Abstract
The Vibrio vulnificus nuclease, Vvn, is a non-specific periplasmic nuclease capable of digesting DNA and RNA. The crystal structure of Vvn and that of Vvn mutant H80A in complex with DNA were resolved at 2.3Å resolution. Vvn has a novel mixed α/β topology containing four disulfide bridges, suggesting that Vvn is not active under reducing conditions in the cytoplasm. The overall structure of Vvn shows no similarity to other endonucleases; however, a known ‘ββα–metal’ motif is identified in the central cleft region. The crystal structure of the mutant Vvn–DNA complex demonstrates that Vvn binds mainly at the minor groove of DNA, resulting in duplex bending towards the major groove by ∼20°. Only the DNA phosphate backbones make hydrogen bonds with Vvn, suggesting a structural basis for its sequence-independent recognition of DNA and RNA. Based on the enzyme–substrate and enzyme–product structures observed in the mutant Vvn–DNA crystals, a catalytic mechanism is proposed. This structural study suggests that Vvn hydrolyzes DNA by a general single-metal ion mechanism, and indicates how non-specific DNA-binding proteins may recognize DNA.
Keywords: crystal structure/DNA-binding protein/DNA hydrolysis/protein–DNA interactions/sequence-independent recognition
Introduction
Protein–nucleic acid interactions play many important cellular roles, including regulating DNA replication, controlling gene expression and DNA housekeeping, such as maintaining a supercoiled state, performing damage and mismatch repair, allowing recombination, restricting foreign DNA, and the processes of DNA ligating, methylating and degrading. A wealth of information regarding how proteins select DNA target sites has been discovered through biochemical, structural and statistical analysis in site-specific DNA-binding proteins, such as transcription factors (Burley, 1994; Jones et al., 1999; Wolberger, 1999; Pabo and Nekludova, 2000). However, non-specific protein–DNA interactions have not been so well studied due to the small number of resolved three-dimensional structures of non-specific protein–DNA complexes.
DNA endonucleases provide good opportunities to investigate sequence-independent recognition between proteins and DNA. The most well studied endonucleases in bacteria are the type II restriction enzymes (Kovall and Matthews, 1999; Pingoud and Jeltsch, 2001). These restriction enzymes bind DNA non-specifically, but cleave only short palindromic DNA sequences in the presence of Mg2+ at specific sites where large DNA conformational changes are triggered upon enzyme binding. The major biological function of restriction enzymes is to cleave foreign unmethylated DNA and thereby protect the host methylated genome (Kobayashi, 2001). In prokaryotic organisms, a variety of other types of endonucleases involved in the protection of bacterial cells have been identified, but differ from restriction enzymes in that they cleave DNA in a sequence-independent manner. Examples include Escherichia coli colicins, such as ColE7 and ColE9 (Ku et al., 2002; Walker et al., 2002), which are secretory endonucleases that digest chromosomal DNA to kill target cells, thereby enabling the host cells to have a better survival advantage during times of stress.
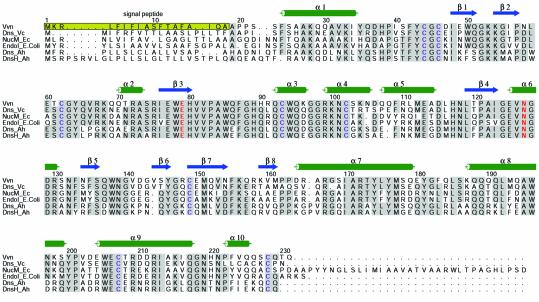
Vvn from Vibrio vulnificus (Wu et al., 2001) belongs to a family of periplasmic or extracellular nucleases that represent another class of non-specific endonucleases which are also involved in protecting bacterial cells. This family of enzymes include Dns from Vibrio cholerae (Focareta and Manning, 1987), NucM from Erwinia chrysanthemi (Moulard et al., 1993), EndoI from Escherichia coli (Jekel and Wackernagel, 1995), and Dns and DnsH from Aeromonas hydrophila (Chang et al., 1992; Dodd and Pemberton, 1996). These endonucleases all contain a signal peptide located at the N-terminus and eight strictly conserved cysteine residues. The signal peptides are cleaved off during the enzymes’ transportation from cytoplasm to periplasm, resulting in a mature protein of an average size of ∼25kDa (see Figure1). All of these endonucleases are capable of digesting both DNA and RNA and are only active in their oxidized form (Wu et al., 2001). Some of them are extracellular endonucleases, such as the Dns from V.cholerae, which is released to degrade DNA in the environment possibly to reduce the rate of DNA transformation (Focareta and Manning, 1987). Periplasmic enzymes, including Vvn and EndoI, guard the cell wall against the uptake of foreign DNA during transformation. Escherichia coli mutants lacking endA (encoding EndoI) or V.vulnificus mutants lacking vvn can be transformed more efficiently, but they resemble wild-type cells with regards to growth rate, conjugational properties and their ability to propagate various phages (Durwald and Hoffmann-Berling, 1968; Wu et al., 2001). The dns-deficient V.cholerae and vvn-deficient V.vulnificus mutants showed no observed virulence effects in the mouse model (Focareta and Manning, 1991; Wu et al., 2001), indicating that Dns and Vvn were not required for the bacterial virulence.
Fig. 1. Sequence alignment of V.vulnificus Vvn (Wu et al., 2001) with several representative extracellular nucleases: Dns from Vibrio cholerae (Focareta and Manning, 1987), NucM from Erwinia chrysanthemi (Moulard et al., 1993), EndoI from Escherichia coli (Jekel and Wackernagel, 1995), and Dns and DnsH from Aeromonas hydrophila (Chang et al., 1992; Dodd and Pemberton, 1996). The N-terminal signal peptide from residues 1 to 18 marked in the yellow-shaded box is cleaved off during transportation. The secondary structural elements of α-helices and β-strands in the crystal structure of Vvn are illustrated above the sequences. The eight cysteine residues making the four disulfide bridges are marked in blue. The residues responsible for metal binding are marked in red.
Here we report the crystal structures of Vvn and a Vvn mutant in complex with an 8bp DNA, both at a resolution of 2.3Å. Vvn represents the first solved structure in this family of extracellular endonucleases and it bears a novel mixed α/β topology. Comparison with other nucleases interestingly reveals a similar structural arrangement in the endonuclease active site containing two β-strands and one α-helix with a centrally located divalent metal ion. In the crystal structure of the Vvn–DNA complex, an enzyme–substrate complex with an intact DNA and an enzyme–product complex with a cleaved DNA are both present, providing an ideal case to elucidate the hydrolysis mechanism. Moreover, the interaction between Vvn and DNA demonstrates how a protein may bind duplex DNA without sequence preference.
Results
Endonuclease activity
The vvn gene of V.vulnificus was subcloned into pTYB2 vector and Vvn was overexpressed in E.coli B834. The purified Vvn had a mol. wt of 24780Da as measured by mass spectrometry, close to the calculated Mr of 24785Da for the matured Vvn containing residues 19–231 without the N-terminal signal peptide. A plasmid DNA was used as the substrate to monitor Vvn endonuclease activity through DNA topological changes in agarose gel electrophoresis. We found that the supercoiled DNA was cleaved into linear and open-circular forms or completely degraded with higher concentrations of enzymes. In identical buffer solutions (50mM Tris–HCl and 10mM MgCl2 at pH8), Vvn appeared to have a slightly better endonuclease activity than DNaseI and ColE7, but weaker activity than Serratia nuclease (see Figure2A).
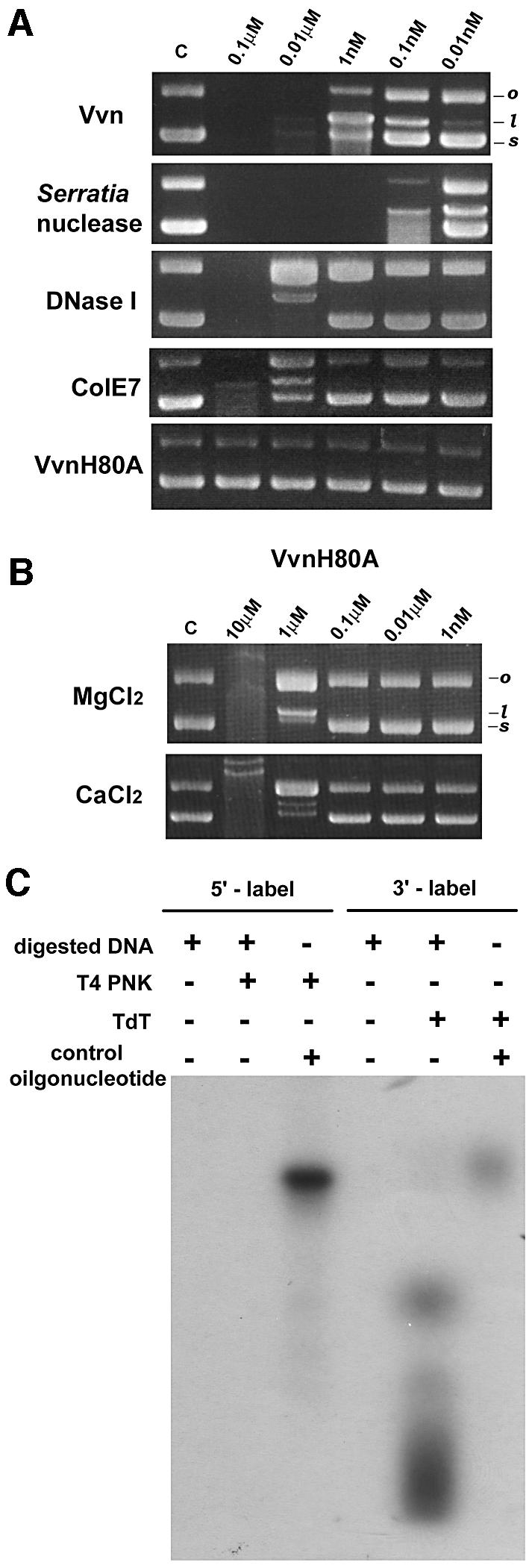
Fig. 2. Characterization of the endonuclease activity of Vvn. (A)Comparison of the endonuclease activity of Vvn with different non-specific endonucleases. Several endonucleases in the concentration range of 0.01nM–0.1µM were each incubated with a plasmid DNA in the same buffer solution containing 10mM MgCl2 and 50mM Tris–HCl at pH8.0. The topological changes of the substrate DNA were monitored by 1% agarose gel electrophoresis. The supercoiled (s) form of the plasmid DNA was digested into open-circular (o) and linear (l) forms. In the reaction buffers used here, Vvn appeared as a weaker DNase than the Serratia nuclease, but more active than DNaseI and ColE7. (B)The endonuclease activity of the Vvn mutant H80A was assayed in the presence of 10mM Mg2+ or Ca2+. Vvn mutant H80A showed no observable DNase activity when its concentration was <1µM. As the concentration of H80A increased to the µM range, ∼0.1% residual activity was detected compared with the wild-type Vvn in the presence of Mg2+ or Ca2+. (C)The Vvn cleaved DNA products were labeled with T4 polynucleotide kinase (T4 PNK) and terminal deoxynucleotidyl transferase (TdT), which can only label 5′-OH and 3′-OH terminal products, respectively. The products were only labeled by TdT, suggesting that Vvn cleaved at the 3′ side of a phosphodiester bond, generating a 5′-phosphate and 3′-OH products.
The DNA cleavage products resulting from Vvn hydrolysis were assayed further using two different enzymes: T4 polynucleotide kinase (T4 PNK), which only labels 5′-hydroxyl groups, and terminal deoxynucleotidyl transferase (TdT), which only labels 3′-hydroxyl groups. Figure2C shows that the DNA hydrolysis products were only labeled by TdT, suggesting that the products were DNA fragments containing a 3′-OH and a 5′-phosphate. Therefore, Vvn cleaves at the 3′ side of a phosphodiester bond, similar to DNaseI, Serratia nuclease and ColE7.
Structure determination
The crystal structure of Vvn was solved by the multiwavelength amorphous diffraction (MAD) method using the anomalous signals from selenomethionine (Se-Met)-labeled protein crystals. A Vvn mutant replacing His80 with alanine was constructed which possessed only ∼0.1% endonculease activity of the wild-type enzyme in the presence of Mg2+ or Ca2+ (see Figure2B). This mutant was co-crystallized with an 8bp double-stranded DNA (dsDNA), 5′-GCGATCGC-3′, to avoid DNA degradation by the enzyme. The crystal structure of the Vvn-H80A–DNA complex was solved by the molecular replacement method using the Se-Met-labeled Vvn structure as the search model. The final model of the Vvn structure contained a single Vvn molecule (residues 19–228), one magnesium ion and 194 water molecules. There were two protein–DNA complexes in one asymmetric unit of the monoclinic crystals of the Vvn-H80A–DNA complex. Therefore, the final model of the Vvn-H80A–DNA complex contained two Vvn molecules (residues 19–231 in the A chain and residues 19–228 in the B chain), two dsDNAs (chains C/D and E/F), two calcium ions and 328 water molecules. Both structures were refined to a resolution of 2.3Å with good geometrical and refinement statistics (see Table I).
Table I. Crystallographic data.
| Data collection and MAD phasing statistics | ||||
| Crystal | Vvn-H80A–DNA | SeMet-Vvn | ||
| Wavelength (Å) | 0.9065 | λ1=0.9794 | λ2=0.9796 | λ3=0.9600 |
| Resolution (Å) | 2.3 | 2.3 | 2.3 | 2.3 |
| Space group | P21 | P21 | P21 | P21 |
| Cell constants (Å) | a=53.7 | a=48.1 | a=48.1 | a=49.1 |
| b=50.1 | b=39.3 | b=39.3 | b=40.1 | |
| c=89.6 | c=49.6 | c=49.6 | c=50.6 | |
| β=97.5° | β=90.5° | β=90.5° | β=90.5° | |
| Total reflections | 157796 | 56334 | 59303 | 62270 |
| Unique reflections | 21165 | 8760 | 8736 | 9127 |
| Completeness (%) | 100 | 96.2 | 96.3 | 96.0 |
| I/σ(I) overall | 20.2 | 23.6 | 24.1 | 24.4 |
| I/σ(I) last shell (2.38–2.30Å) | 4.4 | 13.1 | 12.8 | 9.6 |
| Rsyma (%) overall | 7.3 | 7.1 | 6.7 | 7.5 |
| Rsyma (%) last shell | 28.2 | 11.8 | 12.5 | 14.6 |
| Phasing power (centrics/acentrics) | 2.73/2.46 | |||
| Figure of merit (centrics/acentrics) |
|
|
0.81/0.79 |
|
| Refinement | ||||
| Resolution (Å) | 40–2.3 | 40–2.3 | ||
| Reflections (working/test) | 19113/1616 | 8072/731 | ||
| Rcrystb/Rfree (%) | 19.1/26.0 | 18.6/26.9 | ||
| R.m.s. bond lengths (Å) | 0.011 | 0.012 | ||
| R.m.s. bond angles (°) | 1.35 | 1.28 | ||
| No. of atoms (protein/water/metal/DNA) | (3450/328/2/645) | (1718/194/1/–) | ||
| Average B-factor (Å2) | ||||
| Protein | 31.2 | 23.3 | ||
| Water | 37.2 | 31.8 | ||
| DNA | 59.9 | |||
| Ramachandran plot (%) | ||||
| Most favored | 89.4 | 90.7 | ||
| Additionally allowed | 9.8 | 8.2 | ||
| Generously allowed | 0.8 | 1.1 | ||
| Disallowed | 0 | 0 | ||
aRsym=ΣhklΣi|Ii(hkl)–<I(hkl)>|/Ii(hkl).
bRcryst=Σhkl||Fo(hkl)|–|Fc(hkl)||/Σhkl|Fo(hkl)|.
Overall structure of Vvn
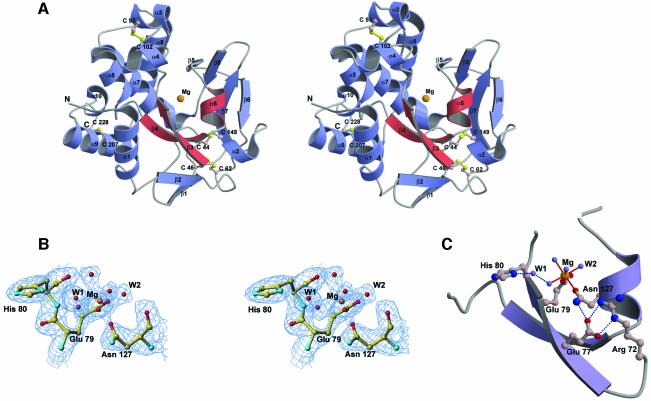
The crystal structure of Vvn revealed a V-shaped mixed α/β fold (see Figure3A). The left half of the structure contains mainly eight α-helices within which the helix α7 is extended from the left to the right arm of the molecule. The right half of Vvn contains mostly β-strands, with a four-stranded β-sheet on the top, another four-stranded β-sheet on the bottom and two small α-helices in between. There are four disulfide bridges within the whole molecule, two in the left side mostly between helices (Cys93–Cys102 and Cys207–Cys228) and two in the right side mostly between loops (Cys44–Cys149 and Cys46–Cys62). Since Vvn is not active under reducing conditions (Wu et al., 2001), these four disulfide bonds are probably required for stable folding of the protein.
Fig. 3. The crystal structure of the magnesium ion-bound Vvn. (A)Stereo view of the ribbon diagram of Vvn structure. Vvn has a novel V-shaped mixed α/β fold. The four disulfide bridges are displayed in a ball-and-stick model. The ββα–metal motif in Vvn is in red with a magnesium ion located in the center. (B)Stereo view of the omitted (2Fo – Fc) electron density map at 2.3Å resolution (contoured at 1.0σ) overlaid with the final structural model of Vvn at the metal-binding site. (C)The endonuclease active site of Vvn. The magnesium ion is bound to Glu79, Asn127 and four water molecules. A water molecule, W1, bridges His80 and a Mg2+-bound water.
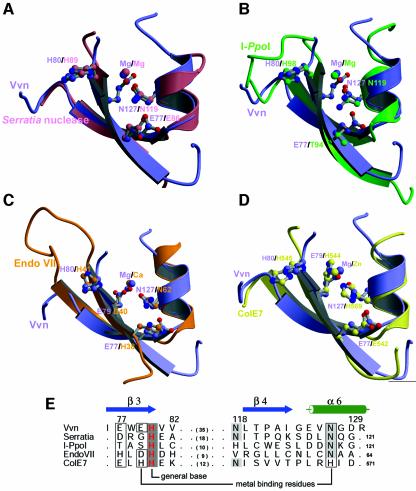
A structural homology search using DALI (Holm and Sander, 1993) revealed that Vvn has a novel fold, different from those of other known folds in the database. However, DALI revealed unexpectedly that a small region in Vvn, containing β3, β4 and α6 (displayed in red in Figure3A), bears some structural similarity to the putative endonuclease active site of phage T4 EndoVII (Raaijmakers et al., 1999). A metal ion was identified further in Vvn, bound directly to the residues located in β3 and α6. Since a calcium ion is located at a similar position in the active site of EndoVII (described in the Discussion), it is very likely that the metal-binding site in Vvn is also an active site for DNA hydrolysis.
Structure of the active site
The six-coordinated metal ion located in the central cleft region of Vvn was assigned as a magnesium ion for the following two reasons. First, magnesium was the only metal ion (10mM MgCl2) present under the crystallization conditions used. Secondly, the coordination and geometry of the metal ion-binding site are best fitted as a magnesium ion (Dudev and Lim, 2001), which is bound to Glu79 (Oε1), Asn127 (Oδ1) and four water molecules in an octahedral geometry, with an average Mg–O bond distance of 2.08Å. Structural refinement gives a reasonable B-factor of 23.9Å2 for the magnesium ion, close to the B-factors of those magnesium-bound oxygen atoms (see the omitted electron density maps around the magnesium-binding site in Figure3B). A water molecule, W1, bridges the magnesium-bound water molecule and His80 (see Figure3C).
The side chain of Asn127 is restrained by the hydrogen bond between Asn127 (Nδ2) and Glu77 (Oε1). Glu77 makes further hydrogen bonds with Arg72. This hydrogen bond network (see Figure3C) fixes the side chain conformation of Asn127 for metal binding. A similar phenomenon has been observed for the catalytic zinc ion-bound histidines which are hydrogen-bonded almost universally to oxygen-containing residues in the second shell (Alberts et al., 1998). In this way, a more rigid framework is established for the binding of the catalytic metal ion.
Overall structure of Vvn–DNA complex
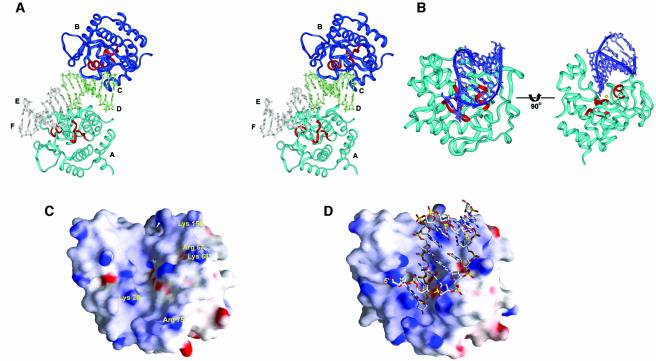
The overall crystal structure of two Vvn molecules (chains A and B) bound to two 8bp DNA (chains C/D and E/F) in one asymmetric unit is shown in Figure4A. The V-shaped cleft of Vvn faces DNA so that the ββα–metal motif is located close to the DNA backbones. Vvn appears to bind DNA as a monomer mainly at its minor groove, resulting in DNA being bent away from the enzyme at an angle of ∼20°, effectively widening the narrow minor groove. The α-helix (α6) in the ββα–metal motif is aligned in the minor groove, and one of the DNA backbones is inserted in the cleft region between α6 and β3. The two DNA molecules appear to stack on top of each other; however, the twist angle between the interfaced base pairs of the two DNAs is –28.7°, indicating that the two DNA molecules are not stacked in a conventional way as a pseudo continuous DNA.
Fig. 4. The crystal structure of the Vvn mutant H80A in complex with DNA. (A)Stereo view of the overall crystal structure of two Vvn (chains A and B) bound to two octanucleotides (chains C/D and E/F) in one asymmetric unit of the monoclinic unit cell. (B)Two different views of the Vvn–DNA (chains B/CD) complex. The orientation of Vvn in the left panel is similar to that in Figure3A. DNA is bound at the cleft region of Vvn close to the ββα motif (colored in red). (C)The molecular surface of the free-form Vvn and (D) the DNA-bound Vvn, color-coded to indicate electrostatic potential. One of the phosphate backbones of the DNA is inserted into the central deep cleft and the other backbone is bound at a shallower cleft on the right.
The structure of the H80A mutant in the protein–DNA complex is almost identical to that of the free-form Vvn, only deviating from it by an average r.m.s.d. of 0.8Å for the A chain and 0.86Å for the B chain (Cα atoms). This indicates that DNA binding does not induce conformational changes in Vvn. The molecular surfaces of the free-form Vvn and DNA-bound Vvn, color-coded with electrostatic potential, are displayed in Figure4C and D. Vvn has a shape-complementary surface for accommodating a dsDNA. A relatively deep cleft located in the central surface of Vvn is bound to one strand (chains C or E) of DNA backbones and a shallower cleft next to it is bound to the other strand (chains D or F). Several basic and polar residues located along the two clefts make direct or water-bridged hydrogen bonds to the phosphates in DNA.
Enzyme–substrate and enzyme–product complexes
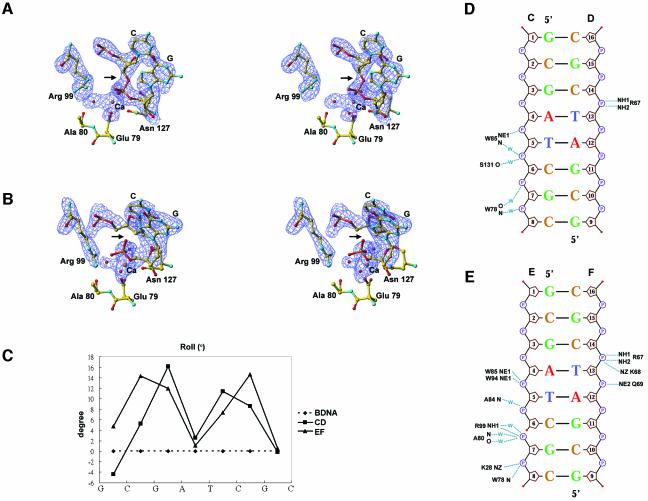
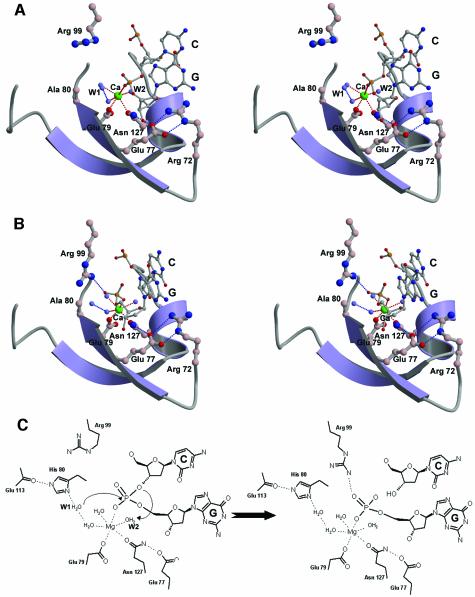
The two protein–DNA complexes (B/CD and A/EF in Figure4A) in the crystals are not crystallographically identical and deviate from each other with an average r.m.s.d. of 0.51Å for Vvn (Cα atoms) and 2.22Å for DNA (P atoms) when the two Vvn molecules were superimposed. The major differences lie in the interface regions where the E strand of DNA is bound closer to the active site of Vvn (chain A) than that of the C strand (to chain B). A closer look at the electron density maps (see Figure5A and B) in the active site region surprisingly shows that the B/CD is the enzyme–substrate complex and the A/EF is the enzyme–product complex in which the phosphate backbone is cleaved. Since the Vvn H80A mutant still contained residual activity in 200mM CaCl2 buffers (data not shown), which was close to the crystallization conditions, the enzyme cleaved part of the DNA molecules in the crystal. The calcium ion assigned in the structure has a reasonable temperature factor of 25Å2, and a good geometry with an average Ca–O distance of 2.5Å. The average temperature factor for DNA atoms is 59.9Å2, significantly higher than the average B-factor of 31.2Å2 for the proteins atoms. However, the electron density for the DNA strands (chains C, D, E and F) were well defined and a gap was clearly shown between the scissile phosphate and sugar group of C6. The Arg99 is located more distantly from the scissile phosphate in the enzyme–substrate complex, but is directly hydrogen-bonded to the cleaved phosphate in the enzyme–product complex. This interaction could explain how the E strand of the DNA duplex moves closer to the enzyme.
Fig. 5. DNA conformation and its interactions with Vvn. (A)Stereo view of the electron density maps at the active site region for the Vvn–DNA complex. An enzyme–substrate structure is observed in one of the complexes in the asymmetric unit (chains B/CD). (B)Stereo view of the electron density maps for an enzyme–product structure observed in one of the complexes (chains A/EF). The difference maps (Fo – Fc) contoured at 3σ were calculated after omitting the active site atoms in the refinement model followed by a full round of positional and simulated annealing refinement. In the enzyme–substrate complex, the electron density was continuous in the scissile phosphate, but in the enzyme–product complex, the electron density was broken at the phosphate group (marked by an arrow), indicating the cleavage of the phosphate backbone. The terminal 5′-phosphate in the enzyme–product complex forms a hydrogen bond with Arg99. (C)The inter-base roll angles are plotted as a function of sequence for DNA bound to Vvn. (D)Schematic diagrams of Vvn–DNA contacts in the enzyme–substrate (chains B/CD) and (E)enzyme–product (chains A/EF) complexes. Hydrogen bonds between Vvn and DNA are indicated by dotted lines.
In the enzyme–substrate complex (chains B/CD), the DNA strand (strand C) bound at the central cleft region makes contacts with Vvn mainly at its 3′ end. The schematic diagram shown in Figure5D lists all the hydrogen bonds between Vvn and DNA in the B/CD complex. The residues from the ββα–metal motif, including Trp78 and Ser131, and also the residue outside the motif, Trp85, make direct or water-bridged hydrogen bonds with DNA phosphate. The second DNA strand (D chain) is also bound mainly at its 3′ side to Arg67. There are altogether seven direct and water-bridged hydrogen bonds in the buried protein surface of 572Å2, which is a considerably lower value than the average buried protein surface of ∼1600Å2 found in other protein–DNA complexes (Jones et al., 1999; Nadassy et al., 1999). This is probably because a shorter DNA octamer was used for crystallization in this study; a longer DNA may contact Vvn over a larger interface. The hydrogen bond pattern between Vvn and DNA is unique in that only the phosphates, but not the sugar groups or the bases, are involved in the hydrogen bond contacts with Vvn. This result explains the sequence-independent recognition of DNA and RNA by Vvn.
In the enzyme–product complex (chains A/EF), the protein–DNA interactions resemble those of the enzyme–substrate complex (B/CD). However, there are more hydrogen bonds, a total of 13, between Vvn and DNA (see Figure5E). This is probably because DNA strands are bound closer to Vvn in the enzyme–product complex than those of the enzyme–substrate complex. A notable difference is that the side chain of Arg99 and the main chain atoms of Ala80 are hydrogen-bonded to the cleaved 5′-phosphate, which are not observed in the enzyme–substrate complex. The buried protein solvent-accessible surface in the enzyme–product complex increases from 572 to 669Å2.
The DNA octamers in both enzyme–substrate and enzyme–product complexes are deformed from a straight B-form DNA. There are two kinks between G3 and A4, and T5 and C6 in the enzyme–substrate complex, as demonstrated by the large positive roll angles at these two base pair steps, resulting in a total bending by ∼20° (Figure5C). This DNA bending widens the minor groove at the central region from 5.6Å in the case of canonical B-DNA to ∼9.5Å, as calculated by Curves (Lavery and Sklenar, 1988). Several hydrophobic residues are located close to the DNA sugar–phosphate backbones near the kink positions, including Phe155 which makes van der Waals contacts with the ribose group of C14, and Trp85 and Trp94 in close contact with the ribose group of T5. These hydrophobic residues do not intercalate into the minor groove, such as the Leu116 in I-PpoI (Galburt et al., 2000), or intercalate between base pairs, such as those found in TBP (Kim,J.L. et al., 1993; Kim,Y. et al., 1993), Sac7d (Robinson et al., 1998) and SRY (Werner et al., 1995). However, the hydrophobic interactions between these residues and DNA probably also stabilize the bent structure of DNA. DNA in a bent conformation allows DNA backbones to align closely to Vvn and places the DNA scissile phosphate group between C6 and G7, near to the active site of the enzyme. The scissile phosphate directly binds to the calcium ion, replacing a water molecule originally bound to the metal ion in the free form of Vvn.
Discussion
Comparison with other endonucleases
The active site of Vvn contains two antiparallel β-strands and one C-terminal α-helix, with a divalent metal ion located in the center. A similar ‘ββα–metal’ topology has been revealed in the active site regions of several endonucleases (Friedhoff et al., 1999b; Kuhlmann et al., 1999; Miller et al., 1999), including the homing endonucleaseI-PpoI (Flick et al., 1998), Serratia nuclease (Miller et al., 1994), phage T4 Endo VII (Raaijmakers et al., 1999), and the H-N-H ColE7 (Ko et al., 1999; Cheng et al., 2002) and ColE9 (Kleanthous et al., 1999). Among these DNases, I-PpoI is the only site-specific endonuclease recognizing and cleaving dsDNA with a specific sequence of 13–15bp (Ellison, 1993). Serratia nuclease (Friedhoff et al., 1996) and ColE7 (Ku et al., 2002) are non-specific endonucleases cleaving ds or ssDNA and RNA with little sequence preferences. Endo VII recognizes structural perturbations in DNA, cleaving a variety of branched DNA structures, such as Holliday junctions, cruciform DNA, single-base mismatches and abasic sites (Greger and Kemper, 1998).
The DALI search revealed a similarity in the active site region of Vvn and Endo VII; however, the ββα–metal motif of Vvn also highly resembles those of the endonucleases described above. The superimposition (see Figure6) between the secondary structure regions of the ββα fold shows that Vvn matches well with Serratia nuclease (r.m.s.d.=0.58Å for Cα-atoms), I-PpoI (r.m.s.d.=1.06Å), Endo VII (r.m.s.d.=1.68Å) and ColE7 (r.m.s.d.=0.86Å). The amino acid residues (listed in Figure6E) used for the superimposition were aligned mainly based on secondary structure similarities. This structurally based sequence alignment shows that a histidine (displayed in red) is strictly conserved among these endonucleases, although they share little sequence homology to each other. This histidine (His98) functions as the general base in I-PpoI, activating a water molecule which then attacks the scissile phosphate in-line (Galburt et al., 1999). Mutation of the corresponding histidine (His80) in Vvn produces an inactive mutant (H80A) and confirms the critical importance of this residue in DNA hydrolysis (see Figure2A). The Nε atom of His80 in Vvn forms a hydrogen bond with the backbone carbonyl oxygen of Glu113, making His80 a stronger base in deprotonating a water molecule. A similar hydrogen bonding arrangement between the putative histidine general base and a carbonyl oxygen has been observed in I-PpoI and Serratia nuclease (Galburt et al., 1999). The Nδ atom of His80 in Vvn appears to make a hydrogen bond to the water molecule, W1, which is the nucleophilic water molecule, likely to be responsible for attacking the scissile phosphate (see Figure3C).
Fig. 6. The superimposition of the endonuclease active sites of Vvn with the ‘ββα–metal’ fold found in several endonucleases. (A)Serratia nuclease (Miller et al., 1994), (B)I-PpoI (Flick et al., 1998), (C)Endo VII (Raaijmakers et al., 1999) and (D)ColE7 (Cheng et al., 2002), respectively. These endonucleases share no sequence homology but all contain a similar folded active site. (E)The structure-based sequence alignment of the ββα–metal motif in several endonucleases. The His80 in Vvn is conserved among these endonucleases. Glu79 and Asn127 in Vvn are responsible for metal ion binding, and the aligned residues in other endonucleases play a similar role (boxed residues). Glu77 in Vvn makes a hydrogen bond to Asn127 to fix its orientation for metal ion binding, and the residues aligned at the same position to Glu77 all function in a similar manner.
The metal ions located in the ββα–metal motif are assigned as a magnesium ion in Vvn, Serratia nuclease and I-PpoI, a calcium ion in Endo VII and a zinc ion in ColE7. The asparagine (Asn127) responsible for metal binding in Vvn is well conserved among this group of endonucleases, except that it is replaced by a histidine (His569) in ColE7. This is probably because ColE7 needs a softer histidine to bind a transition metal ion, instead of alkaline earth metal ions (Sui et al., 2002). The metal-binding residues aligned with Asn127 in Vvn are almost all making a hydrogen bond with a neighboring residue (Glu77 in Vvn). This interaction may restrain the side chain conformation of the metal-binding residue and makes the asparagine more polar to bind to the metal ion. The second metal-ion binding residue in Vvn, Glu79, is not as well conserved among these endonucleases, with only an aspartate (Asp40) in EndoVII and a histidine (His544) in ColE7, aligned in the same position, involved in metal binding.
Proposed hydrolysis mechanism
The crystal structure of the Vvn mutant in complex with DNA, containing an enzyme–substrate and an enzyme–product complex, provides a great opportunity to elucidate the DNA cleavage mechanism catalyzed by Vvn. Based on three rationales, we suggest that the His80 in Vvn acts as a general base by activating the water molecule (W1) to attack the scissile phosphate. First, we found in this study that the mutation of His80 to alanine abolishes the endonuclease activity of Vvn, demonstrating the critical importance of this residue in DNA hydrolysis. Secondly, structural similarities observed in the active site regions of Vvn, I-PpoI and Serratia nuclease indicate that these endonucleases may use a similar mechanism to catalyze DNA hydrolysis (Friedhoff et al., 1999a). The His80 in Vvn can be structurally superimposed on to the general base residues, His98 in I-PpoI and His89 in Serratia nuclease, implying that it may play a similar role. Moreover, the hydrogen bond network surrounding His80 is similar to that observed for the general base histidines in I-PpoI and Serratia nuclease, including the hydrogen bonds between His80 and a backbone carbonyl group and a water molecule, W1. Thirdly, in the Vvn-H80A–DNA–Ca2+ complex, the Ca2+-coordinated outer sphere water molecule W1 is 3.3Å away from the phosphorus atom and the bond angle in W1-P-O3′ is 160° (in the B/CD enzyme–substrate complex), ideal for an in-line attack on the scissile phosphate (see Figure7A).
Fig. 7. The structure of the active sites in the Vvn–DNA complex: (A)enzyme–substrate and (B)enzyme–product complexes. In the enzyme–substrate complex, the scissile phosphate is bound to the calcium ion and Arg99 points away from the phosphate. However, in the enzyme–product complex, the scissile phosphate makes a hydrogen bond with Arg99. (C)Proposed DNA hydrolysis mechanism catalyzed by Vvn. His80 functions as the general base to activate the water molecule, W1, for an in-line attack on the scissile phosphate. The magnesium ion stabilizes the phosphoanion transition state and the magnesium ion-bound water molecule (W2) provides a proton to the 3′-oxygen leaving group.
After nucleophilic attack of the scissile phosphate by a water molecule, a general acid is required to provide a proton for the departing oxygen group in a DNA hydrolysis reaction catalyzed by an endonuclease. What is the general acid in Vvn that protonates the leaving 3′-oxygen group? The magnesium ion in I-PpoI has been shown biochemically (Friedhoff et al., 1999a; Mannino et al., 1999) and structurally (Galburt et al., 1999) to accelerate the reactions in three ways: stabilizing the phosphoanion transition state; positioning and activating a water molecule to donate a proton to the leaving 3′-oxygen; and introducing strain into the substrate complex that is relieved in the product complex. In the Vvn-H80A–DNA complex, the inner sphere metal-coordinated water molecule, W2, is close to the 3′-oxygen atom, positioned appropriately for providing a proton to the leaving group. Therefore, the divalent metal ion in Vvn directly bound to the scissile phosphate may participate not only in stabilization of the phosphoanion transition state, but also in protonating the 3′-oxygen leaving group. The proposed catalytic mechanism for Vvn is thus a typical single-metal ion mechanism as shown in Figure7C, similar to those suggested for I-PpoI and Serratia nuclease.
In the active site of Vvn, the side chain of Arg99 is oriented away from the scissile phosphate in the enzyme–substrate complex; however, the side chain is re-oriented and makes a hydrogen bond to the closer 5′-phosphate in the enzyme–product complex. The Arg61 in I-PpoI seems to behave similarly (Galburt and Stoddard, 2002) in that the cleaved 5′-phosphate moves to form a hydrogen bond with Arg61 in the enzyme–product complex (Galburt et al., 1999). Therefore, we suggest that Arg99 in Vvn plays a role similar to that of Arg61 in I-PpoI in stabilizing the product relative to the transition state to decelerate the reverse reaction.
Non-specific DNA recognition
Non-specific DNA-binding proteins almost always bind dsDNA at the minor groove (Luscombe and Thornton, 2002), including chromosomal architectural proteins, such as the prokaryotic HU (Tanaka et al., 1984), archaeal Sac7d (Robinson et al., 1998) and the eukaryotic HMG1/2 family of proteins (Allain et al., 1999), and the DNA enzymes, DNaseI (Suck et al., 1988) and Taq polymerase (Eom et al., 1996). Deficiency of chemical features presented by the minor groove is considered as the basis of using the narrow groove for sequence-independent recognition in DNA. Moreover, the DNA bending and deformation induced by the protein binding at the minor groove is sometimes required for the biological functioning of these proteins (Bewley et al., 1998). Hence, for non-specific DNA-binding proteins, binding at minor grooves can be advantageous in avoiding sequence-dependent DNA recognition and in inducing DNA deformation.
Vvn is an example of a non-specific endonuclease that binds at the minor groove. Remarkably, Vvn only contacts phosphate backbones of DNA molecules but not their base pairs. This is rare compared with other non-specific DNA-binding proteins, most of which still hydrogen-bond considerably to DNA base pairs in the widened minor groove (Luscombe and Thornton, 2002). Vvn differs from the chromosome-associated proteins in that it does not need to deform DNA significantly to carry out DNA hydrolysis. Therefore, it is possible that non-specific DNA-binding proteins, which are not required to largely deform DNA, may choose to bind DNA at minor grooves mainly to avoid any sequence-dependent contacts at major grooves. Therefore, Vvn may bind DNA at minor grooves but only moderately bend DNA in order to avoid closer contact with DNA base pairs and the higher energy required for DNA deformation. More crystalline structures of non-specific DNA-binding protein–DNA complexes need to be solved to ascertain if this hypothesis can be extended to interactions between sequence-independent proteins and DNA in general, especially for the cases in which DNA is not required to be sharply bent.
Conclusion
This work is the first to report the structure of the periplasmic endonuclease Vvn, of the family of extracellular nucleases that participate in DNA digestion in order to protect bacterial cells. Vvn contains a novel structural fold but bears a known endonuclease active site identified in several site-specific or non-specific endonucleases. This indicates that evolution has preserved the general ββα–metal fold of the active site for endonuclease activity. The crystal structure of Vvn in complex with its DNA substrate further reveals how this family of proteins binds and hydrolyzes DNA. Vvn binds DNA at minor grooves inducing moderate DNA bending, and only contacts the phosphate backbones. The interaction between Vvn and DNA, shown in this study, implicates a mechanism by which non-specific DNA-binding proteins recognize DNA without sequence preference. Finally, a single-metal ion mechanism is proposed for the DNA hydrolysis catalyzed by Vvn, based on the crystal structures of Vvn in complex with an intact and a cleaved DNA, respectively. Our data provide a solid foundation for a better understanding of the molecular mechanisms of non-specific endonucleases involved in the protection of bacterial cells and the recognition of DNA by sequence-independent DNA-binding proteins.
Materials and methods
Cloning, expression and protein purification
Vibrio vulnificus vvn from plasmid pSI026 (Wu et al., 2001) was amplified by PCR, using primers 5′-CTGCTCTAGAATGAAGCGA TTATTC-3′ and 5′-CGCTCTAGATTATTGAGTTTGACA-3′. The PCR product was cloned into the XbaI–NdeI site of pTYB2 (NEB) vector. The pTYB2-vvn expression vector was transformed into E.coli B834(DE3) cells (NEB), cultured using LB medium (100µg/ml ampicillin) to an OD600of 0.6 and then induced with 0.8mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 30°C for 8h. The periplasmic fraction of the cultured cells was prepared by the standard sucrose-EDTA method described previously (Wu et al., 2001). The Vvn-containing fraction was then applied onto a HiTrap-SP column (Pharmacia). After washing with buffer A (50mM Tris–HCl pH7.5), Vvn was eluted using a NaCl gradient (buffer A plus 1M NaCl). The purified Vvn had a homogeneity of ∼99% as analyzed by Coomassie-stained SDS–PAGE. The protein sample was concentrated to 10mg/ml by ultrafiltration on Centricon units (Amicon) and stored at –70°C.
The Se-Met-labeled Vvn was expressed from methionine auxotroph E.coli B834(DE3) in a medium consisting of 1× M10 minimal medium supplemented with 80mg/l each amino acid except methionine and 80mg/l seleno-l-methionine (Mechaly et al., 2000). The induction conditions of Se-Met-Vvn were modified by addition of 1.2mM IPTG to the culture at 30°C for 24h. The purification process was the same as that of the wild-type Vvn. The eluted SeMet-Vvn was concentrated to 10mg/ml via Centricon. The H80A mutant of Vvn was constructed using a ‘QuickChangeTM site-Directed Mutagenesis kit’ (Stratagene) and was expressed and purified in the same manner as wild-type Vvn. Electrospray ionization mass spectrometry confirmed the identities of the wild-type, Se-labeled and mutated Vvn.
Endonuclease activity assay
Plasmid nicking assays (Figure2A and B).
Enzymes used for plasmid nicking assay were quantified by Bio-Rad protein assay. Protein concentrations of Serratia nuclease (Sigma), DNaseI (Sigma), the nuclease domain of ColE7 (Ku et al., 2002), Vvn-H80A and Vvn were adjusted between 0.01nM and 0.1µM by series dilution. The plasmid digestion experiment was performed at 37°C in reaction buffers containing 10mM MgCl2 or 10mM CaCl2 and 50mM Tris–HCl pH8.0. The supercoiled form of the DNA plasmid prepared by CsCl–ethidium bromide gradients was incubated with each enzyme at 37°C for 10min and the reactions were stopped by the sample buffer containing 10mM EDTA. The plasmid nicking results were analyzed on 1% agarose gels.
Terminal labeling experiments (Figure2C).
The plasmid DNA digested by Vvn at 37°C overnight was used for labeling experiments. For 5′ labeling, digested DNA, [γ-32P]ATP and T4 polynucleotide kinase (Roche) were mixed and incubated at 37°C for 30min. For 3′ labeling, digested DNA, [α-32P]ATP and TdT transferase (Roche) were mixed and incubated at 37°C for 30min. Reactions were stopped by sample loading buffers and the labeling results were analyzed by 15% PAGE.
Crystallization and data collection
SeMet-Vvn was crystallized at room temperature by hanging drop vapor diffusion against 30% PEG8000, 0.2M ammonium acetate, 0.01M magnesium acetate and 0.05M sodium cacodylate at pH6.5. The SeMet-Vvn crystallized in a monoclinic P21 space group with one molecule per asymmetic unit. The MAD experiments using synchrotron X-ray radiation were performed at the beamline 18B, KEK Photon Factory, Tsukuba, Japan. Crystals firstly were flash-cooled in mother liquor containing 20% glycerol (v/v) and diffraction data sets were collected at –150°C. After a wavelength scan for selenium absorption in the Se-Met-Vvn crystal, three data sets were collected by a Quantum 4 CCD detector at wavelengths of 0.9795 (peak), 0.9796 (edge) and 0.9600Å (remote). Diffraction data were processed with DENZO and SCALEPACK (Otwinowski and Minor, 1997).
The Vvn-H80A–DNA complex crystals were grown by the hanging drop vapor diffusion method. A protein–DNA solution (9.4µg/µl) was prepared by mixing the 8bp palindromic DNA (5′-GCGATCGC-3′) with Vvn-H80A at a 1:1 molar ratio. The Vvn-H80A–DNA solution was mixed further with the reservoir solution containing 20% PEG3350 and 0.2M calcium acetate. Crystals of Vvn-H80A–DNA complex were grown at room temperature after 3 weeks. Crystals were flash-cooled in reservoir solution supplemented with 20% glycerol for data collection. The diffraction data of VvnH80A–DNA were collected at Taiwan beamline BL12B2 at SPring-8, Japan. The Vvn-H80A–DNA complex crystallized in a monoclinic P21 space group with two protein–DNA complexes per asymmetric unit. Cell parameters and diffraction statistics are listed in TableI.
Structure determination and refinement
The MAD method was used in conjunction with the CNS program (Brünger et al., 1998) to identify five selenium sites and calculate the initial MAD phases of the Vvn structure. Solvent flattening was employed to improve the experimental maps further. Vvn model building was performed using the program TURBO-FRODO. The resulting Vvn model was refined and water molecules were added with a water-pick routine in the CNS program. The quality of the model was cross-validated by using 8% of independent reflections in a test data set. The final Vvn model has an R-factor of 18.6% and an Rfree of 26.9% for 9127 reflections in the resolution range 40.0–2.3Å. Stereochemical analysis of the refined model using PROCHECK revealed that main chain and side chain parameters were better than or within the typical range of values for protein structures determined at corresponding resolutions.
The Vvn-H80A–DNA complex structure was determined by molecular replacement using the program CNS (Brünger et al., 1998). The Se-Met-Vvn structure was used as the search model for the two expected solutions in rotational and translational searches. After the refinement of the two protein molecules, the R-factor decreased to 28.9%. At this stage, the Fourier maps gave a clear continuous electron density of the two DNA octamers, and the DNA structural models were built accordingly. The final complex model has an R-factor of 19.1% and an Rfree of 26.0% for 21165 reflections in the resolution range 40.0–2.3Å. The final refinement statistics are listed in TableI. Atomic coordinates have been deposited in the PDB with accession codes of 1OUO for Vvn and 1OUP for the Vvn–DNA complex.
Acknowledgments
Acknowledgements
We thank Carmay Lim for discussions and careful reading of the manuscript. We gratefully acknowledge access to the synchrotron radiation beamline 18B in the KEK Photon Factory and the 12B Taiwan beamline in SPring-8, Japan. This work was supported by research grants from Academia Sinica and the National Science Council of the Republic of China to H.S.Y.
References
- Alberts I.L., Nadassy,K. and Wodak,S.J. (1998) Analysis of zinc binding sites in protein structures. Protein Sci., 7, 1700–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain F.H.-T., Yen,Y.-M., Masse,J.E., Schultze,P., Dieckmann,T., Johnson,R.C. and Feigon,J. (1999) Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding. EMBO J., 18, 2563–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley C.A., Gronenborn,A.M. and Clore,G.M. (1998) Minor groove-binding architectural proteins: structure, function and DNA recognition. Annu. Rev. Biophys. Biomol. Struct., 27, 105–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system (CNS): a new software system for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Burley S.K. (1994) DNA-binding motifs from eukaryotic transcription factors. Curr. Opin. Struct. Biol., 4, 3–11. [Google Scholar]
- Chang M.C., Chang,S.Y., Chen,S.L. and Chuang,S.M. (1992) Cloning and expression in Escherichia coli of the gene encoding an extracellular deoxyribonuclease (DNase) from Aeromonas hydrophila. Gene, 122, 175–180. [DOI] [PubMed] [Google Scholar]
- Cheng Y.-S., Hsia,K.-C., Doudeva,L.G., Chak,K.-F. and Yuan,H.S. (2002) The crystal structure of the nuclease domain of ColE7 suggests a mechanism for binding to double-stranded DNA by the H-N-H endonucleases. J. Mol. Biol., 324, 227–236. [DOI] [PubMed] [Google Scholar]
- Dodd H.N. and Pemberton,J.M. (1996) Cloning, sequencing and characterization of the nucH gene encoding an extracellular nuclease from Aeromonas hydrophila JMP636. J. Bacteriol., 178, 3926–3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudev T. and Lim,C. (2001) Metal selectivity in metalloproteins: Zn2+ vs. Mg2+. J. Phys. Chem., 105, 4446–4452. [Google Scholar]
- Durwald H. and Hoffmann-Berling,H. (1968) EndonucleaseI-deficient and ribonucleaseI-deficient Escherichia coli mutants. J. Mol. Biol., 34, 331–346. [DOI] [PubMed] [Google Scholar]
- Ellison E.L. (1993) Interaction of the intron-encoded mobility endonucleaseI-PpoI with its target site. Mol. Cell. Biol., 13, 7531–7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom S.H., Wang,J. and Steitz,T.A. (1996) Structure of Taq polymerase with DNA at the polymerase active site. Nature, 382, 278. [DOI] [PubMed] [Google Scholar]
- Flick K.E., Jurica,M.S., Monnat,R.J. and Stoddard,B.L. (1998) DNA binding and cleavage by the nuclear intron-encoded homing endonucleaseI-PpoI. Nature, 394, 96–101. [DOI] [PubMed] [Google Scholar]
- Focareta T. and Manning,P.A. (1987) Extracellular proteins of Vibrio cholerae: molecular cloning, nucleotide sequence and characterization of the deoxyribonuclease (DNase) together with its periplasmic locatlization in Escherichia coli K-12. Gene, 53, 31–40. [DOI] [PubMed] [Google Scholar]
- Focareta T. and Manning,P.A. (1991) Distinguishing between the extracellular DNases of Vibrio cholerae and development of a transformation system. Mol. Microbiol., 5, 2547–2555. [DOI] [PubMed] [Google Scholar]
- Friedhoff P., Meiss,G., Kolmes,B., Pieper,U., Gimadutdinow,O., Urbanke,C. and Pingoud,A. (1996) Kinetic analysis of the cleavage of natural and synthetic substrates by the Serratia nuclease. Eur. J. Biochem., 241, 572–580. [DOI] [PubMed] [Google Scholar]
- Friedhoff P., Franke,I., Krause,K.L. and Pingoud,A. (1999a) Cleavage experiments with deoxythymidine 3′,5′-bis-(p-nitrophenylphosphate) suggest that the homing endonucleaseI-PpoI follows the same mechanism of phosphodiester bond hydrolysis as the non-specific Serratia nuclease. FEBS Lett., 443, 209–214. [DOI] [PubMed] [Google Scholar]
- Friedhoff P., Franke,I., Meiss,G., Wende,W., Krause,K.L. and Pingoud,A. (1999b) A similar active site for non-specific and specific endonucleases. Nat. Struct. Biol., 6, 112–113. [DOI] [PubMed] [Google Scholar]
- Galburt E.A. and Stoddard,B.L. (2002) Catalytic mechanisms of restriction and homing endonucleases. Biochemistry, 41, 13851–13860. [DOI] [PubMed] [Google Scholar]
- Galburt E.A., Chevalier,B., tang, Jurica,M.S., Flick,K.E., Monnat,R.J. and Stoddard,B.L. (1999) A novel endonuclease mechanism directly visualized for I-PpoI. Nat. Struct. Biol., 6, 1096–1099. [DOI] [PubMed] [Google Scholar]
- Galburt E.A., Chadsey,M.S., Jurica,M.S., Chevalier,B.S., Erho,D., Tang,W., Monnat,R.J. and Stoddard,B.L. (2000) Conformational changes and cleavage by the homing endonucleaseI-PpoI: a critical role for a leucine residue in the active site. J. Mol. Biol., 300, 877–887. [DOI] [PubMed] [Google Scholar]
- Greger B. and Kemper,B. (1998) An apyrimidinic site kinks DNA and triggers incision by endonuclease VII of phage T4. Nucleic Acids Res., 26, 4432–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Jekel M. and Wackernagel,W. (1995) The periplasmic endonucleaseI of Escherichia coli has amino-acid sequence homology to the extracellular DNases of Vibrio cholerae and Aeromonas hydrophila. Gene, 154, 55–59. [DOI] [PubMed] [Google Scholar]
- Jones S., Heyningen,P.V., Berman,H.M. and Thornton,J.M. (1999) Protein–DNA interactions: a structural analysis. J. Mol. Biol., 287, 877–896. [DOI] [PubMed] [Google Scholar]
- Kim J.L., Nikolov,D.B. and Burley,S.K. (1993) Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature, 365, 520–527. [DOI] [PubMed] [Google Scholar]
- Kim Y., Geiger,J.H., Hahn,S. and Sigler,P.B. (1993) Crystal structure of a yeast TBP/TATA-box complex. Nature, 365, 512–520. [DOI] [PubMed] [Google Scholar]
- Kleanthous C., Kuhlmann,U.C., Pommer,A.J., Ferguson,N., Radford,S.E., Moore,G.R., James,R. and Hemmings,A.M. (1999) Structural and mechanistic basis of immunity toward endonuclease colicins. Nat. Struct. Biol., 6, 243–252. [DOI] [PubMed] [Google Scholar]
- Ko T.-P., Liao,C.-C., Ku,W.-Y., Chak,K.-F. and Yuan,H.S. (1999) The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein. Structure, 7, 91–102. [DOI] [PubMed] [Google Scholar]
- Kobayashi I. (2001) Behavior of restriction–modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res., 29, 3742–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall R.A. and Matthews,B.W. (1999) Type II restriction endonucleases: structural, functional and evolutionary relationships. Curr. Opin. Chem. Biol., 3, 578–583. [DOI] [PubMed] [Google Scholar]
- Ku W.-Y., Liu,Y.-W., Hsu,Y.-C., Liao,C.-C., Liang,P.-H., Yuan,H.S. and Chak,K.-F. (2002) The zinc ion in the HNH motif of the endonuclease domain of colicin E7 is not required for DNA binding but is essential for DNA hydrolysis. Nucleic Acids Res., 30, 1670–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann U.C., Moore,G.R., James,R., Kleanthous,C. and Hemmings,A.M. (1999) Structural parsimony in endonuclease active site: should the number of homing endonuclease families be redefined? FEBS Lett., 463, 1–2. [DOI] [PubMed] [Google Scholar]
- Lavery R. and Sklenar,H. (1988) The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn., 6, 63–91. [DOI] [PubMed] [Google Scholar]
- Luscombe N.M. and Thornton,J.M. (2002) Protein–DNA interactions: amino acid conservation and the effects of mutations on binding specificity. J. Mol. Biol., 320, 991–1009. [DOI] [PubMed] [Google Scholar]
- Mannino S.J., Jenkins,C.L. and Raines,R.T. (1999) Chemical mechanism of DNA cleavage by the homing endonucleaseI-PpoI. Biochemistry, 38, 16178–16186. [DOI] [PubMed] [Google Scholar]
- Mechaly A., Teplitsky,A., Belakhov,V., Baasov,T., Shoham,G. and Shoham,Y. (2000) Overproduction and characterization of seleno-methionine xylanase T-6. J. Biotechnol., 78, 83–86. [DOI] [PubMed] [Google Scholar]
- Miller M.D., Tanner,J., Alpaugh,M., Benedik,M.J. and Krause,K.L. (1994) 2.1 Å structure of Serratia endonuclease suggests a mechanism for binding to double-stranded DNA. Nat. Struct. Biol., 1, 461–468. [DOI] [PubMed] [Google Scholar]
- Miller M.D., Cai,J. and Krause,K.L. (1999) The active site of Serratia endonuclease contains a conserved magnesium–water cluster. J. Mol. Biol., 288, 975–987. [DOI] [PubMed] [Google Scholar]
- Moulard M., Condemine,G. and Robert-Baudouy,J. (1993) Characterization of the nucM gene coding for a nuclease of the phytopathogenic bacteria Erwinia chrysanthemi. Mol. Microbiol., 8, 685–695. [DOI] [PubMed] [Google Scholar]
- Nadassy K., Wodak,S.J. and Janin,J. (1999) Structural features of protein–nucleic acid recognition sites. Biochemistry, 38, 1999–2017. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Pabo C.O. and Nekludova,L. (2000) Geometric analysis and comparison of protein–DNA interfaces: why is there no simple code for recognition? J. Mol. Biol., 301, 597–624. [DOI] [PubMed] [Google Scholar]
- Pingoud A. and Jeltsch,A. (2001) Structure and function of type II restriction endonucleases. Nucleic Acids Res., 29, 3705–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers H., Vix,O., Toro,I., Golz,S., Kemper,B. and Suck,D. (1999) X-ray structure of T4 endonuclease VII: a DNA junction resolvase with a novel fold and unusual domain-swapped dimer architecture. EMBO J., 18, 1447–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson H., Gao,Y.-G., McCrary,B.S., Edmondson,S.P., Shriver,J.W. and Wang,A.H.-J. (1998) The hyperthermophile chromosomal protein Sac7d sharply kinks DNA. Nature, 392, 202–205. [DOI] [PubMed] [Google Scholar]
- Suck D., Lahm,A. and Oefner,C. (1988) Structure refined to 2 Å of a nicked DNA octanucleotide complex with DNaseI. Nature, 332, 464–468. [DOI] [PubMed] [Google Scholar]
- Sui M.-J., Tsai,L.-C., Hsia,K.-C., Doudeva,L.-G., Chak,K.-F. and Yuan,H.S. (2002) Metal ions and phosphate binding in the H-N-H motif: crystal structures of the nuclease domain of ColE7/Im7 in complex with a phosphate ion and different divalent metal ions. Protein Sci., 11, 2947–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka I., Appelt,K., Dijk,J., White,S.W. and Wilson,K.S. (1984) 3-Å resolution structure of a protein with histone-like properties in prokaryotes. Nature, 310, 376–381. [DOI] [PubMed] [Google Scholar]
- Walker D.C., Georgiou,T., Pommer,A.J., Walker,D., Moore,G.R., Kleanthous,C. and James,R. (2002) Mutagenic scan of the H-N-H motif of colicin E9: implications for the mechanistic enzymology of colicins, homing enzymes and apoptotic endonucleases. Nucleic Acids Res., 30, 3225–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M.H., Huth,J.R., Gronenborn,J.R. and Glore,G.M. (1995) Molecular basis of human 46X, Y sex reversal revealed from the three-dimensional solution structure of the human SRY–DNA complex. Cell, 81, 705–714. [DOI] [PubMed] [Google Scholar]
- Wolberger C. (1999) Multiprotein–DNA complexes in transcription regulation. Annu. Rev. Biophys. Biomol. Struct., 28, 29–56. [DOI] [PubMed] [Google Scholar]
- Wu S.-I., Lo,S.-K., Shao,C.-P., Tsai,H.-W. and Hor,L.-I. (2001) Cloning and characterization of a periplasmic nuclease of Vibrio vulnificus and its role in preventing uptake of foreign DNA. Appl. Environ. Microbiol., 67, 82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]