Abstract
Pozol is an acid beverage obtained from the natural fermentation of nixtamal (heat- and alkali-treated maize) dough. The concentration of mono- and disaccharides from maize is reduced during nixtamalization, so that starch is the main carbohydrate available for lactic acid fermentation. In order to provide some basis to understand the role of amylolytic lactic acid bacteria (ALAB) in this fermented food, their diversity and physiological characteristics were determined. Forty amylolytic strains were characterized by phenotypic and molecular taxonomic methods. Four different biotypes were distinguished via ribotyping; Streptococcus bovis strains were found to be predominant. Streptococcus macedonicus, Lactococcus lactis, and Enterococcus sulfureus strains were also identified. S. bovis strain 25124 showed extremely low amylase yield relative to biomass (139 U g [cell dry weight]−1) and specific rate of amylase production (130.7 U g [cell dry weight]−1 h−1). In contrast, it showed a high specific growth rate (0.94 h−1) and an efficient energy conversion yield to bacterial cell biomass (0.31 g of biomass g of substrate−1). These would confer on the strain a competitive advantage and are the possible reasons for its dominance. Transient accumulation of maltooligosaccharides during fermentation could presumably serve as energy sources for nonamylolytic species in pozol fermentation. This would explain the observed diversity and the dominance of nonamylolytic lactic acid bacteria at the end of fermentation. These results are the first step to understanding the importance of ALAB during pozol fermentation.
Amylolytic lactic acid bacteria (ALAB) have been reported from different tropical amylaceous fermented foods, prepared mainly from cassava and cereals (e.g., maize and sorghum). Amylolytic strains of Lactobacillus plantarum have been isolated from African cassava-based fermented products (26), and the new ALAB species Lactobacillus manihotivorans (23) was isolated from cassava sour starch fermentations carried out in Colombia. ALAB have also been isolated from cereal-based fermented foods. Olympia et al. (27) characterized amylolytic strains of L. plantarum isolated from burong isda, a fermented food made from fish and rice in Philippines. Amylolytic strains of Lactobacillus fermentum were isolated for the first time from Benin maize sourdough (ogi and mawè) by Agati et al. (1). More recently, Sanni et al. (30) described amylolytic strains of L. plantarum and L. fermentum strains in various Nigerian traditional amylaceous fermented foods.
The search for ALAB in fermented amylaceous foods has been justified by the high starch content of the raw material. Their role has yet to be elucidated since mono- and disaccharides, such as glucose and sucrose, which occur naturally in cereals and cassava, are readily available for lactic acid fermentation. The way the raw material is processed may determine the composition of the microbiota and, in particular, the occurrence of ALAB (14). For instance, during cassava processing for sour starch production, starch washing by sieving under running water eliminates most of mono- and disaccharides leaving starch as the main substrate available (21), thus allowing ALAB (L. manihotivorans) to predominate (6).
In addition, pozol may offer another interesting example of the influence of processing conditions on microbial diversity. Pozol is a popular maize sourdough produced in urban and rural areas in Southeastern Mexico (34). Maize processing for pozol production involves a first step known as nixtamalization, which corresponds to the boiling of kernels in a lime suspension for ca. 1.5 h. The grains are then dehulled and washed. This water (called nexayote) is eliminated, the maize kernels (nixtamal) are ground, and the resulting dough is shaped into balls, wrapped in banana leaves, and left to ferment for several days. The main soluble sugar of maize is sucrose, which is present at a concentration of 2 g 100 g of the whole kernel−1 on a dry weight basis (5). It is reduced to 0.1 to 0.7 g 100 g of dry dough−1, after alkaline cooking, soaking, and washing to produce nixtamal (31). Thus, the question is raised as to how a low free sugar content might determine such rich microbial diversity and support a high number of lactic acid bacteria in pozol as reported by Wacher et al. (34) and Escalante et al. (12).
Research on the physiology of nondairy lactic acid bacteria and, in particular, members of the amylolytic group is necessary in order to determine their ecological significance in natural fermentations of amylaceous substrates, to develop new processes, and to improve existing techniques on a more rational basis by using specific starters.
The purpose of the present study was to determine the diversity of ALAB during pozol fermentation and their main physiological characteristics in order to understand their role in this food ecosystem.
MATERIALS AND METHODS
Sample description.
Two freshly ground nixtamal dough samples (A and B) were purchased at Villahermosa market (Tabasco, Mexico). Each sample was divided into 100-g portions, wrapped in banana leaves, and incubated at 30°C for 7 days.
Enumeration of lactic acid bacteria and ALAB.
First, 25 g dough was added to 225 ml of 0.1% peptone water; all further 10-fold dilutions were prepared in this diluent. Lactic acid bacteria and ALAB were enumerated on duplicate, surface-inoculated plates of MRS agar (CM361; Oxoid, Ltd., Hampshire, England) and MRS agar with 1.5% soluble starch (J. T. Baker, Mexico City, Mexico) instead of 2% glucose, respectively. Water contents of pozol doughs were measured by determining loss in weight after being dried at 80°C for 24 h, and counts were expressed as the log CFU g of dry dough−1. Randomly selected colonies were checked by conducting a Gram stain and catalase reaction. For ALAB, colonies with clear hydrolysis zones after being flooded with Gram iodine solution were considered.
Isolation of ALAB.
ALAB were isolated from MRS-starch replica plates by picking at random colonies with different diameters of hydrolysis zones. The strains recovered were purified further by streaking them onto MRS-starch medium. They were tested for Gram stain and catalase reaction. Pure cultures were stored at −70°C on glass beads with 20% glycerol as cryoprotectant (13).
Determination of the metabolic pathway.
Cells were cultured for 18 h in 10 ml of MRS broth (0881-17-5; Difco Laboratories, Detroit, Mich.). Cultures were centrifuged (10 min at 15,300 × g and 4°C), and the supernatants were filtered through a 0.22-μm-pore-size membrane filter (Millipore). The filtrates were analyzed on a Waters (Milford, Mass.) 515 liquid chromatograph equipped with a hydrogen loaded ion exchange column (Bio-Rad Aminex HPX-87H, 300 by 7.8 mm). The mobile phase was 6 mM H2SO4 with a flow rate of 0.8 ml min−1 at 65°C. A Waters 2410 refractive index detector was used. Noninoculated MRS broth served as a control. Homofermentative or heterofermentative pathways were established by determination of end-fermentation products, i.e., lactate or an equimolar mixture of lactate and ethanol-acetate, respectively.
Biochemical identification.
Carbohydrate fermentation profiles were done on API 50CH (bioMérieux, Marcy l'Étoile, France) according to the manufacturer's instructions.
Ribotyping.
Overnight cultures were diluted in fresh MRS broth and incubated for 4 h at 30°C. The cells were harvested by centrifugation (10 min, 4,480 × g, 4°C) and washed twice in sterile TS buffer (50 mM Tris-HCl, pH 7.5; 25% [wt/vol] sucrose). The cells were then suspended in TS buffer to obtain an absorbance value of 3 at 600 nm. The cell suspension was treated with lysozyme (100 mg ml−1) for 1 h at 37°C. Agarose blocks containing the extracted DNA were prepared and digested with EcoRI endonuclease (Eurogentec, Seraing, Belgium) according to the method of McClelland et al. (20). Standard electrophoresis of the DNA samples was performed in a large electrophoresis cell (30 by 20 cm) with 1% (wt/vol) agarose gel with TAE buffer (40 mM Tris, 40 mM acetate, 2 mM EDTA [pH 8]) at 4 V cm−1 and 10°C for 15 h. DNA was transferred to nylon membranes (Hybond-N+; Amersham, Buckinghamshire, United Kingdom) by the alkaline method according to the instructions provided by the manufacturer. The probe used for hybridization was a combination of PCR-amplified 16S rRNA gene fragments of each of the 15136, 15125, and 25421 isolates. Primers 27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 354r (5′-CTGCTGCSYCCCGTAG-3′) were used (17). The amplification conditions were as follows: initial denaturation at 94°C for 11 min and 30 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 50°C, and elongation at 72°C for 30 s. A final extension was carried out at 72°C for 7 min. Amplification products were cleaned (Wizard DNA Clean-Up System; Promega). The amplified product was then labeled with digoxigenin (DIG)-dUTP by using the DIG labeling kit supplied by Roche (Meylan, France). Hybridization and detection experiments were performed with a DIG detection kit (Roche, Meylan, France), according to the instructions provided by the manufacturer. Hybridization band profiles were evaluated for the presence or absence of common unique bands, and a matrix was constructed. It was analyzed by using the TREECON v1.2 program (33). The analysis includes calculation of similarity distance by using the procedure of Link et al. (19), construction of the tree topology by using UPGMA (unweighted pair-group method with arithmetic averages), and generation of the phenogram.
Determination of 16S rRNA gene sequences.
A fragment of the 16S rRNA gene (corresponding to positions 30 to 1521 of the Escherichia coli 16S rRNA gene) was amplified by PCR with conserved primers close to the 3′ and 5′ ends of the gene. The PCR products were directly sequenced (a single strand) by using a Taq dye-deoxy terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.) and an automatic DNA sequencer (model 373; Applied Biosystems). Identifications were based on a partial sequence (corresponding to positions 30 to 338 of the E. coli 16S rRNA gene) or a nearly full sequence (corresponding to positions 30 to 1521 of the E. coli 16S rRNA gene) comparisons. The closest known relatives were determined by performing database searches at GenBank by using the BLAST program.
Starch fermentation by S. bovis strain 25124: growth conditions.
Cultures were grown in 1-liter flasks with 800 ml of MRS medium (9) with soluble potato starch (20 g liter−1; Prolabo-Merck Eurolab) instead of glucose, inoculated with 25 ml of 12 h precultures grown in MRS-starch medium at 30°C. Fermentation broth was sampled every 30 min. The fermentation was performed in triplicate.
Biomass estimation and growth parameters.
A calibration curve between the optical density at 600 nm (OD600) and the cell dry weight was established for the strain. The biomass concentration was determined by measuring the OD600 with Spectronic Genesys 5 spectrophotometer (Milton Roy) and related to dry weight measured after two washing and centrifugation cycles, followed by a drying step at 80°C for 24 h. The maximum specific growth rate was estimated during exponential growth phase by linear regression of the ln(OD600) versus time. Product and growth yields relative to substrate (Ylac/s and Yx/s, respectively) were calculated as the slope of the linear regressions of either lactic acid or biomass versus residual substrate. Lactic acid and amylase yields from biomass (Ylac/x and Yamy/x, respectively) were calculated in a similar manner by plotting the product versus the biomass. All calculations were made with data obtained during the corresponding exponential growth phase. Specific rates of lactic acid or amylase production (qlac and qamy, respectively) and substrate consumption (qs) were calculated as indicated by Pirt (29): qp = Yp/x · μ (“p” for product, i.e., lactic acid or amylase) and qs = μ/Yx/s.
Analytical methods.
Cells were removed by filtering the medium through a membrane filter (0.22-μm pore size; Millipore). To quantify lactic acid, filtrates were analyzed on a Perkin-Elmer 250 liquid chromatograph equipped with a hydrogen loaded ion-exchange column (Bio-Rad Aminex HPX-87H, 300 by 7.8 mm) and a Perkin-Elmer 30 refractive index detector (Perkin-Elmer, Norwalk, Conn.). The mobile phase was 0.01 N H2SO4 with a flow rate of 0.6 ml min−1 at 50°C. l-Lactate (L-2250; Sigma) was used as standard. Hydrolysis products from starch fermentation were examined by high-performance liquid chromatography by using a Prodigy 5 ODS 2 C18 column (250 by 4.6 mm; Phenomenex) with the refractive index detector. The products were eluted with water at a flow rate of 0.8 ml min−1 and a column temperature of 35°C. G2 (maltose) to G7 (maltoheptaose) were used as standards (Sigma). Total and reducing sugars were determined by the methods of Dubois et al. (11) and Miller (22), respectively. The residual starch content was determined by measuring the iodine-starch complex color (24). Glucose and maltose were determined by using a commercial enzymatic test (product 1113950; Boehringer, Mannheim, Germany).
Amylase activity.
To detect cell-bound amylase activity, the cultures (10 ml) were harvested by centrifugation (15 min at 27,200 × g and 4°C); the cell pellets were washed and suspended in 0.1 M phosphate buffer (pH 6.8). Cell-bound amylase activity was assayed at pH 6.8 and 37°C by measuring the iodine-complexing ability of starch as described by Agati et al. (1). One enzyme unit was defined as the amount of enzyme hydrolyzing 10 mg of starch in 30 min.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the partial 16S rRNA gene sequences of strains 25124, 15430, A12203, A45201, A57206, A57103, A56203, A45212, and A36202 reported in the present study are AY184231 to AY184239.
RESULTS
Enumeration and isolation of ALAB.
The initial concentration of lactic acid bacteria was 4.9 log CFU g of dry dough−1 (Table 1). This concentration increased to 9.9 log CFU g of dry dough−1 after 24 h and was kept approximately constant for 72 h of fermentation. The initial number of ALAB was high (4.5 log CFU g of dry dough−1). This level increased during the first 24 h of fermentation to 8.4 log CFU g of dry dough−1 and remained constant until 72 h (8.7 log CFU g of dry dough−1). The pH value decreased from 7.4 to 4.8 in 24 h and to 4.4 in 72 h (Table 1).
TABLE 1.
Concentrations of lactic acid bacteria and ALAB, and pH values during pozol fermentation at 30°Ca
| Fermentation time (h) | pH | Concn (log CFU g of dry weight−1) of:
|
|
|---|---|---|---|
| ALAB | Lactic acid bacteria | ||
| 0 | 7.4 | 4.5 | 4.9 |
| 6 | 5.9 | 7.6 | 8.2 |
| 24 | 4.8 | 8.4 | 9.9 |
| 48 | 4.8 | 8.7 | 10.4 |
| 72 | 4.4 | 8.7 | 10.2 |
Values are the means of two independent fermentations (samples A and B). pH values varied from 0.2 to 1.6 U, and log CFU g of dry weight−1 values varied from 0.1 to 0.6 U.
A total of 257 strains of ALAB were isolated from both samples at different fermentation times. Rod and coccoid morphology were observed at all times. Strains isolated from sample B showed larger hydrolysis diameters (2 to 18 mm) on MRS-starch medium than those from sample A (1 to 9 mm). A starch hydrolysis zone of 7 mm in diameter was evident in a large number of strains (Fig. 1). The largest starch hydrolysis zones (9 to 18 mm) were observed in strains isolated at 24 h in sample B. Similar results were obtained for sample A.
FIG. 1.
Distribution of hydrolysis diameters in MRS-starch medium of ALAB isolated from pozol (sample B) at different fermentation times.
The 40 most amylolytic strains (those with starch hydrolysis diameters larger than 9 mm) were further characterized by using a combination of phenotypic and molecular taxonomic approaches. All strains were homofermentative, since they produced only lactic acid from glucose.
Evaluation of diversity of ALAB by ribotyping and 16S rRNA gene sequence analysis.
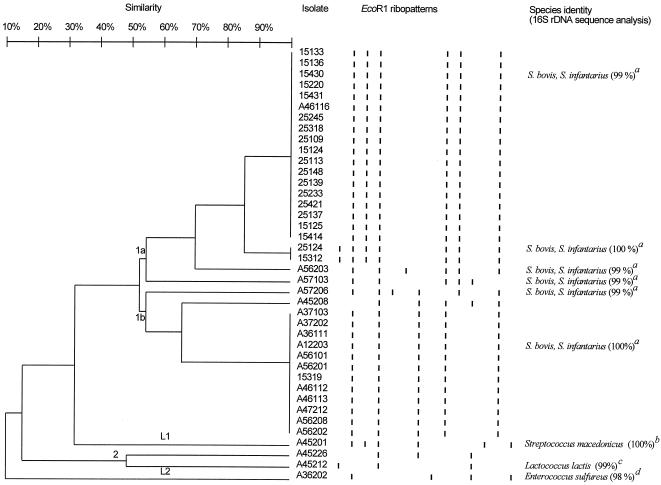
Two clusters and two lines (lines are one-strain clusters) were distinguished by ribotyping (Fig. 2). Cluster 1a included 22 isolates (19 from sample B and 3 from sample A) that merged at a similarity level of 54%. Cluster 1b included 1 isolate from sample B and 13 isolates from sample A that merged at 54% similarity. A line (L1) included one isolate from sample A that merged at 32% similarity. Two isolates from sample A merged at a similarity level of 49% (cluster 2), and a line (L2) with an isolate from sample A merged at a similarity level of 10%.
FIG. 2.
Dendrogram of the 40 most amylolytic of the lactic acid bacteria isolated from pozol (samples A [strains marked with an “A”] and B [strains with no letter in the numbering system]) based on EcoRI ribotyping. Schematic ribopatterns are shown. A similarity analysis was performed by using the procedure of Link et al. (19) and clustering by UPGMA. Identifications of “a,” “b,” and “c” were based on a partial sequence (corresponding to positions 30 to 338 of the Escherichia coli 16S rRNA gene), and identifications of “d” were based on a nearly full sequence (corresponding to positions 30 to 1521 of the E. coli 16S rRNA gene) 16S rRNA gene sequence comparisons. The percent similarity values of the 16S rRNA gene sequence comparison identifications are shown in parentheses.
16S rRNA genes of strains from the main clusters were partially sequenced (positions 30 to 338 of the E. coli 16S rRNA gene). Strains 15430, 25124, A56203, and A57103 from group 1a and strains A57206 and A12203 from group 1b were tentatively identified as S. bovis, since they showed similarity coefficients of between 99 and 100% with this species, but was also close to Streptococcus infantarius subsp. coli. Strain A45201 (a line between clusters lb and 2) showed 100% similarity with S. macedonicus, whereas strain A45212 (group 2) showed 99% similarity with L. lactis. Strain A36202 full sequence (positions 30 to 1521 of the E. coli 16S rRNA gene) showed 98% similarity to E. sulfureus (Fig. 2), but its identity would have to be confirmed, since many different enterococcal species display <1% sequence divergence (10).
Biochemical identification of ALAB.
From API 50CH strips, all strains fermented d-glucose, d-fructose, d-mannose, N-acetylglucosamine, esculin, salicin, maltose, sucrose, glycogen, and starch. Sugars with variable fermentation profiles are presented in Table 2. None of the other sugars tested were fermented. Most strains from ribotyping clusters 1a and 1b shared the same biochemical profile, with the exception of trehalose fermentation, which was negative for most strains from cluster 1a and positive for most strains from cluster 1b. Strains from both lines and cluster 2 showed variations in α-methyl-d-glucoside, amygdalin, arbutin, cellobiose, d-turanose, melibiose, glycerol, β-gentobiose, galactose, and d-raffinose compared to clusters 1a and 1b.
TABLE 2.
Carbohydrate fermentation profiles of ALAB from pozol (samples A and B)
| Strain | Fermentationa of:
|
Cluster no.b | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GLY | RIB | GAL | MDG | NAG | AMY | ARB | CEL | MEL | TRE | RAF | GEN | TUR | ||
| 15133 | − | − | + | − | + | − | − | − | + | − | + | − | − | 1a |
| 15136 | − | − | + | − | + | − | − | − | + | − | + | − | − | 1a |
| 15430 | − | − | + | − | + | − | − | − | + | − | + | − | − | 1a |
| 15220 | − | − | + | − | + | − | + | − | + | + | + | − | + | 1a |
| 15431 | − | − | + | − | + | − | − | + | + | − | + | − | − | 1a |
| A46116 | − | − | + | − | + | − | − | − | + | + | + | − | − | 1a |
| 25245 | − | − | + | − | + | − | − | − | + | − | + | − | − | 1a |
| 25318 | − | − | + | − | + | − | − | + | + | − | + | − | − | 1a |
| 25109 | − | − | + | − | + | − | − | − | + | − | + | − | − | 1a |
| 15124 | − | − | + | − | + | − | − | − | + | − | + | − | − | 1a |
| 25113 | − | − | + | − | + | − | − | − | + | − | + | − | − | 1a |
| 25148 | − | − | + | − | + | − | − | − | + | − | + | − | − | 1a |
| 25139 | − | − | + | − | + | − | − | − | + | − | + | − | − | 1a |
| 25233 | − | − | + | − | + | − | − | − | + | − | + | − | − | 1a |
| 25421 | − | − | + | − | + | − | − | − | + | − | + | − | − | 1a |
| 25137 | − | − | + | − | + | − | − | − | + | − | + | − | − | 1a |
| 15125 | − | − | + | − | + | − | − | − | + | − | + | − | − | 1a |
| 15414 | − | − | + | − | + | − | − | + | + | − | + | − | − | 1a |
| 25124 | − | − | + | − | + | − | − | − | + | − | + | − | − | 1a |
| 15312 | − | − | + | − | + | − | − | + | + | − | + | − | − | 1a |
| A56203 | + | − | − | − | + | − | + | + | + | − | + | + | − | 1a |
| A57103 | − | − | + | − | + | + | + | + | + | − | + | + | − | 1a |
| A57206 | − | − | + | − | + | − | − | − | + | − | + | − | − | 1b |
| A45208 | − | − | + | − | − | − | − | − | + | + | + | − | − | 1b |
| A37103 | − | − | + | − | + | − | − | − | + | + | + | − | − | 1b |
| A37202 | − | − | + | − | + | − | − | − | + | + | + | − | − | 1b |
| A36111 | − | − | + | − | + | − | − | − | + | + | + | − | − | 1b |
| A12203 | − | − | + | − | + | − | + | + | + | − | + | + | − | 1b |
| A56101 | − | − | + | + | + | + | − | − | + | + | + | − | − | 1b |
| A56201 | − | − | + | + | + | − | − | − | + | + | + | − | − | 1b |
| 15319 | − | − | + | − | + | − | − | + | + | − | + | − | − | 1b |
| A46112 | − | + | + | − | + | + | + | + | + | + | + | + | − | 1b |
| A46113 | − | − | − | − | + | − | − | − | + | − | + | − | − | 1b |
| A47212 | − | − | + | − | + | − | − | − | + | + | + | − | − | 1b |
| A56208 | − | − | + | − | + | − | − | − | + | + | + | − | − | 1b |
| A56202 | − | − | + | + | + | − | − | − | + | + | + | + | − | 1b |
| A45201 | − | − | + | + | + | + | + | + | + | + | + | + | − | L1 |
| A45226 | − | − | + | + | + | − | − | + | + | + | + | − | + | 2 |
| A45212 | − | − | + | + | + | − | − | − | − | + | + | − | + | 2 |
| A36202 | + | − | − | − | + | + | + | + | − | + | − | + | − | L2 |
+, Fermentation; −, no fermentation; Abbreviations: GLY, glycerol; RIB, ribose; GAL, galactose; MDG, α-methyl-d-glucoside; NAG, N-acetyl glucosamine; AMY, amygdalin; ARB, arbutin; CEL, cellobiose; MEL, melibiose; TRE, trehalose; RAF, d-raffinose; GEN, β-gentobiose; TUR, d-turanose.
Cluster numbers correspond to those obtained by ribotyping (Fig. 2).
Starch fermentation by S. bovis 25124.
Starch fermentation by a strain from the dominant species, S. bovis, was studied in MRS-starch medium (Fig. 3). The initial medium contained small amounts (<0.5 g liter−1) of glucose, maltose, and maltooligosaccharides (maltotriose [G3], maltopentaose [G5], maltohexaose [G6], and maltoheptaose [G7]) and 0.82 g of maltotetraose (G4) liter−1. After a lag phase of 1.5 h (Fig. 3A), biomass and lactic acid concentrations increased at specific rates of 0.94 h−1 and 2.4 g of lactic acid g of cell dry weight−1 h−1, respectively. Amylase activity, found to be only associated with cells (unpublished data), increased at a maximum specific rate of 130.7 U g of cell dry weight−1 h−1 (Table 3), whereas the pH decreased to 4.9 (Fig. 3A). Ylac/s and Yx/s were, respectively, 0.31 and 0.78 (g of lactic acid or biomass g of substrate−1), and product yields relative to biomass Ylac/x and Yamy/x were 2.56 g of lactic acid g of cell dry weight−1 and 139 U g of cell dry weight−1, respectively (Table 3). The concentrations of starch and total sugars were reduced to 9.7 and 14.8 g liter−1, respectively. With strain 25124, the low initial concentration of reducing sugars decreased from 0.25 to 0.05 g liter−1 (Fig. 3B). As for maltooligosaccharides, the G3 and G7 concentrations were reduced after 2 to 2.5 h fermentation, and the G5 and G6 concentrations were reduced after 4 h. Maltotetraose (G4) was the main intermediary maltooligosaccharide produced during starch hydrolysis (Fig. 3C); the peak concentration was noted between 4 and 5 h and corresponded approximately to the difference between the total sugars and starch. Glucose was only detected during the first 1.5 h, and the maltose concentration was variable. Maltooligosaccharides concentrations decreased to less than 0.2 g liter−1 at 8.5 h. After 10.5 h of fermentation, the strain had fermented only 26.6% total sugars and hydrolyzed 50% of the initial starch concentration. These values were constant after 26 h of fermentation.
FIG. 3.
Fermentation of starch by S. bovis strain 25124 at 30°C without pH control. (A) Biomass (◊), lactate (▵), amylase activity (□), and pH values (○); (B) total sugars (□), reducing sugars (▵), and starch (⋄); (C) glucose (⧫), maltose (▪), G3 (▴), G4 (○), G5 (∗), G6 (•), and G7 (+). The results are the means of three independent replicates. Variation coefficients were lower than 10% except for biomass (0.5 to 44%), lactate (0.2 to 25%), and maltose (5.3 to 47%).
TABLE 3.
Yields and metabolic quotients for S. bovis 25124, L. manihotivorans 18010T, L. plantarum A6, and L. fermentum OgiE1
| Parametera | Valueb obtained for:
|
|||
|---|---|---|---|---|
| S. bovis 25124c | L. manihotivorans 18010Td | L. plantarum A6d | L. fermentum OgiE1e | |
| Total sugar consumed (%) | 25.5 | 45 | ND | 78 |
| Yx/s (g g−1) | 0.31 | 0.09 (0.15) | ND (0.18) | 0.1 |
| Ylac/s (g g−1) | 0.78 | 0.71 (0.67) | ND (0.84) | 0.33 |
| Ylac/x (g g−1) | 2.56 | 7.89 (4.47) | ND (4.7) | 3.3 |
| Yamy/x (U g−1) | 139 | 2,400 (4,900) | 2,300 (5,700) | 5,400 |
| μ (h−1) | 0.94 | ND (0.36) | 0.43 (0.41) | 0.35 |
| qlac (g g of cell dry wt−1 h−1) | 2.4 | ND (1.0) | 3.1 (3.0) | 1.1 |
| qs (g g of cell dry wt−1 h−1) | 3.0 | ND (2.4) | ND (2.3) | 3.5 |
| qamy (U g of cell dry wt−1 h−1) | 130.7 | ND (1,764) | 989 (2,337) | 1,890 |
Yx/s, growth yield relative to substrate; Ylac/s, lactic acid yield relative to substrate; Ylac/x, lactic acid yield relative to biomass; Yamy/x, amylase yield relative to biomass; μ, maximum specific growth rate; qlac, specific rate of lactic acid production; qs, specific rate of substrate consumption; qamy, specific rate of amylase production.
Fermentation values obtained at a controlled pH (6.0) are given in parentheses. ND, not determined.
As determined in this study.
Guyot et al. (14).
Calderon et al. (7).
DISCUSSION
Importance of ALAB during fermentation.
Lactic acid bacteria are the dominant group during all stages of pozol fermentation, as has been shown previously by classical culture methods (25, 34) and culture-independent methods (2, 12). Because of the low concentration of sugars remaining in maize after nixtamalization (0.1 to 0.7 g of sucrose 100 g of dry nixtamalized dough−1) (31), starch must be an important substrate for lactic fermentation. Forty percent of lactic acid bacteria present in the initial dough were amylolytic, and this ratio decreased to 3% after 72 h of fermentation (Table 1). The high initial concentration of ALAB indicates their importance during pozol fermentation, mainly during the first 24 h, when a high number of ALAB with a 7- to 18-mm hydrolysis zone diameter on agar plates were present. This suggests that conditions such as high initial pH values (7.4) could have favored the growth of bacteria with higher amylolytic activity. In contrast, a relatively high number of nonamylolytic lactic acid bacteria were observed at the end of fermentation that might be explained by an efficient use of mono- and disaccharides initially present and other sugars made available from starch hydrolysis by ALAB.
Diversity of ALAB.
An integrated vision of the role and the diversity of ALAB in pozol, together with their main physiological characteristics, is reported in this investigation. All of the 40 amylolytic isolates were homofermentative lactic acid bacteria. Frequently, ALAB isolated from foods belong to the genus Lactobacillus and are commonly homofermentative such as L. manihotivorans (23) or facultative heterofermentative such as L. plantarum (16, 26, 27). Strictly heterofermentative ALAB strains have also been isolated, such as Leuconostoc mesenteroides from Nigerian ogi (16), Leuconostoc sp. from fish silage (18), and L. fermentum from fermented maize dough (mawe and ogi) (1).
From the API 50CH profiling results, all ALAB isolated from pozol were able to ferment sucrose, maltose, glucose, and fructose, which are the main soluble sugars of maize (5), reflecting adaptation to their habitat. Furthermore, all ALAB (except strain A36202) were able to ferment raffinose, an α-galactoside contained in cereals and legumes that is responsible for digestive disorders.
The isolates belong to the closely related Streptococcus, Enterococcus, and Lactococcus genera, with S. bovis as the dominant species during fermentation. Ribotyping and fermentation profiles indicate strain diversity among pozol isolates belonging to the group S. bovis. The presence of S. bovis in pozol sourdoughs has previously been detected by denaturing gradient gel electrophoresis fingerprints and molecular probes (3), but the organism was not isolated. S. bovis comprises strains capable of growth in the presence of 6.5% NaCl and at pH 9.6, and some strains are thermoduric (15). Considering these properties, alkaline cooking of maize to prepare nixtamal would have contributed in selecting these bacteria. However, high survival rates could not be expected, since rapid inhibition of growth of this species linked to pH fall has been reported (15). This could account for the decrease in overall ALAB population at the end of pozol fermentation; however, strains belonging to S. bovis species were isolated throughout pozol fermentation, despite the increasing acid conditions. These results seem to contradict previous studies (2, 3) that showed an intense denaturing gradient gel electrophoresis band corresponding to S. bovis over the entire period of 96 h of pozol fermentation, which accounted for 50% of the total active microbiota. These data could have been overestimated since the authors of that study emphasized that their results could be biased by the techniques used to determine relative abundance of species and by possible comigration of fragments from different species (3). However, a high DNA concentration is not contradictory with low plate counts, since the growth ability of the species could have been altered by pH conditions developed during fermentation, leading to viable but nonculturable cells. Another aspect that cannot be neglected is the potential presence of higher pH microenvironments (maintained after the alkali treatment) that could explain S. bovis survival in the acidified dough.
Three other species of ALAB were found: S. macedonicus, L. lactis, and E. sulfureus. To our knowledge, ours is the first report of the amylolytic S. macedonicus and E. sulfureus strains. S. macedonicus was isolated from Greek Kasseri cheese (32). L. lactis has been reported previously in pozol (3, 12, 25), a starch-fermenting L. lactis subsp. lactis strain was isolated from the Thai product plaa-som (28). Most of the ALAB isolated from other cereal- or cassava-based fermented foods belong to the genus Lactobacillus (1, 16, 30). In comparison to African cereal-based fermented foods, pozol thus appears to be a food ecosystem in which other types of ALAB have developed. The absence of Lactobacillus among the most amylolytic of the lactic acid bacteria in pozol might be linked to conditions imposed by process conditions (i.e., alkaline cooking).
Starch fermentation by S. bovis.
Unlike starch fermentation by L. manihotivorans and L. fermentum Ogi E1 (14, 7), starch is only poorly hydrolyzed by the dominant species S. bovis, and a high concentration of total sugars remains at the end of fermentation. Poor starch hydrolysis could be explained by the low level of amylase produced but, as shown for L. manihotivorans and L. fermentum Ogi E1, even at low amylase activity starch can be completely degraded (14, 7). Furthermore, the difference between total sugar and reducing sugar concentrations, together with the low concentration of low-molecular-weight maltooligosaccharides indicates that higher-molecular-weight dextrins were produced.
In the present study, the main strain was characterized by a low yield of lactic acid relative to biomass (Ylac/x), an extremely low amylase yield relative to biomass (Yamy/x), and a specific rate of amylase production (qamy) (Table 3) and by a high specific growth rate and biomass yield (Yx/s). These characteristics are very different from those described for ALAB such as L. manihotivorans or amylolytic strains of L. plantarum and L. fermentum (Table 3). Streptococcus strains isolated from bovine rumen grew faster than other lactic acid bacteria on a wide variety of mono- and disaccharides, with specific growth rates higher than 1.5 h−1 (4). In particular, S. bovis JB1 presented higher specific growth rate on starch (1.98 h−1) than on maltose (1.38 h−1) (8). The specific growth rate determined for the pozol S. bovis strain (0.94 h−1) could give it a competitive advantage over the other nonamylolytic lactic acid bacterium species of pozol. In addition, although substrate consumption and lactate production parameters are in the classical range observed for other ALAB (Table 3), a relatively high value of Yx/s indicates efficient energy conversion into biomass, which would also confer an additional competitive advantage, in spite of a very low efficiency in starch hydrolysis and total sugar consumption. Nevertheless, a low tolerance at acidic pH of S. bovis strains will also have to be taken into account to explain the dynamics of microbial populations.
Since early and drastic reduction in the content of mono- and disaccharides in nixtamal, microbial diversity and the high bacterial concentration reported in pozol cannot be supported by these carbohydrates. However, the presence of ALAB from the first stage of the fermentation could explain the diversity of microbial population through trophic relationships. Based on the specific properties and intrinsic limitations of S. bovis strains, it can be assumed that the major role of S. bovis could be to provide low-molecular-weight maltooligosaccharides to the nonamylolytic microflora during the early steps of nixtamal dough fermentation. This assumption opens new fields of investigations on the microbial ecology of starchy fermented foods.
Acknowledgments
G.D.-R. acknowledges CONACYT (México) for providing a personal Ph.D. grant, PAEP UNAM for financial support through doctoral projects 103313 and 202309, and IRD, Montpellier, France, for short research stays.
We thank Judith Espinosa and Dora Centurión from UJAT, Tabasco, Mexico, for providing samples and laboratory facilities during field studies and Rocío Santillana for technical assistance. We are grateful to Matthew D. Collins and Paul A. Lawson for help with 16S rRNA gene sequencing of the strains.
REFERENCES
- 1.Agati, V., J. P. Guyot, J. Morlon-Guyot, P. Talamond, and D. J. Hounhouigan. 1998. Isolation and characterization of new amylolytic strains of Lactobacillus fermentum from fermented maize doughs (mawe and ogi) from Benin. J. Appl. Microbiol. 85:512-520. [Google Scholar]
- 2.Ampe, F., N. Ben Omar, C. Moizan, C. Wacher, and J. P. Guyot. 1999. Polyphasic study of the spatial distribution of microorganisms in mexican pozol, a fermented maize dough, demonstrates the need for cultivation-independent methods to investigate traditional fermentations. Appl. Environ. Microbiol. 65:5464-5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben Omar, N., and F. Ampe. 2000. Microbial community dynamics during production of the Mexican fermented maize dough Pozol. Appl. Environ. Microbiol. 66:3664-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bond, D. R., B. M. Tsai, and J. B. Russell. 1998. The diversion of lactose carbon through the tagatose pathway reduces the intracellular fructose 1,6-biphosphate and growth rate of Streptococcus bovis. Appl. Microbiol. Biotechnol. 49:600-605. [DOI] [PubMed] [Google Scholar]
- 5.Boyer, C. D., and J. C. Shannon. 1987. Carbohydrates of the kernel, p. 253-272. In S. A. Watson and P. E. Ramstad (ed.), Corn: chemistry and technology. American Association of Cereal Chemists, Inc., St. Paul, Minn.
- 6.Brabet, C., D. Dufour, M. Raimbault, and J. Giraud. 1996. Improving casava sour starch quality in Colombia, p. 241-246. In D. Dufour, G. M. O'Brien, and R. Best (ed.), Cassava flour and starch. CIAT, Cali, Colombia.
- 7.Calderon, M., G. Loiseau, and J. P. Guyot. 2001. Nutritional requirements and simplified cultivation medium to study growth and energetics of a sourdough lactic acid bacterium Lactobacillus fermentum Ogi E1 during heterolactic fermentation of starch. J. Appl. Microbiol. 90:508-516. [DOI] [PubMed] [Google Scholar]
- 8.Cotta, M. A. 1988. Amylolytic activity of selected species of ruminal bacteria. Appl. Environ. Microbiol. 54:772-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Man, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 10.Devriese L. A., and B. Pot. 1995. The genus Enterococcus, p. 327-367. In B. J. B. Wood and W. H. Holzapfel (ed.), The lactic acid bacteria. The genera of lactic acid bacteria, vol. 2. Blackie Academic and Professional, Glasgow, United Kingdom.
- 11.Dubois, M., K. A. Gilles, J. K. Hamilton, P. A. Rebers, and F. Smith. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 28:350-356. [Google Scholar]
- 12.Escalante, A., C. Wacher, and A. Farrés. 2001. Lactic acid bacterial diversity in the traditional Mexican fermented dough pozol as determined by 16S rDNA sequence analysis. Int. J. Food Microbiol. 64:21-31. [DOI] [PubMed] [Google Scholar]
- 13.Feltham, R. K. A., A. K. Power, P. A. Pell, and P. H. A. Sneath. 1978. A simple method for storage of bacteria at −76°C. J. Appl. Bacteriol. 44:313-316. [DOI] [PubMed] [Google Scholar]
- 14.Guyot, J. P., M. Calderon, and J. Morlon-Guyot. 2000. Effect of pH control on lactic acid fermentation of starch by Lactobacillus manihotivorans LMG 18010T. J. Appl. Microbiol. 88:176-182. [DOI] [PubMed] [Google Scholar]
- 15.Hardie, J. M., and R. A. Whiley. 1995. The genus Streptococcus, p. 55-124. In B. J. B. Wood and W. H. Holzapfel (ed.), The lactic acid bacteria: the genera of lactic acid bacteria, vol. 2. Blackie Academic and Professional, Glasgow, United Kingdom.
- 16.Johansson, M. L., A. Sanni, C. Lönner, and G. Molin. 1995. Phenotypically based taxonomy using API 50CH of lactobacilli from Nigerian ogi, and the occurrence of starch fermenting strains. Int. J. Food Microbiol. 25:159-168. [DOI] [PubMed] [Google Scholar]
- 17.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley and Sons, Chichester, United Kingdom.
- 18.Lindgren, S., and O. Refai. 1984. Amylolytic lactic acid bacteria in fish silage. J. Appl. Bacteriol. 57:221-228. [Google Scholar]
- 19.Link, W., C. Dixhens, M. Singh, M. Schwall, and A. E. Melchinger. 1995. Genetic diversity in European and Mediterranean faba bean germ plasm revealed by RAPD markers. Theor. Appl. Genet. 90:27-32. [DOI] [PubMed] [Google Scholar]
- 20.McClelland, M., R. Jones, Y. Patel, and M. Nelson. 1987. Restriction endonucleases for pulsed field mapping of bacterial genomes. Nucleic Acids Res. 15:5985-6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mestres, C., and X. Rouau. 1997. Influence of natural fermentation and drying conditions on the physicochemical characteristics of cassava starch. J. Sci. Food. Agric. 74:147-155. [Google Scholar]
- 22.Miller, G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugars. Anal. Chem. 31:426-428. [Google Scholar]
- 23.Morlon-Guyot, J., J. P. Guyot, B. Pot, I. Jacobe de Haut, and M. Raimbault. 1998. Lactobacillus manihotivorans sp. nov., a new starch-hydrolyzing lactic acid bacterium isolated from cassava sour starch fermentation. Int. J. Syst. Bacteriol. 48:1101-1109. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura, L. K. 1981. Lactobacillus amylovorus, a new starch-hydrolyzing species from cattle waste-corn fermentations. Int. J. Syst. Bacteriol. 31:56-63. [Google Scholar]
- 25.Nuraida, L., C. Wacher, and J. D. Owens. 1995. Microbiology of pozol, a mexican fermented maize dough. World J. Microbiol. Biotechnol. 11:567-571. [DOI] [PubMed] [Google Scholar]
- 26.Nwankwo, D., E. Anadu, and R. Usoro. 1989. Cassava fermenting organisms. Mircen J. 5:169-179. [Google Scholar]
- 27.Olympia, M., H. Fukuda, H. Ono, Y. Kaneko, and M. Takano. 1995. Characterization of starch-hydrolyzing lactic acid bacteria isolated from a fermented fish and rice food, “Burong Isda,” and its amylolytic enzyme. J. Ferment. Bioeng. 80:124-130. [Google Scholar]
- 28.Østergaard, A., P. K. B. Embarek, C. Wedell-Neergaard, H. H. Huss, and L. Gram. 1998. Characterization of anti-listerial lactic acid bacteria isolated from Thai fermented fish products. Food Microbiol. 15:223-233. [Google Scholar]
- 29.Pirt, S. J. 1985. Principles of microbe and cell cultivation. Blackwell Scientific Publications,Oxford, United Kingdom.
- 30.Sanni, A., J. Morlon-Guyot, J. P. Guyot. 2002. New efficient amylase-producing strains of Lactobacillus plantarum and L. fermentum isolated from different Nigerian traditional fermented foods. Int. J. Food Microbiol. 72:53-62. [DOI] [PubMed] [Google Scholar]
- 31.Santillana, R. 1995. Desarrollo de un método por cromatografía líquida de alta eficiencia para el análisis químico de nixtamal y pozol. MSc. thesis. Universidad Nacional Autónoma de México, Mexico D.F., Mexico.
- 32.Tsakalidou, E., E. Zoidou, B. Pot, L. Wassill, W. Ludwig, L. A. Devriese, G. Kalantzopoulos, K. H. Schleifer, and K. Kersters. 1998. Identification of streptococci from Greek Kasseri cheese and description of Streptococcus macedonicus sp. nov. Int. J. Syst. Bacteriol. 48:519-527. [DOI] [PubMed] [Google Scholar]
- 33.van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 34.Wacher, C., A. Cañas, E. Bárzana, P. Lappe, M. Ulloa, and J. D. Owens. 2000. Microbiology of Indian and Mestizo pozol fermentations. Food Microbiol. 17:251-256. [Google Scholar]