Thus, in all cases, substantial frameshifting was promoted under conditions where only the A-site tRNALys could re-pair in the –1 frame. In other aspects, the frameshift was typical of a traditional –1 ribosomal frameshift event. First, point mutations that changed the slippery sequence so as to reduce the predicted stability of the post-slippage A-site mRNA codon–anticodon complexes either greatly reduced (pMM41, slippery sequence U-UUG-AAG; 2%) or abolished frameshifting (pMM30, pMM40; slippery sequence U-UUA-CAG). Secondly, frameshift efficiency was comparably higher in those variants where the stimulatory RNA was a pseudoknot (pMM2, 29, 31–34) as opposed to a stem–loop (pMM7, 35–38, 40), indicating that the event is responsive to the nature of the downstream structure.
Table I. Details of the post-slippage P-site mRNA–tRNA contacts predicted to form following –1 ribosomal frameshifting in E.coli.
| Slippery sequence | P-site tRNA | Post-slip contactsin P-site (codon:anticodon) | Wobble basemodificationa | Supposed stability ofpost-slip complexb | % frameshifting(pseudoknot) | % frameshifting(stem–loop) |
|---|---|---|---|---|---|---|
| UUUAAAG | tRNALeu | 5′-UUU-3′ | cmnm5Um | Stable | 37 | 29 |
| ∗∗ | ||||||
| 3′-AAU-5′ | ||||||
| CUUAAAG | tRNALeu | 5′-CUU-3′ | cmnm5Um | Unstable | 32 | 26 |
| ∗ | ||||||
| 3′-AAU-5′ | ||||||
| UAUAAAG | tRNAIle | 5′-UAU-3′ | Lysidine | Unstable | 29 | 22 |
| 3′-UAL-5′ | ||||||
| UCUAAAG | tRNALeu | 5′-UCU-3′ | Probably modified | Unstable | 37 | nd |
| ∗ | ||||||
| 3′-GAU-5′ | ||||||
| UGUAAAG | tRNAVal | 5′-UGU-3′ | cmo5U | Unstable | 36 | 35 |
| 3′-CAU-5′ | ||||||
| UUCAAAG | tRNASer | 5′-UUC-3′ | cmo5U | Stable | 26 | 22 |
| ∗∗ | ||||||
| 3′-AGU-5′ |
aFor a complete description of the modifications, see Inokuchi and Yamao (1995).
bWithin the context of a programmed –1 ribosomal frameshift.
nd, not determined.
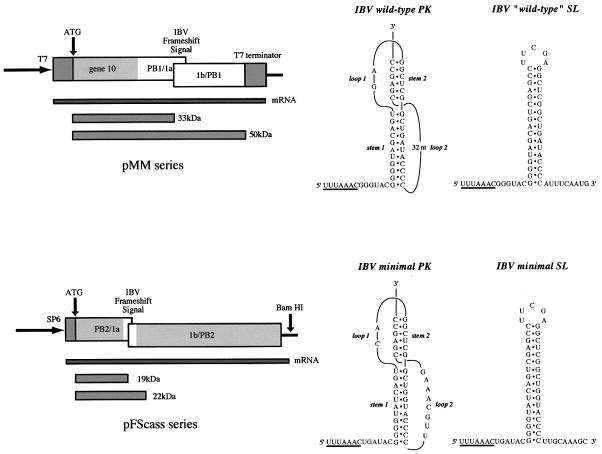
Fig. 1. Plasmids used in this study. Frameshifting in E.coli BL21 cells was studied using the pMM series. Induction of T7 RNA polymerase with IPTG (see Materials and methods) leads to the expression of a 33 kDa non-frameshifted product, corresponding to ribosomes that terminate at the IBV 1a stop codon and a 50 kDa frameshift product from ribosomes that frameshift prior to encountering the stop codon and continue to translate the 1b ORF in the –1 frame. Frameshifting in tRNA-depleted WG extracts employed synthetic mRNAs transcribed from the pFScass plasmid series. Here, the non-frameshifted and frameshifted species are 19 and 22 kDa, respectively. The frameshift signal present in each plasmid contained either a pseudoknot (PK) or a related stem–loop structure (SL). In the pMM series, these were the wild-type IBV PK and a related SL (Brierley et al., 1991). In the pFScass series, the minimal IBV PK was employed (Brierley et al., 1992) along with a related SL (this study).
Fig. 2. Frameshift assays in E.coli cells. Purified proteins expressed from the relevant frameshift plasmids were separated on 15% SDS–polyacrylamide gels and detected by staining with Coomassie Brilliant Blue R as described in Materials and methods. Unless indicated (E.coli TH78, 79, 159, 160), E.coli BL21 cells were employed. The non-frameshifted (stop) and frameshifted (fs) products are indicated. Lysozyme (lyso) carried over from the purification procedure is indicated. The relevant mutant number is indicated at the bottom of each track and the frameshift efficiency (%FS) at the top of each track. The slippery sequence and stimulatory RNA tested are shown above or below each track with relevant changes in bold. XXXAAAG, variant slippery sequence where the base composition of XXX is indicated above the relevant track; PK, wild-type IBV pseudoknot; SL, ‘wild-type’ stem–loop. *pMM29 and pMM30 were reported previously (Brierley et al., 1997).
Previously, Weiss et al. (1989) assessed the primary sequence requirements for frameshifting in E.coli of a variant of the mouse mammary tumour virus (MMTV) gag/pro frameshift signal with slippery sequence A-AAA-AAG. In contrast to what is observed with the IBV system, even a modest reduction in the potential for post-slippage P-site mRNA–tRNA pairing (e.g. a change from A-AAA-AAG to G-AAA-AAG) reduced frameshift efficiency markedly with the MMTV signal, indicating that a dual slippage event was occurring. The most obvious explanation for the discrepancy between the two studies is that the identity of the P-site tRNA is influencing the slippage process. To test this, we compared variant IBV signals with slippery sequences A-AAA-AAG and G-AAA-AAG together with either pseudoknot or hairpin stimulator. As shown in Figure 2, the frameshift efficiencies seen were consistent with the study of Weiss et al. (1989). The A-AAA-AAG signal promoted efficient frameshifting with a pseudoknot (pMM44, 43%) or a stem–loop stimulator (pMM45, 37%), but a change to G-AAA-AAG reduced frameshifting markedly (pMM46, 18%; pMM47, 7%). Thus, the ability of tRNALys to induce a single tRNA slip can be influenced by the adjacent P-site tRNA. Certainly, when this tRNA is also tRNALys, a dual slippage event appears to be the most likely outcome.
We also tested whether the strength of the interaction between the anticodon of tRNALys and the mRNA codon prior to slippage could influence the selection of a single- or dual-slippage mechanism by assessing frameshifting in constructs where the A-site codon was AAA. It is known that the frameshift efficiency elicited in E.coli by a combination of U-UUA-AAA and the IBV pseudoknot is considerably lower than that seen when the slippery sequence is U-UUA-AAG. It is believed that this is a consequence of the increased recognition of the AAA codon by tRNALys (Lustig et al., 1981; Yokoyama et al., 1985; Tsuchihashi and Brown, 1992; Brierley et al., 1997). Here, comparing U UUA AAA (pMM9) and U CUA AAA (pMM42), we found that the P-site change resulted in a reduction in frameshifting (from 14 to 7%; Figure 2), suggesting that a dual-slippage mechanism operates during frameshifting at U-UUA-AAA. A similar pattern was seen in comparisons of U-UUA-AAC (pMM1; 2%) and U-GUA-AAC (pMM43; <1%), indicating that a simultaneous slippage process also occurs when the A-site codon is decoded by tRNAAsn.
The potential role of the mnm5s2 modifications of tRNALys U34 in the single-tRNA slippage mechanism was also examined using two mutant E.coli strains (mnmA and mnmE) unable to fully modify tRNALys (Brierley et al., 1997; Hagervall et al., 1998). In these strains, tRNALys contains mnm5U34 (mnmA) or s2U34 (mnmE). Frameshifting at four different slippery sequences (U-UUA-AAG, C-UUA-AAG, U-CUA-AAG or U-GUA-AAG) was tested in each strain and the results are shown in Figure 2. For all slippery sequences tested, very little effect on frameshifting was seen in the mnmA strain (there was a very small, although consistent reduction), but in the mnmE background, frameshifting was noticeably stimulated in comparison to the isogenic wild-type strain. Thus, the individual modifications do not appear to be required for the frameshift process. We cannot rule out that they are required in combination, but an mnmA, mnmE double mutant strain remains to be isolated.
Single- and dual-tRNA slippage events during ribosomal frameshifting in a eukaryotic in vitro translation system
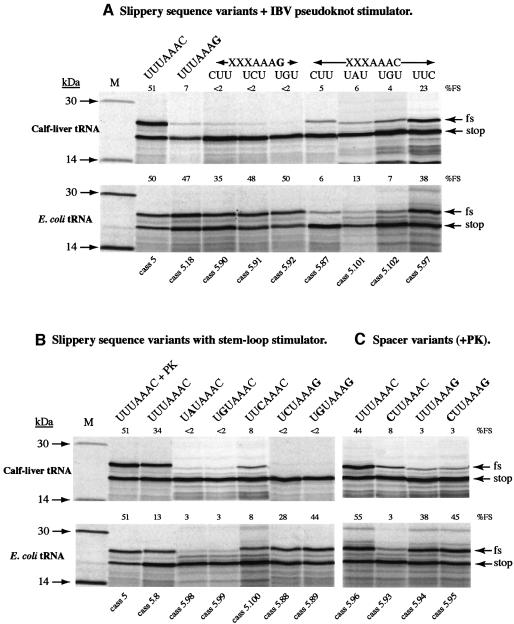
We next addressed whether this unusual single tRNA slippage event was restricted to the prokaryotic translational apparatus or could also occur in a eukaryotic system. To test this, we programmed a tRNA-dependent in vitro translation system derived from WG with reporter mRNAs containing variants of the IBV frameshift signal, following addition of either eukaryotic (calf liver) or prokaryotic (E.coli) tRNA populations. The mRNAs employed in this analysis—derived from plasmid pFScass 5, which contains the minimal IBV pseudoknot (Brierley et al., 1992) and pFScass 5.8, in which the pseudoknot was replaced by a related hairpin (Figure 1)—specify the synthesis of an 18 kDa non-frameshift product and a 22 kDa –1 frameshift product. The prokaryotic tRNAs were derived from E.coli JM101 cells and purified under acidic conditions to preserve their amino-acyl groups (see Materials and methods). As expected (Pfitzinger et al., 1989), we found that translation in depleted WG extracts could be rescued by aminoacetylated E.coli tRNAs, although translation efficiencies were somewhat reduced in comparison to eukaryotic tRNA populations. The tRNA content and tRNA dependence of the depleted WG used in this experiment are shown in Figure 3. Translational activity was entirely dependent upon the addition of exogenous tRNA. Slippery sequence variants of pFScass 5 and 5.8 were translated in depleted WG extracts supplemented with either calf-liver or E.coli tRNAs and the results are shown in Figure 4. With calf-liver tRNAs, the pattern of frameshifting seen was consistent with previous studies, although the efficiencies were uniformly slightly higher, indicating that the depleted WG is somewhat more promiscuous. In combination with the pseudoknot (Figure 4A), the wild-type slippery sequence (U-UUA-AAC, pFScass 5) stimulated efficient frameshifting (51%), a variant with slippery sequence U-UUA-AAG (pFScass 5.18) had greatly reduced frameshifting (7%), and variants with point mutations in the first part of the slippery sequence (U-UUA-AAC) showed reduced frameshifting, consistent with the dual-tRNA slippage model of frameshifting. P-site variants of the slippery sequence U-UUA-AAG also showed reduced levels of frameshifting (<2%), and we attribute this to an inability of the calf-liver tRNALys species decoding the A-site codon (presumably an isoacceptor with anticodon 3′UUC5′) to slip. The situation was different in translations programmed with E.coli tRNAs. Under these conditions, frameshift signals with a slippery sequence ending with AAG (pFScass 5.18, 5.90, 5.91, 5.92), decoded by prokaryotic tRNALys, showed much greater activity (47, 35, 48 and 50%, respectively), strongly supporting the view that an A-site single tRNALys slippage event also occurs in the plant system. In contrast, the frameshift efficiency of slippery sequences ending in AAC (i.e. A-site decoded by prokaryotic tRNAAsn) was highly dependent on the P-site tRNA and its ability to re-pair in the –1 phase. In constructs in which P-site re-pairing was predicted to be impaired (cass 5.87, C-UUA-AAC; 5.101, U-AUA-AAC; 5.102, U-GUA-AAC), the frameshift efficiencies were considerably lower (6, 13 and 7%, respectively) than those in which such re-pairing was likely (cass 5, U-UUA-AAC; 5.97, U-UCA-AAC), where frameshifting occurred efficiently (50 and 38%, respectively). This pattern closely resembled that seen with calf-liver tRNAs and supports the view that prokaryotic tRNAAsn is not ‘inherently unslippery’ and is capable of promoting efficient dual-tRNA frameshifting within the context of a eukaryotic ribosome. Figure 4B shows the translation products synthesized from a set of frameshift reporter mRNAs in which the stimulatory structure was a hairpin–loop. The pattern of frameshifting observed was qualitatively similar to that seen with the pseudoknot in that the lysyl- and asparaginyl-tRNAs decoding the slippery sequences are associated with single- and dual-tRNA slippage events, respectively. Thus, the nature of the stimulatory RNA does not appear to influence the mechanism of frameshifting. In quantitative terms, frameshifting occurred uniformly less efficiently with the hairpin stimulator, consistent with previous studies (Brierley et al., 1991, 1997). However, the presence of the hairpin or pseudoknot was required for frameshifting; the introduction of a destabilizing mutation into the stem of the hairpin or stem 1 of the pseudoknot (in constructs with slippery sequence U-GUA-AAG) greatly reduced frameshifting in translations supplemented with E.coli tRNAs (to 2%, data not shown).
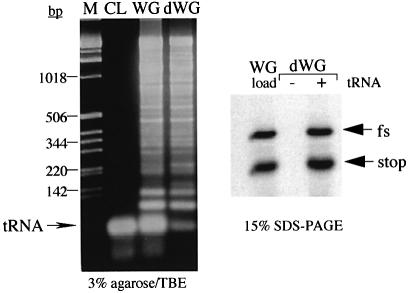
Fig. 3. Nucleic acid content and translational activity of depeleted WG. Commercial WG extracts (Promega) were depleted of tRNAs by ethanolamine–Sepharose chromatography. The load material (WG) and pooled, depleted fractions (dWG) were tested for their ability to translate a synthetic mRNA derived from BamHI-digested plasmid pFScass 5 in the absence (–) or presence (+) of added calf-liver tRNA (to 50 µg/ml) (right) and for nucleic acid content (left). Translation products were labelled with [35S]methionine, separated on SDS–15% polyacrylamide gels and detected by autoradiography. The pFScass 5 mRNA directs the synthesis of a 19 kDa non-frameshift (stop) and a 22 kDa –1 frameshift (fs) product (Brierley et al., 1992). Translation was entirely dependent upon the added tRNAs. Nucleic acids were separated on a 3% agarose–TBE gel, stained with ethidium bromide and photographed. Size markers (M) were kb ladder (Gibco-BRL). The CL lane was loaded with 1 µg of calf-liver tRNAs (Sigma). Some 95% of the endogenous WG tRNAs was removed by the chromatography step.
Fig. 4. Frameshift assays in tRNA-depleted WG extracts. (A–C) In vitro translations were performed in tRNA-dependent WG in the presence of tRNAs from calf-liver or E.coli JM101 cells. Translations were programmed with mRNAs derived from pFScass 5 or mutant derivatives. Translation products were labelled with [35S]methionine, separated on SDS–15% polyacrylamide gels and detected by autoradiography. The non-frameshifted (stop) and frameshifted (fs) products are indicated by arrows. The relevant mutant number is indicated at the bottom of each track and the frameshift efficiency (%FS) at the top of each track. The slippery sequence and stimulatory RNA tested is indicated with relevant changes in bold. M, low molecular weight 14C protein size standards (Amersham Pharmacia Biotech); XXXAAAC and XXXAAAG, variant slippery sequences where the base composition of XXX is indicated above the relevant track; PK, minimal IBV pseudoknot. In (C), the spacer sequence between the slippery sequence and pseudoknot was 5′-UACUGA-3′.
Given the published evidence that in E.coli HIV-1 frameshifting appears to involve the E- and P- sites (Horsfield et al., 1995), we wished to rule out any influence of the termination codon present immediately downstream of the slippery sequence in the pFScass 5 series (UGA, Figure 1). To do this, we changed the spacer region of four of the constructs (with slippery sequences U-UUA-AAC, C-UUA-AAC, U-UUA-AAG and C-UUA-AAG and a downstream pseudoknot) from 5′-UGAUAC-3′ to 5′-UACUGA-3′ to move the stop codon to a position 3 nt downstream. As can be seen in Figure 4C, the pattern of frameshifting seen with all four constructs was largely unaffected by the spacer change, arguing against involvement of the E-site in the frameshift mechanism.
Discussion
Here, we tested the influence of host translational environment and the identity of the slippery sequence-decoding tRNAs on the mechanism of programmed –1 ribosomal frameshifting. By expressing candidate frameshifting signals in E.coli and in WG extracts depleted of endogenous tRNAs and supplemented with prokaryotic or eukaryotic tRNA populations, we found that when present in the ribosomal A-site, E.coli tRNALys promotes a highly unusual single-tRNA slippage event in both prokaryotic and eukaryotic ribosomes. The efficiency of the process is influenced both by the A-site codon (AAG or AAA) and the identity of the P-site tRNA. Asparaginyl-tRNA, on the other hand, promoted a dual slippage event in either system. The type of stimulatory RNA, stem–loop or pseudoknot, did not appear to influence the choice of frameshift mechanism, although it did modulate the efficiency of the process. Thus, the tRNAs themselves are the main determinants in the selection of single- or dual-tRNA slippage mechanisms. The tRNA-dependent translation system enabled us to confirm, for the first time, that prokaryotic asparaginyl tRNA is not inherently ‘unslippery’ and is capable of inducing highly efficient frameshifting in the context of a eukaryotic translation system.
Before discussing the implications of these findings, it is first necessary to consider the supportive evidence that the single-tRNA slip is indeed taking place in the ribosomal A-site. Heelprint analysis of RRL ribosomes paused at the IBV pseudoknot indicate that the ribosomal P- and A-sites are positioned over the slippery sequence during the pause (Kontos et al., 2001) and the same is true for WG ribosomes (Kontos, 1997). Furthermore, the response of both prokaryotic (Brierley et al., 1997) and eukaryotic (Brierley et al., 1989) ribosomes to changes in the length of the spacer region between slippery sequence and stimulatory RNA is the same, i.e. reduction to 3 (nt) or expansion to 9 nt (from the normal 6 nt) abolishes frameshifting in either system. The decoding sites of the prokaryotic and eukaryotic ribosomes employed here are, therefore, highly likely to interact with the pseudoknot or stem–loop at the same relative position on the mRNA. The differential responses to pseudoknot- or stem–loop-stimulated frameshifting observed in the present study are consistent with this belief. It seems highly unlikely, therefore, that the single-tRNA slippage event occurs in the P-site, since this would require movement of the ribosome (hence unwinding of the pseudoknot) by an additional codon. Furthermore, the differential responses in frameshifting seen when the ‘A-site’ of the slippery sequence is decoded by tRNALys or tRNAAsn are unaffected by the presence of a stop codon immediately downstream of the slippery sequence, arguing against a slippage mechanism involving the P- or P/E-sites.
Our experimental data are strongly supportive of an A-site single tRNALys slippage event. However, the nature of the P-site tRNA can also influence the process. Although most of the P-site tRNAs tested (tRNALeu, tRNAIle, tRNAVal, tRNASer) allowed efficient single-tRNA frameshifting, the presence of tRNALys in the P-site (when decoding G-AAA-AAG) was quite inhibitory. Another important factor is the nature of the wobble pair formed between the A-site tRNALys and the mRNA. It is generally accepted that the strength of the interaction between the codon and anticodon can influence frameshifting efficiency and, in this light, the role of the mnm5s2 modifications of tRNALys U34 is significant. In normal translation, these modifications are thought to restrict wobble, preventing misreading of near-cognate codons ending in U, but at the cost of greatly reduced recognition of G (Yokoyama et al., 1985). The high frameshift efficiency elicited by E.coli tRNALys at the AAG codon is consistent with a weak wobble pair, and it was perhaps surprising to find that in mnmA and mnmE strains, the absence of mnm5 or s2 did not lead to a notable reduction in frameshifting (there was a very small reduction in the case of mnmA; Figure 2). However, these data are consistent with more recent studies on these modifications (Brierley et al., 1997; Hagervall et al., 1998; Krüger et al., 1998) and support the idea that the 5-methylaminomethyl group facilitates base-pairing with G and in its absence (in the mnmE strain) the interaction is weaker, leading to increased frameshifting. In contrast, the absence of the 2-thio group (mnmA) does not appear to affect recognition of G. It would be interesting to test frameshifting in a strain defective in the synthesis of both modifications, should such a strain be viable. Recognition of the AAG codon by wild-type tRNALys, however, is still sufficiently poor to allow efficient frameshifting. A change of this codon to AAA, which is bound more tightly to the anticodon (at least in vitro) (Lustig et al., 1981; Yokoyama et al., 1985), gave the expected reduction in frameshift efficiency at U-UUA-AAA (pMM9), but alteration to U-CUA-AAA (pMM42; Figure 2) depressed the efficiency further, indicating that a single tRNA slip was less favoured under these circumstances. However, it will be necessary to test additional P-site tRNAs in the context of an A-site AAA codon before the significance of this observation can be fully assessed.
The capacity of E.coli tRNALys to promote a single tRNA slip does not appear to be restricted to prokaryotic ribosomes as the event was quantitatively and qualitatively similar in a eukaryotic translational environment. Escherichia coli tRNAAsn, on the other hand, promoted a simultaneous slippage event in either system, consistent with previous studies of the closely related MMTV frameshift signal (Weiss et al., 1989; Chamorro et al., 1992). This suggests that it is the tRNA rather than the host translational environment that is the main determinant in the selection of single- or dual-tRNA slippage mechanisms. The extent of the frameshift however was variable between host systems. Although the prokaryotic asparaginyl- and lysyl-tRNAs promoted similar levels of frameshifting in the WG system, this was not the case in E.coli. It has been known for some time that in prokaryotes, frameshifting at slippery sequences that end in AAAC is generally very inefficient in comparison to eukaryotic systems (Weiss et al., 1989). The prokaryotic and eukaryotic asparaginyl-tRNAs employed in this study have identical anticodon loops and it is known that the presence or absence of the queuosine modification of the wobble base does not influence frameshifting (Brierley et al., 1997; Marczinke et al., 2000) except for yeast tRNAAsn (Carlson et al., 2000). It has thus been speculated that differences in frameshifting efficiencies seen between the two systems are a consequence of either sequence variation elsewhere in the tRNAs or the differing translational environments (Marczinke et al., 2000). Here we show for the first time that prokaryotic tRNAAsn is not inherently ‘unslippery’ and is able to promote efficient frameshifting in the WG system to a level similar to that stimulated by WG tRNAAsn. Thus, prokaryotic ribosomes somehow restrict frameshifting by this tRNA. It is clear, therefore, that the two translation systems are not identical from a frameshifting perspective, a point that must be borne in mind when attempting mechanistic comparisons.
Current models of ribosomal frameshifting seem inadequate to explain a single-tRNA slippage event of the kind described here. One can speculate that the A-site tRNALys slips during translocation at a time in the cycle when the P-site tRNA detaches from the mRNA as it moves fully into the E-site (Weiss et al., 1989). Under these circumstances, there would be no requirement for P-site tRNA re-pairing (Brierley et al., 1997). This is consistent with an earlier suggestion that pseudoknot-induced ribosomal pausing may occur during translocation (Kontos et al., 2001), but not with studies of antibiotics that influence frameshifting, which indicate that the event takes place prior to translocation (see Plant et al., 2003). A perhaps more compelling model can be devised based on the –1 frameshifting signal of the B.subtilis cytidine deaminase gene (cdd; CGA-AAG) (Mejlhede et al., 1999). Frameshifting at this site has been suggested to occur without P-site slippage, with the A-site tRNALys repairing from AAG to AAA, and the first base of the re-pairing A-site tRNA displacing the weak wobble pair (A:I) of the P-site tRNAArg (anticodon 3′GCI5′). The CGA codon is rare, yet replacing it with another rare arginine codon (AGA) leads to a great reduction in frameshifting, suggesting that the important aspect of the CGA codon is the weak pair it forms with the decoding tRNAArg rather than tRNA abundance (Mejlhede et al., 1999). The single tRNA slippage event detailed in the present study has similarities to the cdd signal and could conceivably occur in the same manner with the first base of the repairing tRNALys displacing the wobble base of the adjacent P-site tRNA. In almost all of the examples tested, the wobble base that would be displaced comprises an A in the mRNA and a modified U of some form in the anticodon. If the modification were to weaken the base-pair in a manner akin to the aforementioned A:I pair, this may promote displacement and the stimulation of a –1 single-tRNA slip. Indeed, in the one case tested where the mRNA base was G (in pMM41, U-UUG-AAG), frameshifting was greatly reduced (<2%). In the case of this site, the decoding tRNA in the P-site, tRNALeu, possesses an unmodified anticodon (3′AAC5′) that would presumably have a stable wobble pair (G:C). However, a more systematic analysis is required before this hypothesis can be generally accepted. Another issue that requires resolution is the role of the stimulatory RNAs in frameshifting. The single-tRNA slippage event described in the present study requires the participation of a downstream stimulatory element (pseudoknot or stem–loop), whereas frameshifting at the cdd site is promoted by an interaction between the anti-Shine–Dalgarno (anti-SD) sequence close to the 3′-end of 16S rRNA and an SD-like sequence 14 nt upstream of the recoding site (Mejlhede et al., 1999). An additional question is how such movement could be accommodated by the ribosome. Recent crystallographic studies indicate that there is a considerable gap (10 Å) between the anticodon loops of the P- and A-site tRNAs that could potentially allow some reorganization of the A-site tRNA without P-site tRNA slippage (Yusupov et al., 2001). However, whether there is sufficient freedom of movement to allow the formation of a post-slippage pair across the mRNA kink present between the P- and A-site codons is unknown.
Materials and methods
Transfer RNAs
Calf-liver transfer RNAs were purchased from Sigma or Boehringer. Charged E.coli tRNAs were prepared from E.coli JM101 according to the procedure of Varshney et al. (1991). Inhibitory acidic polysaccharides were removed by DE52 column chromatography; 4 ml columns were equilibrated with 140 mM KAc pH 4.5, 100 µg of tRNA loaded, the column washed with 10 vols of equilibration buffer and the tRNAs eluted with the same buffer containing 0.3 M NaCl. Charged tRNAs were stored at –70°C in 10 mM sodium acetate pH 4.5.
Preparation of a tRNA-dependent WG in vitro translation system
The specific depletion of tRNAs from commercial (Promega) WG extracts was achieved by ethanolamine–Sepharose column chromatography under defined salt conditions (105 mM KCl, 10 mM NaCl, 1.6 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol, 10 mM HEPES–KOH pH 7.4) according to the method of Jackson et al. (2001). WG was diluted (to 60–70% v/v) to adjust the K+ content to match that of the column buffer and supplemented with amino acids to 1 mM (except methionine, which was not added). One millilitre columns were loaded with 1–2 ml lysate and eluate fractions tested for their ability to translate a reporter mRNA in the presence or absence of added tRNAs (at 50 µg/ml final concentration). Active fractions with good tRNA dependence (<5% residual activity in the absence of added tRNA) were retained and stored under liquid nitrogen prior to use.
Site-specific mutagenesis
Site-directed mutagenesis was carried out as described (Brierley et al., 1989) or using the Stratagene ‘Quik Change’ kit.
Construction of plasmids
Plasmids for frameshift assays in E.coli were derived from pFS7, pFS8 or mutant derivatives (Brierley et al., 1989, 1991). These plasmids contain the IBV ORF 1a/1b frameshift signal flanked by portions of the influenza virus A/PR8/34 PB1 gene (Young et al., 1983). Following site-directed mutagenesis to introduce changes in the IBV slippery sequence or spacer region, 585bp NheI–EcoRI fragments encompassing the frameshift region were subcloned from the mutated plasmids into BamHI-cleaved pET3xc, an E.coli expression vector (Studier et al., 1990). Both fragments and vector were end-filled using the Klenow fragment of DNA polymerase I prior to ligation with T4 DNA ligase. The resulting plasmids, part of the pMM series (Brierley et al., 1997), are shown in Figure 1. The PB1:1a/1b:PB1 fragment is located downstream of, and in-frame with, the first 783 bp of coding sequence of the bacteriophage T7 gene 10 and the ensemble is under the control of the bacteriophage T7 promoter. In BL21 cells (see below), the constructs express a 33 kDa non-frameshifted and a 50 kDa –1 frameshift product. Frameshifting assays in WG employed plasmid pFScass 5 and mutant derivatives (Brierley et al., 1992). This plasmid contains the minimal IBV frameshifting signal flanked by portions of the influenza virus A/PR8/34 PB2 gene (Young et al., 1983). Messenger RNAs derived from BamHI-cleaved pFScass 5 direct the synthesis of a 19 kDa non-frameshifted and a 22 kDa –1 frameshift product. Some of the variants of pFScass 5 employed in this study were described earlier (Brierley et al., 1992).
Frameshift assays in E.coli cells
Frameshifting in E.coli BL21 cells was assessed by expressing pMM plasmids in E.coli BL21/DE3/pLysS cells (Figure 1; Table I) as described (Studier et al., 1990; Brierley et al., 1997). These cells contain the gene for T7 RNA polymerase, which is induced by addition of isopropyl-β-d-thiogalactopyranoside (IPTG). In frameshift assays in E.coli TH79 [ara Δ (lac-proB) nalA rif thi metB argEam valR supB], TH78 (as TH79 but trmE), TH160 [ara Δ (lac-proB) nalA rif thi metB argEam supB fadR::Tn10] and TH159 (as TH160 but asuE107), T7 RNA polymerase was introduced by infection with λCE6 as described (Brierley et al., 1997). The purified expression products of the pMM plasmids were dissolved in Laemmli’s sample buffer and aliquots analysed on SDS–15% (w/v) polyacrylamide gels. Proteins were stained with Coomassie Brilliant Blue R (0.1% w/v) in 10% (v/v) acetic acid, 50% (v/v) methanol and destained in 10% (v/v) acetic acid, 30% methanol. The relative abundance of non-frameshifted or frameshifted products was estimated (Adobe Photoshop and NIH Image software) by scanning densitometry and adjusted to take into account the differential size of the products. Scans were performed on gels whose proteins were stained to an intensity at the centre of the dynamic range of the scanner (Epson Expression 1600 Pro). The frameshift efficiencies quoted in the text are the average of three independent measurements which varied by <5%, i.e. a measurement of 40% frameshift efficiency was between 38 and 42%.
Frameshift assays in depleted WG
In vitro transcription of pFScass 5-based plasmids (Brierley et al., 1992) employed the bacteriophage SP6 RNA polymerase and included the synthetic cap structure 7meGpppG (New England Biolabs) to generate capped mRNA. In ribosomal frameshift assays, serial dilutions of purified mRNAs were translated in WG as described (Brierley et al., 1987). Depleted lysates were supplemented with calf-liver or E.coli tRNAs at a final concentration of 50 µg/ml. Translation products were analysed on SDS–15% (w/v) polyacrylamide gels. The relative abundance of non-frameshifted or frameshifted products on the gels was determined by direct measurement of [35S]methionine incorporation using a Packard Instant Imager 2024. Frameshift efficiencies were calculated from those dilutions of RNA where translation was highly processive (RNA concentrations of 10–25 µg RNA/ml of WG). The frameshift efficiencies quoted are the average of at least three independent measurements which varied by <10%, i.e. a measurement of 40% frameshift efficiency was between 36 and 44%. The calculations of frameshift efficiency take into account the differential methionine content of the various products. Translation experiments were also performed with depleted RRL, but upon addition of E.coli tRNAs, translation was not restored sufficiently well to determine frameshifting efficiencies accurately.
Acknowledgments
Acknowledgements
We are grateful to Dr Tord Hagervall for the gift of E.coli strains. This work was supported by grants from the Biotechnology and Biological Sciences Research Council, UK and the Medical Research Council, UK.
References
- Atkins J.F. et al. (2001) Overriding standard decoding: implications of recoding for ribosome function and enrichment of gene expression. Cold Spring Harbor Symp. Quant. Biol., 66, 217–232. [DOI] [PubMed] [Google Scholar]
- Blinkowa A.L. and Walker,J.R. (1990) Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III γ subunit from within the τ subunit reading frame. Nucleic Acids Res., 18, 1725–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I. (1995) Ribosomal frameshifting on viral RNAs. J. Gen. Virol., 76, 1885–1892. [DOI] [PubMed] [Google Scholar]
- Brierley I., Boursnell,M.E.G., Binns,M.M., Bilimoria,B., Blok,V.C., Brown,T.D.K. and Inglis,S.C. (1987) An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J., 6, 3779–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Digard,P. and Inglis,S.C. (1989) Characterisation of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell, 57, 537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Rolley,N.J., Jenner,A.J. and Inglis,S.C. (1991) Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol., 220, 889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Jenner,A.J. and Inglis,S.C. (1992) Mutational analysis of the ‘slippery sequence’ component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol., 227, 463–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Meredith,M.R., Bloys,A.J. and Hagervall,T.G. (1997) Expression of a coronavirus ribosomal frameshift signal in Escherichia coli: influence of tRNA anticodon modification on frameshifting. J. Mol. Biol., 270, 360–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson B.A., Kwon,S.Y., Lee,B.J. and Hatfield,D. (2000) Yeast asparagine (Asn) tRNA without Q base promotes eukaryotic frameshifting more efficiently than mammalian Asn tRNAs with or without Q base. Mol. Cells, 10, 113–118. [DOI] [PubMed] [Google Scholar]
- Chamorro M., Parkin,N. and Varmus,H.E. (1992) An RNA pseudoknot and an optimal heptameric shift site are required for highly efficient ribosomal frameshifting on a retroviral messenger RNA. Proc. Natl Acad. Sci. USA, 89, 713–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M. and Fayet,O. (1993) Translational frameshifting in the control of transposition in bacteria. Mol. Microbiol., 7, 497–503. [DOI] [PubMed] [Google Scholar]
- Condron B.G., Atkins,J.F. and Gesteland,R.F. (1991) Frameshifting in gene 10 of bacteriophage T7. J. Bacteriol., 173, 6998–7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czworkowski J. and Moore,P.B. (1996) The elongation phase of protein synthesis. Prog. Nucleic Acid Res. Mol. Biol., 54, 293–332. [DOI] [PubMed] [Google Scholar]
- den Boon J.A., Snijder,E.J., Chirnside,E.D., De Vries,A.A.F., Horzinek,M.C. and Spaan,W.J.M. (1991) Equine arteritis virus is not a togavirus, but belongs to the coronaviruslike superfamily. J. Virol., 65, 2910–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P.J. (1996) Programmed translational frameshifting. Microbiol. Rev., 60, 103–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P.J. (2000) Translational frameshifting: implications for the mechanism of translational frame maintenance. Prog. Nucleic Acid Res. Mol. Biol., 64, 131–170. [DOI] [PubMed] [Google Scholar]
- Farabaugh P.J. and Bjork,G.R. (1999) How translational accuracy influences frame maintenance. EMBO J., 18, 1427–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower A.M. and McHenry,C.S. (1990) The gamma subunit of DNA Polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc. Natl Acad. Sci. USA, 87, 3713–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futterer J. and Hohn,T. (1996) Translation in plants—rules and exceptions. Plant Mol. Biol., 32, 159–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland R.F. and Atkins,J.F. (1996) Recoding: dynamic reprogramming of translation. Annu. Rev. Biochem., 65, 741–768. [DOI] [PubMed] [Google Scholar]
- Gramstat A., Prüfer,D. and Rohde,W. (1994) The nucleic acid binding zinc-finger protein of potato virus M is translated by internal initiation as well as by ribosomal frameshifting involving a shifty stop codon and a novel mechanism of P-site slippage. Nucleic Acids Res., 22, 3911–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagervall T.G., Pomerantz,S.C. and McCloskey,J.A. (1998) Reduced misreading of asparagine codons by Escherichia coli tRNALys with hypomodified derivatives of 5-methylaminomethyl-2-thiouridine in the wobble position. J. Mol. Biol., 284, 33–42. [DOI] [PubMed] [Google Scholar]
- Horsfield J.A., Wilson,D.N., Mannering,S.A., Adamski,F.M. and Tate,W.P. (1995) Prokaryotic ribosomes recode the HIV-1 gag-pol –1 frameshift sequence by an E/P site post-translocation simultaneous slippage mechanism. Nucleic Acids Res., 23, 1487–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuchi H. and Yamao,F. (1995) Structure and expression of prokaryotic tRNA genes. In Söll,D. and RajBhandary,U.L. (eds), tRNA: Structure, Biosynthesis and Function. ASM Press, Washington, DC, pp. 17–30. [Google Scholar]
- Jacks T., Madhani,H.D., Masiarz,F.R. and Varmus,H.E. (1988) Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell, 55, 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.J., Napthine,S. and Brierley,I. (2001) Development of a tRNA-dependent in vitro translation system. RNA, 7, 765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontos H. (1997) The role of ribosomal pausing in the process of ribosomal frameshifting. PhD thesis, University of Cambridge, Cambridge, UK.
- Kontos H., Napthine,S. and Brierley,I. (2001) Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol. Cell. Biol., 21, 8657–8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger M.K., Pedersen,S., Hagervall,T.G. and Sorenson,M.A. (1998) The modification of the wobble base of tRNAGlu modulates the translation rate of glutamic acid codons in vivo. J. Mol. Biol., 284, 621–631. [DOI] [PubMed] [Google Scholar]
- Larsen B., Wills,N.M., Gesteland,R.F. and Atkins,J.F. (1994) rRNA-mRNA base-pairing stimulates a programmed –1 ribosomal frameshift. J. Bacteriol., 176, 6842–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M.E., Hendrix,R.W. and Casjens,S.R. (1993) A programmed translational frameshift is required for the synthesis of a bacteriophage lambda tail assembly protein. J. Mol. Biol., 234, 124–139. [DOI] [PubMed] [Google Scholar]
- Lustig F., Elias,P., Axberg,T., Samuelsson,T., Tittawella,I. and Lagerkvist,U. (1981) Codon reading and translational error. Reading of glutamine and lysine codons during protein synthesis in vitro. J. Biol. Chem., 256, 2635–2643. [PubMed] [Google Scholar]
- Marczinke B., Hagervall,T. and Brierley,I. (2000) The Q-base of asparaginyl-tRNA is dispensible for efficient –1 ribosomal frameshifting in eukaryotes. J. Mol. Biol., 295, 179–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejlhede N., Atkins,J.F. and Neuhard,J. (1999) Ribosomal –1 frameshifting during decoding of Bacillus subtilis cdd occurs at the sequence CGA AAG. J. Bacteriol., 181, 2930–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfitzinger J.H., Weil,J.H., Pillay,D.T.N. and Guillemaut,P. (1989) Preparation of a tRNA-dependent wheat germ protein-synthesizing system. Plant Mol. Biol., 12, 301–306. [DOI] [PubMed] [Google Scholar]
- Plant E.P., Jacobs,K.L., Harger,J.W., Meskauskas,A., Jacobs,J.L., Baxter,J.L., Petrov,A.N. and Dinman,J.D. (2003) The 9Å solution: How mRNA pseudoknots promote efficient programmed –1 ribosomal frameshifting. RNA, 9, 168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rettberg C.C., Prère,M.F., Gesteland,R.F., Atkins,J.F. and Fayet,O. (1999) A three-way junction and constituent stem-loops as the stimulator for programmed –1 frameshifting in bacterial insertion sequence IS911. J. Mol. Biol., 286, 1365–1378. [DOI] [PubMed] [Google Scholar]
- Sekine Y. and Ohtsubo,E. (1992) DNA sequences required for translational frameshifting in production of the transposase encoded by IS1. Mol. Gen. Genet., 235, 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine Y., Eisaki,N. and Ohtsubo,E. (1994) Translational control in production of transposase and in transposition of insertion sequence IS3. J. Mol. Biol., 235, 1406–1420. [DOI] [PubMed] [Google Scholar]
- Shigemoto K., Brennan,J., Walls,E,. Watson,C.J., Stott,D., Rigby,P.W. and Reith,A.D. (2001) Identification and characterisation of a developmentally regulated mammalian gene that utilises –1 programmed ribosomal frameshifting. Nucleic Acids Res., 29, 4079–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F.W., Rosenberg,A.H., Dunn,J.J. and Dubendorff,J.W. (1990) Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol., 185, 60–89. [DOI] [PubMed] [Google Scholar]
- ten Dam E.B., Pleij,C.W.A. and Bosch,L. (1990) RNA pseudoknots: translational frameshifting and readthrough on viral RNAs. Virus Genes, 4, 121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchihashi Z. (1991) Translational frameshifting in the Escherichia coli dnaX gene in vitro. Nucleic Acids Res., 19, 2457–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchihashi Z. and Brown,P.O. (1992) Sequence requirements for efficient translational frameshifting in the Escherichia coli dnaX gene and the role of an unstable interaction between tRNALys and an AAG lysine codon. Genes Dev., 6, 511–519. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi Z. and Kornberg,A. (1990) Translational frameshifting generates the gamma subunit of DNA polymerase III holoenzyme. Proc. Natl Acad. Sci. USA, 87, 2516–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney U., Lee,C.P. and RajBhandary,U.L. (1991) Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem., 266, 24712–24718. [PubMed] [Google Scholar]
- Weiss R.B., Dunn,D.M., Shuh,M., Atkins,J.F. and Gesteland,R.F. (1989) E. coli ribosomes re-phase on retroviral frameshift signals at rates ranging from 2 to 50 percent. New Biol., 1, 159–169. [PubMed] [Google Scholar]
- Yokoyama S., Watanabe,T., Murao,K., Ishikura,H., Yamaizumi,Z., Nishimura,S. and Miyazawa,T. (1985) Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc. Natl Acad. Sci. USA, 82, 4905–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J.F., Desselberger,U., Graves,P., Palese,P., Shatzman,A. and Rosenberg,M. (1983) Cloning and expression of influenza virus genes. In Laver,W.G. (ed.), The Origin of Pandemic Influenza Viruses. Elsevier Science, Amsterdam, The Netherlands, pp. 129–138. [Google Scholar]
- Yusupov M.M., Yusupova,G.Z., Baucom,A,. Lieberman,K., Earnest,T.N., Cate,J.H. and Noller,H.F. (2001) Crystal structure of the ribosome at 5.5Å resolution. Science, 292, 883–896. [DOI] [PubMed] [Google Scholar]