Abstract
Xanthophyllomyces dendrorhous is one of the relevant sources of the carotenoid astaxanthin. In this paper, we describe for the first time cloning of unexpected cDNAs obtained from the crtI and crtYB genes of X. dendrorhous strain UCD 67-385. The cDNA of the crtI gene conserves 80 bp of the first intron, while the cDNA of the crtYB gene conserves 55 bp of the first intron and lacks 111 bp of the second exon. The crtI and crtYB RNAs could be spliced in alternative splice sites, which produced alternative transcripts which could not be translated to active CRTI and CRTYB proteins since they had numerous stop codons in their sequences. The ratio of mature mRNA to alternative mRNA for the crtI gene decreased as a function of the age of the culture, while the cellular content of carotenoids increased. It is possible that splicing to mature or alternative transcripts could regulate the cellular concentrations of phytoene desaturase and phytoene synthase-lycopene cyclase proteins, depending on the physiological or environmental conditions.
Carotenoids are terpenoid pigments that are synthesized in bacteria, algae, fungi, and green plants (5). They provide protection against photooxidation and inactivate free radicals due to their highly conjugated double-bond systems (11, 28, 29). Astaxanthin, produced primarily by phytoplankton, is the principal carotenoid responsible for the orange-red color of marine invertebrates, fish, and birds. Since animals are not capable of astaxanthin synthesis, this xanthophyll must be added to the feed of aquacultured organisms like salmonids to obtain the appropriate pigmentation for consumer appeal (19, 38). To date, one of the few microbial sources of astaxanthin is the red basidiomycetous yeast Xanthophyllomyces dendrorhous (3, 20, 24).
As in other organisms, the condensation of two molecules of geranylgeranyl pyrophosphate (GGPP) to phytoene is the first specific step in the biosynthesis of astaxanthin. Subsequently, the pathway to lycopene involves the introduction of double bonds by four desaturations of phytoene. Finally, after cyclization of lycopene to β-carotene, hydroxylation occurs at the 3 and 3′ carbons, and keto groups are added at positions 4 and 4′ of β-rings. Recently, the phytoene desaturase-encoding gene (crtI) was isolated by heterologous complementation in an Escherichia coli strain accumulating phytoene. To do this, a cDNA library from X. dendrorhous was introduced into an E. coli strain carrying genes encoding GGPP synthase (crtE) and phytoene synthase (crtB) from Erwinia uredovora (40). By using a similar approach, the first lycopene cyclase gene (crtYB) described in a fungus was isolated from X. dendrorhous (41). The product of the crtYB gene is a bifunctional enzyme with two catalytic activities, as it converts GGPP into phytoene and lycopene into β-carotene. A related phytoene synthase-lycopene cyclase has been cloned from other fungi belonging to the taxonomic groups ascomycota and zygomycota (6, 7, 39).
Additionally, numerous studies on the expression at the mRNA level of carotenogenic genes in ascomycetes and zygomycetes have established that expression increases in response to environmental conditions, such as blue light illumination (12, 27, 31, 33, 34). The synthesis of alternative spliced mRNAs is a well-known process in eukaryotic organisms like the fruitfly Drosophila melanogaster, the nematode Caenorhabditis elegans, and mammals (22, 26, 35). With regard to yeasts and filamentous fungi, there have been some reports of alternative splicing (8, 9, 15, 17, 37, 44, 45), but as far as we know, alternative splicing has not been described in carotenogenic genes yet. In X. dendrorhous, neither the presence of alternative spliced transcripts of carotenogenic genes nor the expression of these genes in relation to the culture conditions has been described. The main goal of this study was isolation of crtI and crtYB transcripts which could be processed in alternative spliced sites in X. dendrorhous strain UCD 67-385. Also, the levels of transcripts for both genes were determined as a function of the culture age.
MATERIALS AND METHODS
DNA techniques and sequence analysis.
General procedures for plasmid DNA purification, gel electrophoresis, cloning, and other standard molecular biology techniques were carried out as described by Sambrook et al. (32). Restriction endonuclease digestion and ligation with T4 DNA ligase were performed as recommended by the enzyme suppliers (New England Biolabs and Gibco-BRL). Reverse transcription (RT)-PCR products for cloning were recovered from agarose gels and purified by using glassmilk as described previously (10). Plasmid pBluescript SK was used in cloning experiments. This vector was digested with EcoRV, and nucleotide T overhangs were added at the 3′ ends to facilitate cloning of RT-PCR products. X. dendrorhous DNA was isolated as described previously (23) from protoplasts prepared from cells grown at 22°C for 4 days in liquid MMv medium (30). Nucleotide sequences were determined by using a Big Dye Terminator v3.0 DNA sequencing kit (Applied Biosystems). The sequence data were analyzed by using the University of Wisconsin Genetics Computer Group package, version 10.0 (16), and the program CLUSTAL W, version 1.8 (36).
Strains and plasmids.
All strains and plasmids used in this study are listed in Table 1. The atxS2 strain was obtained after treatment of UCD 67-385 with N-methyl-N′-nitro-nitrosoguanidine (40 μg/ml) under the conditions described previously (14). A highly pigmented colony was selected by visual inspection, and it was resuspended in distilled water. This suspension was plated on YM medium (2), and a highly pigmented colony, designated strain atxS2, was analyzed for carotenoid production and pigment composition. The atxS2 cultures produced about 2.230 μg of total carotenoid per g (dry weight) of yeast, and high-performance liquid chromatography analysis of carotenoids indicated that the principal pigment was astaxanthin.
TABLE 1.
Plasmids and strains used in this study
| Strain or plasmid | Genotype or relevant features | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F− φ80d lacZΔM15Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rk−mk+) phoA supE44λ−thi-l gyrA96 relAl | 32 |
| X. dendrorhous (Phaffia rhodozyma) UCD 67-385 or ATCC 24230 | Wild type | |
| atxS2 | Astaxanthin-overproducing strain after N-methyl-N′-nitro-nitrosoguanidine treatment of strain UCD 67-385 | Unpublished data |
| Plasmids | ||
| pL25 | pBluescript carrying a 3.6-kb PCR product containing the complete crtI gene | 21 |
| pC13 | pBluescript with a 18.5-kb BamHI fragment which contains the crtI gene, isolated from a genomic library of X. dendrorhous | 21 |
| pl43 | Clone carrying a crtl cDNA insert which conserves 80 bp of the first intron in pBluescript | This study |
| pl41 | Contains a cDNA insert encoding phytoene desaturase of X. dendrorhous in pBluescript | This study |
| pYBm | Contains a cDNA insert encoding phytoene synthase-lycopene cyclase of X. dendrorhous in pBluescript | This study |
| pYBa | Clone carrying a crtYB cDNA insert which conserves 55 bp of the first intron and 96 bp of the second exon in pBluescript | This study |
E. coli growth conditions.
Electrocompetent E. coli cells were transformed with DNA from ligation reactions of RT-PCR products and pBluescript. The transformed cells were plated on selective Luria-Bertani agar plates (32) and incubated at 37°C overnight. These plates contained ampicillin (100 μg/ml) for selection of the plasmid and 40 μl of a 2% solution of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) per plate to select for white colonies.
X. dendrorhous growth conditions.
X. dendrorhous wild-type and mutant strains were grown in fermentor batch cultures, and samples were withdrawn from each culture at different times to collect cells for carotenoid extraction, biomass determination, and total RNA isolation. For each strain, a preculture was prepared in a 1-liter baffled flask containing 400 ml of YM medium (2) by adding 4 ml of a 2 day-culture in YM medium. All cultures were grown with shaking at 100 rpm for baffled flasks and at 150 rpm for nonbaffled flasks in an orbital shaker at a constant temperature of 22°C. A 12-liter jar fermentor (New Brunswick) containing 8.8 liters of YM medium with 450 μl of silicone antifoam agent (1520 EU; Dow Corning) was inoculated with 200 ml of a 2-day preculture of the wild-type or mutant strain. The temperature was controlled at 22 ± 1°C, and the culture was agitated at 300 rpm. Sterile air was injected at a flow rate of 8 liters/min. The antifoam agent was automatically added when it was needed. Samples for carotenoid and RNA extraction were centrifuged at 1,300 × g for 10 min to obtain cell pellets. The cell pellets were immediately frozen in liquid nitrogen and then stored at −70°C until they were processed.
Purification of total RNA from X. dendrorhous.
RNA extraction was performed by using the method described by Chomczynski and Sacchi (13) and modified for X. dendrorhous as follows. To the cellular material, 3 to 5 ml of Chomczynski solution in phenol (Ch-P solution) was added, and then 1 volume of glass beads (diameter, 425 to 600 μm; Sigma) was added. Cells were broken by shaking with a vortex at the maximum speed for 10 min. The mixture was incubated for 10 min at room temperature, and this was followed by addition of 0.2 ml of chloroform per ml of Ch-P solution with shaking and incubation at room temperature for 5 min. After centrifugation at 12,100 × g, the RNA in the aqueous phase was transferred to a sterile tube, and 1 volume of isopropanol was added. After incubation for 10 min at room temperature, the RNA was precipitated by centrifugation at 12,100 × g for 10 min at 4°C. The RNA pellet was washed with 1 ml of 75% ethanol. The RNA was resuspended in water (diethyl pyrocarbonate treated) and then stored at −20°C. The total RNA concentration was determined spectrophotometrically at 260 nm, and aliquots of the extracts were subjected to denaturing agarose gel electrophoresis to check RNA integrity.
RT.
The RNA samples were treated with 1 U of DNase I (RNase free; Roche) per μl in 2.5 mM MgCl2 for 30 min at 25°C. The reaction was stopped by addition of EDTA at a final concentration of 2.5 mM and heating at 65°C for 15 min. The RT reaction was performed in a 25-μl (final volume) mixture containing 3 μg of total RNA, 75 pmol of oligo(dT15-18), each deoxynucleoside triphosphate (dNTP) at a concentration of 0.5 mM, and 200 U of Moloney murine leukemia virus reverse transcriptase (Promega). The reaction mixture was incubated for 60 min at 42°C and then heated for 10 min at 65°C.
PCR amplification.
The sequences of all the primers used in this study are shown in Table 2, and their locations in gene sequences are indicated in Table 2 and Fig. 1. To clone crtI and crtYB cDNAs, specific primers upstream of translation initiation sites and downstream of translation termination sites were designed by using the sequences of strains CBS 6938 (accession numbers Y15007 and AJ133646, respectively) and UCD 67-385 (1, 21). PCR amplification was performed with 1 U of VentR DNA polymerase (New England Biolabs) in a 25-μl (final volume) mixture containing 2.5 μl of 10× VentR DNA polymerase buffer, 0.5 μl of a solution containing each dNTP at a concentration of 10 mM, 1 μl of a solution containing each primer at a concentration of 25 μM, 2 μl of an RT reaction mixture containing the single-stranded cDNA, and 18 μl of water. PCR was performed with a 2400 DNA thermal cycler (Perkin-Elmer) by using the following program: 95°C for 3 min, 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 3 min, and a final extension at 72°C for 10 min. The PCR products were electrophoresed on 0.7 or 1% agarose gels and stained with ethidium bromide. To quantify RT-PCR products, 28 cycles of amplification were employed with annealing temperatures of 60°C for amplification of crtI cDNAs and 55°C for amplification of crtYB cDNAs. PCR amplification was performed with 2 U of Taq polymerase (Promega) in a 25-μl (final volume) mixture containing 2.5 μl of 10× Taq buffer, 0.5 μl of a solution containing each dNTP at a concentration of 10 mM, 1 μl of a solution containing 50 mM MgCl2, 1 μl of a solution containing each primer at a concentration of 25 μM, 2 μl of an RT reaction mixture containing the single-stranded cDNA, and water. All PCR amplifications were performed at least by duplicate and were standardized for the concentration of single-stranded cDNA used and the number of amplification cycles. Equal volumes containing the PCR products were loaded on 3% agarose gels containing ethidium bromide for the quantification of RT-PCR products from the crtI gene, while 4.5% polyacrylamide gels were used in the case of the crtYB gene. After agarose gel electrophoresis, the masses of the bands were quantified by using a 100-bp DNA ladder containing known concentrations of compounds (Fermentas) and Kodak Digital Science 1D image analysis software. Only those bands whose intensity was not oversaturated were used for quantification. To normalize for sample-to-sample variation due to RT and PCR efficiency, relative values were obtained by comparing the intensities of the carotenogenic gene amplification bands with the intensity of the actin (act) amplification product. The primers used for amplification of the act gene were designed by using previously described sequences (43). The level of expression of the act gene is constant throughout the yeast growth cycle.
TABLE 2.
Primers used in this study
| Gene | Primer | Directiona | Sequence (5′→3′) | Location |
|---|---|---|---|---|
| act | ACT3 | F | ACTCCTACGTTGGTGACGAG | Spanning exon 4 and exon 5 |
| ACT4 | R | TCAAGTCTCGACCGGCCAAG | Exon 5 | |
| crtI | 1 | F | CCTCGCCGAATCTAACTTGA | Upstream translation initiation |
| 2 | F | AGCTATCATCGTGGGATGTGGb | Spanning exon 1 and exon 2 | |
| 3 | F | GTATCGGTGGAATCGCCACT | Exon 2 | |
| 4 | F | AGCTATCATCGTGGTTTAATCCc | Spanning exon 1 and intron 1 | |
| 5 | R | AACGAATAAAAAAGATGATGAACA | Downstream translation termination | |
| 6 | R | GACCCAATCTTCCATCTTCTCT | Exon 5 | |
| 7 | R | TTCTCGAACACCGTGACCT | Exon 2 | |
| 8 | R | AACGGATCGAGCGATCACGG | Exon 12 | |
| crtYB | 9 | F | CCGATCTCGGATAGACATCA | Upstream translation initiation |
| 10 | F | TACCCAACTCGTATCATCCC | Upstream translation initiation | |
| 11 | F | GCATATTACCAGATCCATCTGb | Spanning exon 1 and exon 2 | |
| 12 | F | GTGTGCATATGTGTTGCAACCc | Spanning intron 1 and exon 2 | |
| 13 | R | AGGAAGATGGGGGGAAGA | Downstream translation termination | |
| 14 | R | AGTCTTTATGGTCTATAACCT | Downstream translation termination | |
| 15 | R | TCTAGAAACGTTCCAAACACG | Exon 2 |
F, forward; R, reverse.
Specific for mmRNA.
Specific for amRNA.
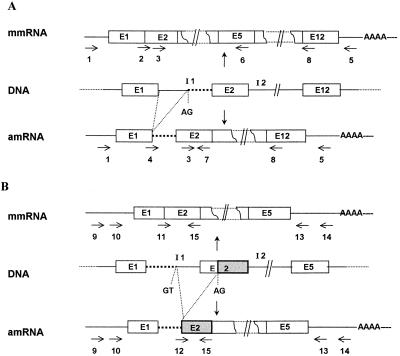
FIG. 1.
Structure of the genomic DNA, mmRNA, and amRNA of the crtI gene (A) and the crtYB gene (B). E, exon; I, intron; AG, alternative splicing acceptor site; GT, alternative splicing donor site. The horizontal arrows indicate the locations of the primers used in PCR, which are described in Table 2. (A) The dotted lines indicate the 80 bases of the first intron present in the crtI amRNA. (B) The dotted lines indicate the 55 bases of the first intron present in the crtYB amRNA, while the 96 bases of the second exon conserved in this transcript are highlighted. The diagram is not to scale.
Biomass determination and carotenoid extraction.
The cell concentration was measured by determining the turbidity of the culture at 560 nm or by cell counting in a Neubauer chamber. For dry cell weight determinations, cells from 3- to 5-ml cultures were pelleted, washed with distilled water, and then dried at 80°C overnight. The acetone extraction method described by An et al. (2) was employed to extract carotenoids from cellular pellets. Total carotenoid concentrations were calculated by using the absorption coefficient (A1% = 2,100) (2). All analyses were carried out in duplicate or triplicate, and the average values and standard deviations are reported below.
Nucleotide sequence accession numbers.
The cDNA nucleotide sequences of the mature mRNA of the crtI gene (crtI mmRNA), the alternative mRNA of the crtI gene (crtI amRNA), the mature mRNA of the crtYB gene (crtYB mmRNA), and the alternative mRNA of the crtYB gene (crtYB amRNA) from X. dendrorhous strain UCD 67-385 have been deposited in the GenBank database under accession numbers AY177424, AY177425, AY177204, and AY174117, respectively.
RESULTS
Isolation of alternative spliced mRNAs of crtI and crtYB genes.
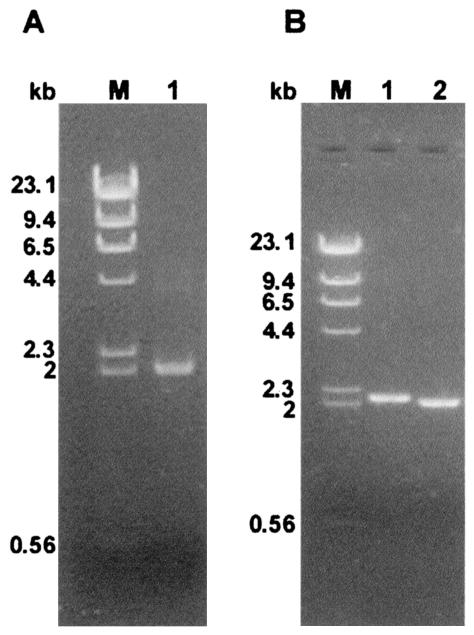
To clone the cDNA from the crtI and crtYB genes, we carried out RT-PCR assays with total RNA samples isolated from stationary-phase cultures of X. dendrorhous grown in YM medium. Primers upstream from the translation initiation codon and downstream from the stop codon were designed for each gene to amplify the entire coding region (Table 2 and Fig. 1). The first-strand cDNA synthesis was performed with total RNA samples previously treated with DNase by using oligo(dT). Then crtYB and crtI cDNAs were amplified by using a Taq DNA polymerase with proofreading activity. RT-PCR with primers 1 and 5 for the crtI gene gave rise to a product of approximately 2 kb (Fig. 2A), which was extracted from the agarose gel and cloned in pBlueskript SK. Restriction analysis of 28 recombinant clones indicated that 16 of them had cDNA inserts of about 2.1 kb, while 12 clones had inserts of about 2.0 kb (Fig. 2B, lanes 1 and 2). The nucleotide sequences of three clones, two with ∼2.1-kb inserts and one with a ∼2.0-kb insert, were determined. Comparison of these nucleotide sequences with the previously published genomic sequence revealed that one cDNA product had been synthesized from an mRNA which had conserved 80 bp of the first intron (crtI amRNA). The other cDNA insert corresponded to the mmRNA of the crtI gene without any introns (crtI mmRNA). Nucleotide sequence translation of the crtI amRNA resulted in stop codons along the entire sequence.
FIG. 2.
(A) Amplification of the crtI mRNA by RT-PCR with primers 1 and 5. (B) The amplification band was cloned, and ampicillin-resistant transformants had plasmids with different inserts (lanes 1 and 2). Lane M contained the λ/HindIII molecular size marker.
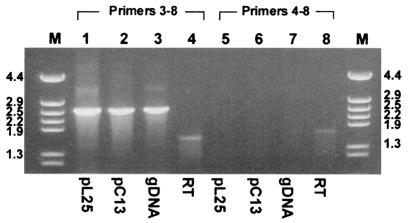
The crtI amRNA could have been synthesized from an RNA which followed an alternative splicing pathway by using an unexpected acceptor site (AG in Fig. 1A). Another possibility is that a crtI gene with the new structure may have been present in the genome of X. dendrorhous. To test the latter proposal, we designed a forward primer (primer 4 in Fig. 1) specific for crtI amRNA spanning the first exon and the first intron sequence absent in the crtI mmRNA. As templates for PCR we employed genomic DNA, RT reaction mixtures, and DNA from plasmids pL25 and pC13, both of which carry the crtI gene (Table 1). Figure 3 shows that when genomic or plasmid DNA was used as the template, PCR performed with primers 3 and 8 (Fig. 1A) resulted in a principal product whose expected size was about 2.6 kb (lanes 1, 2, and 3). In contrast, when PCR was performed with genomic or plasmid DNA and primers 4 and 8, there was no amplification product (Fig. 3, lanes 5, 6, and 7). In the same experiment, RT-PCR performed with primers 3 and 8 resulted in an approximately 1.7-kb product due to the absence of introns, while an approximately 1.8-kb product was amplified by using primers 4 and 8 (Fig. 3, lanes 4 and 8). These results suggest that the crtI amRNA could have been the result of an alternative splicing pathway and not the transcription product of a new crtI gene with a different structure, as has been described previously (40).
FIG. 3.
Amplification of the crtI gene with primers 3 and 8 (lanes 1, 2, 3, and 4) and with primers 4 and 8 (lanes 5, 6, 7, and 8). The substrates used for PCR were DNA of plasmid pL25 (lanes 1 and 5), DNA of plasmid pC13 (lanes 2 and 6), genomic DNA (gDNA) (lanes 3 and 7), and RT reaction products (lanes 4 and 8). Lanes M contained the φ29/HindIII DNA ladder.
Next, we carried out RT-PCR assays to amplify crtYB cDNA, and as a result, two different products were cloned in pBlueskript SK. Nucleotide sequence analysis indicated that one of these products was synthesized from crtYB mmRNA. The other cloned cDNA product had an unexpected sequence as it had 55 bp of the first intron conserved and had lost 111 bp of the second exon. As shown in Fig. 1B, the new isolated cDNA could have been synthesized from an mRNA which followed a splicing pathway in alternative 5′ and 3′ splice sites (GT and AG in Fig. 1B), producing crtYB amRNA. Similarly, as established for the crtI amRNA, translation of the crtYB amRNA yielded several stop codons along the complete sequence.
Expression of crtI and crtYB messengers as a function of the age of the culture.
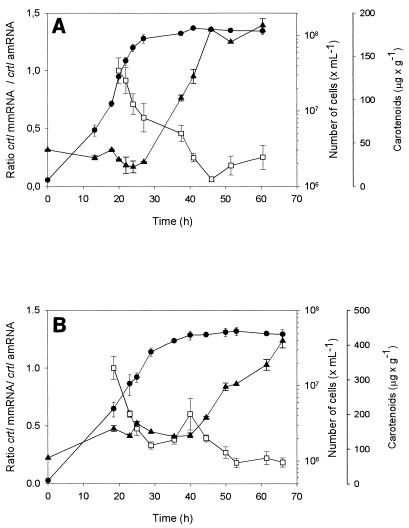
X. dendrorhous strains UCD 67-385 and atxS2 (atxS2 was an astaxanthin-overproducing mutant derived from the wild type) were grown in fermentor batch cultures, and the levels of crtI and crtYB gene transcripts were determined by RT-PCR. Primers 2 and 6 were employed for analysis of the crtI mmRNA levels, while for crtI amRNA we used primers 4 and 7 (Table 1). To analyze crtYB mmRNA levels, primers 11 and 15 were used, while crtYB amRNA levels were analyzed with primers 12 and 15 (Table 1). In addition, we designed primers for amplification of the act gene, which was used as an internal standard in RT-PCR assays. The levels of crtI messengers in relation to the act mRNA level for each time point were quantified, and the ratio of crtI mmRNA to crtI amRNA (M/A ratio) was calculated. The M/A ratio decreased with the age of culture for the wild-type strain (Fig. 4A). However, the cellular concentration of carotenoids began to increase after 27 h of culture. After 13 h of growth, crtI amRNA was detectable, but the level was too low to be determined, while the level of crtI mmRNA was high enough to be quantified. This means that in the early stage of the growth cycle a high proportion of crtI RNA was processed to mmRNA, which could be translated to phytoene desaturase protein, but the level of mmRNA decreased with the age of the culture. With the atxS2 strain, the M/A ratio also decreased with the age of culture (Fig. 4B). As in the wild-type strain, the cellular concentration of carotenoids increased during the stationary phase even though the proportion of mmRNA was less than the proportion in the exponential phase.
FIG. 4.
Kinetics of expression of crtI mmRNA and crtI amRNA for X. dendrorhous strains UCD 67-385 (A) and atxS2 (B). The level of each crtI messenger was normalized by using the level of the act messenger. Each value is relative to the highest value of the curve (which was defined as 1). Symbols: •, number of cells; ▴, carotenoid concentration; □, M/A ratio.
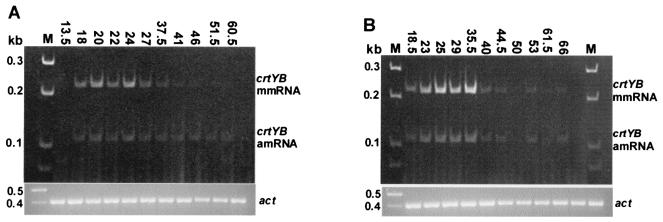
With regard to the crtYB gene, Fig. 5 shows that the mmRNA and the amRNA were detected at different culture times and that there were variable levels for both strains. In this case, the levels of amRNA and mmRNA at many time points were too low to be quantified by the methodology employed previously.
FIG. 5.
Amplification by RT-PCR of act mRNA, crtYB mmRNA, and crtYB amRNA of X. dendrorhous strains UCD 67-385 (A) and atxS2 (B). Total RNA samples were isolated from cells of cultures at various times (in hours), as indicated at the top of each gel. The same RT reaction mixtures were amplified with primers for the act gene and primers for the crtYB gene. The same reaction volume was loaded in each lane. In the case of the crtYB gene, the total reaction mixture was loaded.
DISCUSSION
In this paper we describe isolation of amRNAs of the crtI and crtYB genes, whose predicted translation yielded numerous stop codons. However, the correct reading frame of these transcripts was restored downstream from the proper initial AUG. In this way, a phytoene desaturase without 81 amino acids of the N-terminal part of the protein could be synthesized from the crtI amRNA if translation could be initiated at an internal in-frame AUG. Similarly, the crtYB amRNA could be translated to a phytoene synthase-lycopene cyclase without 153 amino acids of the N-terminal part of the protein. Vittorioso et al. (42) reported al-3 mRNA translation from two internal in-frame AUG codons, which produced a shortened but still active version of GGPP synthase protein as it conserved the domains shared by prenyltransferase enzymes. In the phytoene desaturase from X. dendrorhous, the N-terminal part of the protein has a dinucleotide [flavin adenine dinucleotide or NAD(P)] binding motif that is conserved in phytoene desaturases from bacteria, algae, plants, and fungi (4, 18, 40). This indicates that it is unlikely that a shortened phytoene desaturase lacking 81 N-terminal amino acids would still be active. Similarly, a phytoene synthase-lycopene cyclase enzyme without 153 N-terminal amino acids would be inactive because truncation at the 5′ end of the crtYB cDNA at nucleotide position 514 not only resulted in a total loss of lycopene cyclization activity but also affected about 70% of the phytoene synthesis activity (41).
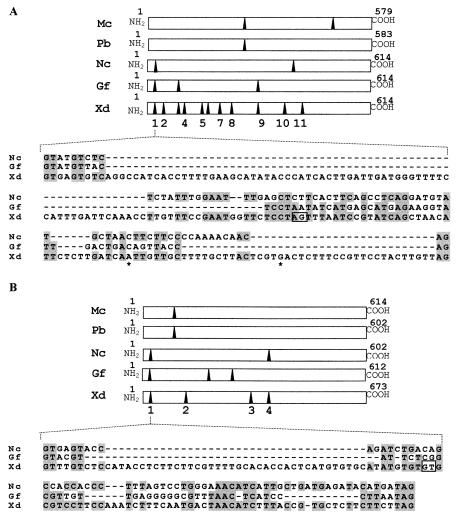
Analysis of the intron positions in phytoene desaturase- and phytoene synthase-lycopene cyclase-encoding genes from fungi indicates that the first intron of these genes is located in the same relative sequence position in basidiomycetes (X. dendrorhous) and ascomycetes (Neurospora crassa and Gibberella fujikuroi) (Fig. 6). The first intron in both genes could have been acquired by the ancestor of the basidiomycetes and ascomycetes after separation of the zygomycetes, since the fungi Mucor circinelloides and Phycomyces blakeesleanus lack these introns. Additionally, a nucleotide sequence comparison of the first intron showed that the alternative splicing sites of the crtI and crtYB genes of X. dendrorhous are absent in N. crassa and G. fujikuroi (Fig. 6). Therefore, phytoene desaturase and phytoene synthase-lycopene cyclase RNAs from ascomycetes would not follow an alternative splicing pathway, as described in this study. The nucleotide sequence of the first crtI intron from strain UCD 67-385 has two mutations, C→A and T→G, compared to the nucleotide sequence of strain CBS 6938 (Fig. 6A). There are also nucleotide changes in a genomic DNA clone derived from strain UCD 67-385 (21). These mutations could be responsible for the splicing in the alternative 3′ site and the high proportion of clones with alternative cDNA compared to clones with proper cDNA (16 and 12 clones, respectively). It remains to be determined whether strain CBS 6938 synthesizes a crtI amRNA and, if it does, what the proportion of alternative and productive mRNAs is.
FIG. 6.
Phytoene desaturase protein (A) and phytoene synthase-lycopene cyclase protein (B) from the fungi M. circinelloides (Mc), P. blakesleeanus (Pb), X. dendrorhous (Xd), N. crassa (Nc), and G. fujikuroi (Gf). The arrowheads indicate the relative positions of introns in the nucleotide sequences. In each panel alignment of the first intron for both genes is shown. Identical bases in X. dendrorhous and/or N. crassa and G. fujikuroi are shaded. The alternative splice signals inside the introns are enclosed in boxes. The asterisks indicate the C→A and T→G mutations in the X. dendrorhous UCD 67-385 sequence compared with the previously published sequence of strain CBS 6938 (accession number Y15007).
Analysis of expression of the crtI and crtYB genes indicated that crtI mRNA and crtYB mRNA levels decreased during the stationary phase. In addition, the level of the crtI mRNA spliced in a proper form in relation to the amRNA decreased as a function of the age of the culture. It could be hypothesized that splicing of the crtI RNA to an unproductive form could be a way to descrease the concentration of the phytoene desaturase protein even though carotenoid synthesis increases after the late exponential phase (Fig. 5). In the nematode C. elegans splicing of the RNA encoding the ribosomal protein RPL-12 is a regulated process for productive or unproductive transcripts (25). Therefore, protein RPL-12 appears to autoregulate its own rpl-12 RNA splicing to an unproductively spliced mRNA depending on the cellular concentration of the protein. Recently, two cytochrome P450 monooxygenease genes (pc-1 and pc-2) and two splice variants of pc-1 in the basidiomycetous fungus Phanerochaete chrysosporium have been cloned and sequenced (44). Translation of one of the splice products indicated that there was a frameshift and that stop codons were present. The physiological significance of these splice variants in P. chrysosporium is unknown. In X. dendrorhous, the M/A ratios for both genes could vary depending on environmental or physiological conditions, such as the age of the culture, as found with the crtI gene. It remains to be established whether production of unproductive transcripts is a regulated process and, if it is, what the effect on carotenoid biosynthesis is.
Acknowledgments
This work was supported by grant DID ENL-2000/17 from the University of Chile. Deutscher Akademischer Austanschdienst provided a graduate scholarship to P. Lodato.
We thank Carlos Medina for discussions concerning the structure and origin of the first intron of the crtI and crtYB genes, Carlos Jerez for critical reading of the manuscript, and Antonio Jiménez for providing primers.
REFERENCES
- 1.Alcaíno, J. 2002. Engineer in Biotechnology thesis. University of Chile, Santiago.
- 2.An, G.-H., D. B. Schuman, and E. Johnson. 1989. Isolation of Phaffia rhodozyma mutants with increased astaxanthin content. Appl. Environ. Microbiol. 55:116-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrewes, A. G., H. J. Phaff, and M. P. Starr. 1976. Carotenoids of Phaffia rhodozyma, a red-pigmented fermenting yeast. Phytochemistry 15:1003-1007. [Google Scholar]
- 4.Armstrong, G. A. 1994. Eubacteria show their true colors: genetics of carotenoid pigment biosynthesis from microbes to plants. J. Bacteriol. 176:4795-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armstrong, G. A. 1997. Genetics of eubacterial carotenoid biosynthesis: a colorful tale. Annu. Rev. Microbiol. 51:628-659. [DOI] [PubMed] [Google Scholar]
- 6.Arrach, N., R. Fernández-Martín, E. Cerdá-Olmedo, and J. Avalos. 2001. A single gene for lycopene cyclase, phytoene synthase, and regulation of carotene biosynthesis in Phycomyces. Proc. Natl. Acad. Sci. USA 98:1687-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arrach, N., T. J. Schmidhauser, and J. Avalos. 2002. Mutants of the carotene cyclase domain of al-2 from Neurospora crassa. Mol. Genet. Genomics 266:914-921. [DOI] [PubMed] [Google Scholar]
- 8.Birch, P. R. J., P. F. G. Sims, and P. Broda. 1995. Substrate-dependent differential splicing of introns in the regions encoding the cellulose binding domains of two exocellobiohydrolase I-like genes in Phanerochaete chrysosporium. Appl. Environ. Microbiol. 61:3741-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boel, E., I. Hjort, B. Svensson, F. Norris, K. E. Norris, and N. P. Fiil. 1984. Glucoamylases G1 and G2 from Aspergillus niger are synthesized from two different but closely related mRNAs. EMBO J. 3:1097-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyle, J. S., and A. M. Lew. 1995. An inexpensive alternative to glassmilk for DNA purification. Trends Genet. 11:8. [DOI] [PubMed]
- 11.Canfield, L. M., J. W. Forage, and J. G. Valenzuela. 1992. Carotenoids as cellular antioxidants. Proc. Soc. Exp. Biol. Med. 200:260-265. [DOI] [PubMed] [Google Scholar]
- 12.Carattoli, A., N. Romano, P. Ballario, G. Morelli, and G. Macino. 1991. The Neurospora crassa carotenoid biosynthetic gene (Albino 3) reveals highly conserved regions among prenyltransferases. J. Biol. Chem. 266:5854-5859. [PubMed] [Google Scholar]
- 13.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 14.Cifuentes, V., G. Hermosilla, C. Martínez, R. León, G. Pincheira, and A. Jiménez. 1997. Genetics and electrophoretic karyotyping of wild-type and astaxanthin mutant strains of Phaffia rhodozyma. Antonie Leeuwenhoek 72:111-117. [DOI] [PubMed] [Google Scholar]
- 15.Davis, C. A., L. Grate, M. Spingola, and M. Ares, Jr. 2000. Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res. 28:1700-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Habara, Y., S. Urushiyama, T. Tani, and Y. Ohshima. 1998. The fission yeast prp10+ gene involved in pre-mRNA splicing encodes a homologue of highly conserved splicing factor, SAP155. Nucleic Acids Res. 26:5662-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirschberg, J. 1998. Molecular biology of carotenoid biosynthesis, p. 149-191. In G. Britton, S. Liaaen-Jensen, and H. Pfander (ed.), Carotenoids: biosynthesis and metabolism. Birkhäuser Verlag, Basel, Switzerland.
- 19.Johnson, E., D. E. Conklin, and M. J. Lewis. 1977. The yeast Phaffia rhodozyma as dietary pigment source for salmonids and crustaceans. J. Fish. Res. Board Can. 34:2417-2421. [Google Scholar]
- 20.Johnson, E., and M. Lewis. 1979. Astaxanthin formation by the yeast Phaffia rhodozyma. J. Gen. Microbiol. 115:173-183. [Google Scholar]
- 21.León, R. 2000. Ph.D. thesis. University of Chile, Santiago.
- 22.Lopez, J. A. 1998. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 32:279-305. [DOI] [PubMed] [Google Scholar]
- 23.Martínez, C. 1995. Ph.D. thesis. University of Chile, Santiago.
- 24.Miller, M. W., M. Yoneyama, and M. Soneda. 1976. Phaffia, a new yeast genus in the Deuteromycotina (Blastomycetes). Int. J. Syst. Bacteriol. 26:286-291. [Google Scholar]
- 25.Mitrovich, Q. M., and P. Anderson. 2000. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev. 14:2173-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Modrek, B., A. Resch, C. Grasso, and C. Lee. 2001. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 29:2850-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Navarro, E., V. L. Ruiz-Pérez, and S. Torres-Martínez. 2000. Overexpression of the crgA gene abolishes light requirement for carotenoid biosynthesis in Mucor circinelloides. Eur. J. Biochem. 267:800-807. [DOI] [PubMed] [Google Scholar]
- 28.Nigoyi, K. N., O. Björkman, and A. R. Grossman. 1997. The roles of specific xanthophylls in photoprotection. Proc. Natl. Acad. Sci. USA 94:14162-14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palozza, P., and N. I. Krinsky. 1992. Astaxanthin and cantaxanthin are potent antioxidants in a membrane model. Arch. Biochem. Biophys. 297:291-295. [DOI] [PubMed] [Google Scholar]
- 30.Retamales, P., G. Hermosilla, R. León, C. Martínez, A. Jiménez, and V. Cifuentes. 2002. Development of the sexual reproductive cycle of Xanthophyllomyces dendrorhous. J. Microbiol. Methods 48:87-93. [DOI] [PubMed] [Google Scholar]
- 31.Ruiz-Hidalgo, M. J., E. P. Benito, G. Sandmann, and A. P. Eslava. 1997. The phytoene dehydrogenase gene of Phycomyces: regulation of its expression by blue light and vitamin A. Mol. Gen. Genet. 253:734-744. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Schmidhauser, T. J., F. R. Lauter, V. E. A. Russo, and C. Yanofsky. 1990. Cloning, sequence and photoregulation of al-1, a carotenoid biosynthetic gene of Neurospora crassa. Mol. Cell. Biol. 10:5064-5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidhauser, T. J., F. R. Lauter, M. Schumacher, W. Zhou, V. E. A. Russo, and C. Yanofsky. 2066. 1994. Characterization of al-2, the phytoene synthase gene of Neurospora crassa. J. Biol. Chem. 269:12060-12061. [PubMed] [Google Scholar]
- 35.Spike, C. A., J. E. Shaw, and R. K. Herman. 2001. Analysis of smu-1, a gene that regulates the alternative splicing of unc-52 pre-mRNA in Caenorhabditis elegans. Mol. Cell. Biol. 21:4985-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompsom, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thornewell, S. J., R. B. Peery, and P. L. Skatrud. 1997. Cloning and characterization of CneMDR1: a Cryptococcus neoformans gene encoding a protein related to multidrug resistance proteins. Gene 201:21-29. [DOI] [PubMed] [Google Scholar]
- 38.Torrissen, O. J., R. W. Hardy, and K. D. Shearer. 1989. Pigmentation of salmonids carotenoid deposition and metabolism. Aquat. Sci. 1:209-225. [Google Scholar]
- 39.Velayos, A., A. P. Eslava, and E. A. Iturriaga. 2000. A bifunctional enzyme with lycopene cyclase and phytoene synthase activities is encoded by the carRP gene of Mucor circinelloides. Eur. J. Biochem. 267:5509-5519. [DOI] [PubMed] [Google Scholar]
- 40.Verdoes, J. C., N. Misawa, and A. J. J. van Ooyen. 1999. Cloning and characterization of the astaxanthin biosynthetic gene encoding phytoene desaturase of Xanthophyllomyces dendrorhous. Biotechnol. Bioeng. 63:750-755. [DOI] [PubMed] [Google Scholar]
- 41.Verdoes, J. C., P. Krubasik, G. Sandmann, and A. J. J. van Ooyen. 1999. Isolation and functional characterisation of a novel type of carotenoid biosynthetic gene from Xanthophyllomyces dendrorhous. Mol. Gen. Genet. 262:453-461. [DOI] [PubMed] [Google Scholar]
- 42.Vittorioso, P., A. Carattoli, P. Londei, and G. Macino. 1994. Internal translational initiation in the mRNA from the Neurospora crassa albino-3 gene. J. Biol. Chem. 269:26650-26654. [PubMed] [Google Scholar]
- 43.Wery, J., M. J. M. Dalderup, J. ter Linde, and A. J. J. van Ooyen. 1996. Structural and phylogenetic analysis of the actin gene from the yeast Phaffia rhodozyma. Yeast 12:641-651. [DOI] [PubMed] [Google Scholar]
- 44.Yadav, J. S., M. B. Soellner, J. C. Loper, and P. K. Mishra. 2003. Tandem cytochrome P450 monooxygenase genes and splice variants in the white rot fungus Phanerochaete chrysosporium: cloning, sequence analysis, and regulation of differential expression. Fungal Genet. Biol. 38:10-21. [DOI] [PubMed] [Google Scholar]
- 45.Ye, D., C.-H. Lee, and S. F. Queener. 2001. Differential splicing of Pneumocystis carinii f. sp. carinii inosine 5′-monophosphate dehydrogenase pre-mRNA. Gene 263:151-158. [DOI] [PubMed] [Google Scholar]