Abstract
This article provides a brief overview of some of the major concepts and molecular features of plant and animal innate immune systems. The rice pathogen recognition receptor, XA21, confers resistance to Xanthomonas oryzae pv. oryzae strains producing the AvrXa21 elicitor. Xa21 codes for a receptor-like kinase consisting of an extracellular leucine-rich repeat domain, a transmembrane domain, and a cytoplasmic kinase domain. We show that AvrXa21 activity requires the presence of rax (required for AvrXa21) A, raxB, and raxC genes that encode components of a type one secretion system. In contrast, an hrpC− strain deficient in type three secretion maintains AvrXa21 activity. Xanthomonas campestris pv. campestris can express AvrXa21 activity if raxST, encoding a putative sulfotransferase, and raxA are provided in trans. Expression of rax genes depends on population density and other functioning rax genes. This and other data suggest that the AvrXa21 pathogen-associated molecule is involved in quorum sensing. Together these data suggest that AvrXa21 represents a previously uncharacterized class of Gram-negative bacterial signaling molecules. These results from our studies of the XA21/AvrXa21 interaction call for some modifications in the way we think about innate immunity strategies.
Keywords: XA21, pathogen-associated molecule pattern, type I secretion, quorum sensing, rice
Animals and plants both have well developed immune systems for protection against pathogen challenges. Adaptive immunity, specific to animals, depends on somatic gene rearrangements for generation of antigen receptors with random specificities. In contrast, innate immunity is common to metazoans and plants and involves perception of pathogen-associated molecular patterns (PAMPs) by pathogen recognition receptors (PRRs) (1). PAMPs have been defined as microbe-associated molecules that are conserved among diverse species and required for the microbe's lifecycle (2, 3). Representative PAMPs recognized by plants and/or animals that have been identified to date are flagellin, a proteinaceous component of bacterial polar flagella (4), the peptidoglycan of Gram-positive bacteria (5), lipopolysaccharide of Gram-negative bacteria (6), single-stranded viral RNA (7), and oomycete transglutaminase (8).
Detection of Pathogens by Plant and Animal Hosts
In animals, recognition of PAMPs in extracellular compartments largely is carried out by the Toll-like receptor (TLR) family, which contains extracellular leucine-rich repeats (LRRs) that act in ligand recognition and an intracellular Toll-interleukin 1 (TIR) domain (9). Although TLRs recognize diverse molecules, they activate a common signaling pathway to induce a core set of defense responses (10). Intracellular recognition largely is carried out by the cytoplasmic nucleotide-binding oligomerization domain (NOD) protein family. The NOD family contains a large number of proteins from animals, plants, fungi, and bacteria (11).
Evidence has accumulated that plants also detect PAMPs but, unlike animal systems, which have PRRs for cytoplasmic and extracellular perception of PAMPs, all biochemically characterized phytopathogen PAMPs are active at the cell surface (12). With notable exceptions, another general characteristic of PAMP recognition by plants is that, although it promotes expression of pathogenesis-related proteins and other characteristics of pathogen response, it does not lead to clear disease resistance or a related hypersensitive response, which includes localized plant cell death. For example, treatment of tobacco with lipopolysaccharide leads to increases in expression of genes associated with defense response but has no immediate effect on infection with virulent strains (6).
Surprisingly little is known about PAMP receptors in plants. The best characterized plant PRR for a PAMP is the Arabidopsis thaliana receptor-like kinase (RLK), FLS2 (flagellin sensing 2), which includes an extracellular LRR ligand-binding domain and an intracellular serine/threonine kinase and directly recognizes a conserved N-terminal fragment of bacterial flagellin (13, 14). Stimulation of FLS2 by flagellin activates pathogenesis-related gene expression, and pretreatment with it leads to resistance (15, 16). Recent studies have shown that the presence of FLS2 decreases host susceptibility to a pathovar of Pseudomonas syringae when the pathogen is applied to leaves by spraying but not infiltration (16, 17). This finding demonstrates a direct link between disease resistance and PAMP perception that previously had been lacking in plants. Another recently characterized plant receptor that perceives a PAMP is EFR (elongation factor Tu receptor) (18), which recognizes elf18, consisting of the first 18 aa of EF-Tu (elongation factor Tu) (19).
In addition to recognition of conserved PAMPs, plant PRRs also recognize strain-specific molecules produced by phytopathogens, termed pathogen avirulence factors (20, 21). This specific recognition generally triggers a strong defense response, often including the hypersensitive response. To explain the observation of interactions between dominant PRR genes and bacterial avr genes, H. H. Flor proposed the gene-for-gene hypothesis in which a single plant-gene product recognizes a single bacterial avr gene product and blocks disease formation (22). Twenty-one rice loci that confer genetically dominant resistance to bacterial blight disease caused by Xanthomonas oryzae pv. oryzae (Xoo) have been identified, and four of these genes have been cloned (reviewed in ref. 23).
The majority of plant PRR genes cloned to date code for cytoplasmically localized NOD family members, with a domain that contains a nucleotide-binding site (NBS) and a domain with LRRs (NBS-LRRs). All characterized NBS-LRRs that block bacterial pathogenesis recognize Avr proteins from pathogenic bacteria that are secreted through a large bacterial complex called a type three secretion system (TTSS). The TTSS is thought to function by transporting proteins, called effectors, directly into the cytoplasm of the host cell, and it is known to be an essential transport system for disease development and bacterial multiplication (24, 25). As recently reviewed in refs. 26 and 27, many Avr molecules rely on the TTSS for secretion and act as type III effectors. These molecules include AvrRpt2, AvrB, AvrRpm1, and AvrPto from P. syringae; XopD and AvrBsT from Xanthomonas campestris; and the three cloned Avr factors from Xoo, AvrXa7, AvrXa10, and AvrXa27. However, only a few type III effectors have known biochemical functions. Of those effectors that have been characterized, several are enzymes, such as proteases and phosphatases, that act on host protein substrates to interfere with, suppress, and manipulate host defense and signaling by defense hormones such as salicylic and jasmonic acid (28). A recent elegant example of this comes from the human pathogen Yersinia, in which the type III effector, YopJ, acetylates a mitogen-activated protein kinase (MAPK) kinase, blocking the site of its activation by phosphorylation (29). Others encode proteins with a nuclear localization signal and more directly modulate host transcription. For example, the AvrBs3/PthA family has a nuclear localization signal and an acidic transcription activation domain that is required for AvrBs3-dependent hypersensitive response (30). Few direct interactions have been reported between NBS-LRR PRRs and their corresponding effectors (31). Instead, the defense response often is triggered by interaction between the NBS-LRR and another plant protein, which is targeted or modified by the type III effector (32).
In addition to the cytoplasmic NBS-LRRs, two other classes of plant PRR that recognize Avr proteins have been described. These are the RLKs, composed of various putative ligand-binding extracellular domains and an intracellular kinase, and receptor-like proteins (RLPs), composed only of a membrane-anchored extracellular domain (e.g., tomato CF9) or a presumed secreted extracellular domain (e.g., rice XA21D) (33, 34). Relatively few plant RLKs and RLPs have been cloned and characterized to date, although, like NBS-LRRs, sequence analyses indicate that there are a large number of genes of these classes in plants (33). The best studied PRR RLK that confers a race-specific response is the rice XA21 protein, which recognizes Xoo strains carrying AvrXa21 activity (35). The three other dominant RLKs cloned and characterized to date are the rice Xa26 and Pi-d2 proteins and barley RPG1 (36–38). The potential ligand-binding capabilities of the extracellular domains of the RLK PRRs, and the facts that XA21 is present in microsomal fractions (39) and that Pi-d2 localizes to the cell membrane (37), suggest a simple model in which XA21 and other RLKs recognize Avr proteins in the extracellular space, directly or indirectly, as the tomato Cf RLPs do (40).
Recent discoveries, including the results described here, call for a blurring of the distinctions between PRR proteins that recognize PAMPs and those that recognize Avr proteins and other conceptual dichotomies that have been established in plant pathology. Rather, a continuum of classes seems to exist, with plants making use of a diversity of strategies for innate immunity (41). Although the AvrXa21 molecule(s) itself has not yet been identified, we show here that AvrXa21 activity (i) depends on a bacterial type one secretion system (TOSS) not on a TTSS, (ii) is regulated by a two-component regulatory system that responds to Xoo cell population density, and (iii) may be conserved in most Xanthomonas spp. Furthermore, we show that a high-pressure liquid chromatography (HPLC)-purified fraction carrying AvrXA21 activity can induce raxST gene expression at low cell density. These data suggest that AvrXA21 represents a previously uncharacterized class of signaling molecules, which is used by Xoo for quorum sensing (QS) and which does not fall into any of the previously described classes of PAMPs or Avr factors.
PRR XA21 Represents a Large Class of Kinases Predicted to be Involved in Innate Immunity
In our recent survey of kinases in yeast, fly, worm, human, Arabidopsis, and rice, a correlation was found between a function in innate immunity and the absence of a conserved arginine (R) adjacent to a conserved asparatate (D) in the activation loop in domain VII of interleukin 1 receptor-associated kinase (IRAK) family kinases (42). Of the 38 characterized IRAK family receptor kinases in plants, all 6 RLKs associated with pathogen recognition fall into this “non-RD” class, as do 6 of the 9 kinases associated with early PRR signal transduction in animal innate immunity, IRAK and receptor-interacting protein kinases. Although there are only 7 non-RD IRAK-family kinases in humans, there are 47 in Arabidopsis and 371 in rice (42). Some of these may, like XA21, represent RLK PRRs that confer dominant and strong resistance to important diseases. Characterization of the molecule that XA21 recognizes could have significant impact toward understanding the mode of action of this large but poorly understood class of non-RD associated receptors.
AvrXa21 Activity Requires a TOSS
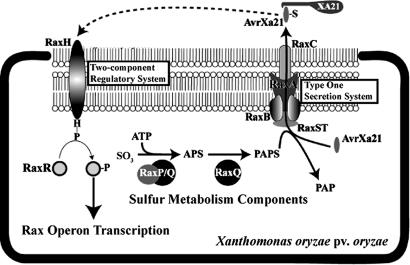
As previously described, we have cloned the raxC gene and seven additional genes in three operons (raxSTAB, raxPQ, and raxRH), that are required for AvrXa21 (rax) activity of Xoo, Philippine race 6 (strain PXO99) (43–45). Mutations in any of the eight rax genes allow this normally avirulent strain to form lesions when inoculated onto XA21-containing rice plants. Fig. 1 shows our working model for the action of the rax gene products in producing AvrXa21 activity. Based on sequence analysis and functional studies, the rax gene products can be grouped into three functional classes as follows: RaxP, RaxQ, and RaxST are sulfur metabolism enzymes; RaxA, RaxB, and RaxC form a TOSS; and RaxH and RaxR form a two-component regulatory system (43–45). Because phylogenetic analysis suggests that the RaxB protein belongs to a specific family of ABC transporters that secretes peptides (44), we hypothesize that the AvrXa21 molecule is a type one secreted peptide.
Fig. 1.
Working model for the synthesis, regulation, and function of AvrXa21. Functions assigned to each of the rax gene products based on sequence homology and/or functional studies are as follows (43–45): RaxH, histidine kinase; RaxR, response regulator; RaxP, ATP sulfurylase; RaxQ, adenosine-5′-phosphosulfate kinase; RaxST, sulfotransferase; RaxA, membrane fusion protein, spanning the inner membrane and the periplasmic space; RaxB, ATP-binding cassette transporter; and RaxC, outer-membrane protein. See text for elaboration.
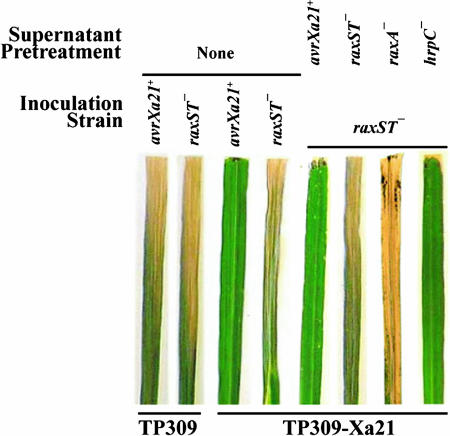
To test the model that AvrXA21 is produced and secreted because of the action of the rax genes, we have developed a bioassay that detects AvrXA21 activity, consisting of cutting the tips off of rice leaves and pretreating the leaves by dipping them into supernatant prepared from media in which Xoo has been grown. Pretreated leaves then are inoculated by cutting immediately below the first cut site with scissors that have been dipped in a suspension of Xoo (46). We used this bioassay to test the effect of mutations in each of the rax genes on AvrXa21 activity. Without pretreatment (first four leaves from left in Fig. 2), an Xoo wild-type strain carrying AvrXa21 activity (AvrXa21+) and a raxST knockout (raxST−) strain lacking AvrXa21 activity were inoculated onto leaves of japonica rice lines TP309 and TP309-XA21. Long, water-soaked lesions show that TP309 is susceptible to both strains, whereas the XA21 plants are susceptible to the raxST− strain (Fig. 2). In contrast, XA21 leaves inoculated with the wild-type strain expressing AvrXa21 activity are resistant (see necrosis at leaf tip). The next four leaves in Fig. 2 show the effects of pretreatment with supernatants from various Xoo genotypes. Pretreatment with wild-type supernatant of the leaves of XA21 plants prevents subsequent infection by the raxST− strain. In contrast, pretreatment with supernatant from AvrXa21−, rax gene knockout mutants (including raxST− and raxA−; see Fig. 2), and raxB−, raxC−, raxP−, and raxQ− strains (data not shown), does not prevent disease by the raxST− strain. However, a hrpC−, TTSS-deficient strain (47) has no effect on AvrXa21 activity because pretreatment with supernatant from this strain blocks lesion development by the raxST− strain. These results indicate that AvrXa21 activity is dominant and is secreted by a TOSS but not the TTSS.
Fig. 2.
Bioassay showing that AvrXa21 activity is present in medium of PXO99 wild-type and TTSS-deficient mutant (hrpC−) strains but not in a TOSS-deficient mutant (raxA−) strain or a sulfuryltransferase-deficient (raxST−) strain. Leaves from 6-week-old TP309 and TP309 transgenic for XA21 (TP309-XA21) rice were inoculated with PXO99 wild-type (first and third leaves from left) or raxST− (second and fourth leaves from left) Xoo strains by using the standard clipping method (46). To measure AvrXa21 activity, TP309-XA21 leaves were pretreated for 5 h with cell-free supernatant of PXO99 wild-type, raxST−, raxA−, and hrpC− strains followed by inoculation with the raxST− strain. Shown are representative leaves 2–3 weeks after inoculation from one of three independent experiments.
We also investigated whether the AvrXa21 elicitor(s) has characteristics of a protein. Supernatant from wild-type Xoo was treated with a variety of proteases, including proteinase K and trypsin, and/or heat (95°C for 15 min). XA21 rice leaves pretreated with protease- or heat-treated supernatant showed strong resistance, similar to rice leaves pretreated with wild-type supernatant (Fig. 4, which is published as supporting information on the PNAS web site). Only supernatant prepared with a combination of heat denaturation before proteinase K treatment showed a slight decrease in AvrXa21 activity (Fig. 4). Although these results do not provide strong evidence that the AvrXa21 molecule is proteinaceous, there are many examples of small peptides that are unaffected by heat denaturation or protease digestion, such as the PAMPs, flg22, and elf18 (13). In addition, our preliminary mass spectrometry data of two bioassay-active fractions isolated from cell-free supernatant by reverse-phase HPLC-C18 show that these fractions are enriched with peptides with masses of <1.5 kD (S.-W.L. and P.C.R., unpublished results).
The data from our previous genetic studies coupled with those from the bioassay represent the discovery of a type I secreted factor that can trigger plant innate immunity. Although there are five systems (types I to V) for protein secretion in Gram-negative bacteria (24), only the TTSS previously has been shown to secrete Avr factors, which then are detected intracellularly. We hypothesize that other members of the large complement of plant non-RD IRAK-family receptor kinases (42) will be found to detect molecules secreted by non-TTSS systems.
The Core AvrXa21 Molecule Is Conserved in X. campestris pv. campestris (Xcc)
XA21 confers resistance to 29 of 32 tested Xoo strains, which suggests that all 29 strains carry AvrXa21 activity (48). In our previous report, the Xoo strain KR1, which lacks AvrXa21 activity, acquired this activity when the raxSTAB operon was provided on a plasmid (44). We hypothesized that other Xanthomonas species carry the cognate molecules that confer AvrXa21 activity but lack the appropriate sulfation and secretion systems. To test this possibility, we introduced the PXO99 raxSTAB region into a bacterium that is nonpathogenic on rice, Xcc (ATCC 33913). We chose this strain that carries closely related homologs of all of the rax genes except for the raxSTAB operon.
We carried out the AvrXa21 activity assay with supernatant prepared from Xcc transformed with the PXO99 raxSTAB genes and compared the lesion lengths (Table 1). Pretreatment with the supernatants of the Xoo wild-type strain and the Xcc strain carrying the raxSTAB genes induces resistance against infection by raxST− Xoo (lesion lengths of 1.3 ± 0.4 cm and 1.8 ± 0.8 cm, respectively), whereas leaves pretreated with the supernatants of the Xoo raxST− and the Xcc wild-type strains have long lesions (10.1 ± 5.2 cm and 10.0 ± 4.8 cm, respectively). Thus, the supernatant of Xcc carrying the raxST, raxA, and raxB genes possesses AvrXa21 activity. Furthermore, Xcc carrying Xoo raxST and raxA genes showed full AvrXa21 activity but not a strain with raxST alone (Table 1).
Table 1.
Comparison of lesion lengths on rice leaves indicating that the backbone of the AvrXa21 PAM is conserved in Xcc
| Supernatant pretreatment | Inoculation | Lesion length, cm |
|---|---|---|
| PXO99 | raxST− | 1.3 ± 0.43 |
| raxST− | raxST− | 10.1 ± 5.2 |
| Xcc | raxST− | 10.0 ± 4.8 |
| Xcc (raxSTAB) | raxST− | 1.8 ± 0.8 |
| Xcc (raxSTA) | raxST− | 2.0 ± 1.0 |
| Xcc (raxST) | raxST− | 12.7 ± 7.3 |
Data are average ± SD from 10 scored leaves in three time repeats.
Although we have yet to test more diverse bacteria, these results suggest that the core AvrXa21 molecule is conserved between at least two species. Conservation across species is a key component of the definition of a PAMP. Nonetheless, the requirement for raxST and raxA suggests that XA21-mediated recognition of the AvrXa21 molecule requires a specific TOSS inner membrane protein (RaxA) and/or a posttranslational modification, sulfation, that is likely catalyzed by the putative sulfotransferase RaxST (44). The requirement for both raxA and raxST suggests that AvrXa21 modification and secretion may be sequential processes, requiring both the raxST and raxA gene products, or that RaxST activity or stability is altered in the absence of RaxA. There are many examples of posttranslational modification affecting extracellular recognition. Typically, sulfated molecules are directed outside the cell to serve as modulators of cell–cell interactions (49). A notable example pertinent to agriculture is sulfation of the Sinorhizobium meliloti Nod factor that is required for specific recognition by its host, alfalfa (50).
Another element of the definition of a PAMP is that it is essential for the pathogen. The fact that the core of AvrXa21 appears to be conserved in Xcc suggests that the molecule(s) has a function that makes it selectively advantageous. In addition, although we did not observe differences in virulence in the raxSTAB and raxC knockout strains under controlled conditions, in a field study, 37 Xoo Korean strains lacking AvrXa21 activity appeared to have reduced fitness (51). These results suggest both that the core AvrXa21 molecule is conserved between Xoo and Xcc and that loss of AvrXa21 results in a fitness cost to Xoo. Therefore, AvrXa21, which is detected by the dominant, resistance-conferring XA21 PRR, shares characteristics of PAMPs and Avr proteins.
Cell Density-Dependent Expression of rax Genes
What is the function of AvrXa21? Like other living organisms, bacteria have developed multiple systems for responding to environmental variation, such as changes in temperature, osmolarity, pH, nutrient availability, and even population size. Two-component systems, composed of histidine kinases and response regulators, have an important role in sensing the environment (52). Phosphorylation of a response regulator by a histidine kinase regulates gene expression and governs the response to an environmental stimulus (53). Bacteria themselves produce some stimuli. In a process called “quorum sensing” (QS), small molecules serve as signals to recognize cell population size, leading to changes in expression of specific genes when a signal has accumulated to some threshold concentration (54). QS molecules regulate their own expression and are called autoinducers (55). Interestingly, recent findings demonstrate that the QS signal molecules are not only active in communication between bacteria but also can affect host immune responses. A QS molecule from Pseudomonas aeruginosa stimulates phagocytic activity in human macrophages through a MAPK pathway (56, 57).
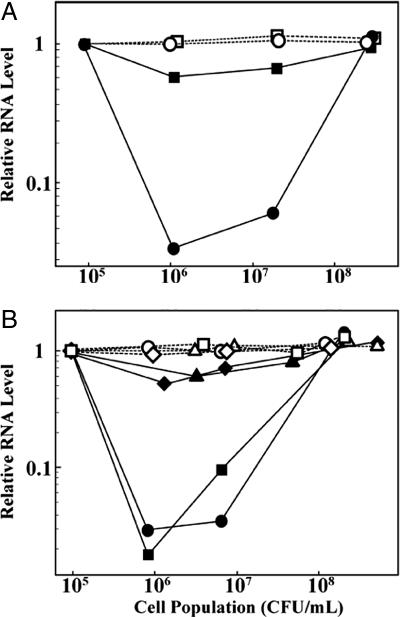
To test whether rax gene expression depends on cell population, rax gene expression in wild-type Xoo was measured by using real-time quantitative PCR with three primer sets specific for each of the following four rax genes, one in each operon: raxST, in raxSTAB (Fig. 3A); raxP, in raxPQ; raxR, in raxRH; and raxC (data not shown). Remarkably, the expression of all four rax genes was observed only in the wild-type strain at high cell densities, whereas levels of a ribosomal RNA were unaffected (Fig. 3A). We also measured rax gene expression in a strain constitutively expressing RaxR. Density dependence of rax gene expression in the raxR constitutive expressing strain is mostly lost (Fig. 3A). This result indicates that RaxR is involved in regulation of the density-dependent expression of the rax genes, including itself.
Fig. 3.
Cell density and AvrXa21-dependent expression of the raxST gene (filled symbols) and rRNA (open symbols). Xoo bacteria were cultured for 72 h and then diluted with fresh media to ≈105 cfu/ml. The diluted cultures were returned to the incubator, and, as they grew, aliquots were removed for RNA isolation at different cell population densities. Equal amounts (1 μg) of bacterial RNA extracted with TRIzol reagent (Invitrogen, Carlsbad, CA) were used for real-time quantitative PCR. Three primer sets for each rax gene were designed to amplify ≈300-bp segments corresponding to three different parts of each gene. Primer sequences are provided in Table 3, which is published as supporting information on the PNAS web site. Expression levels are reported on a log scale and normalized to the copy number in the samples immediately after dilution. Data are from one of set of primers and two independent experiments. Similar trends were seen with all of the primer pairs. Expression is high at 105 cfu/ml because the bacteria in this sample behave as if they are still at high density. (A) Wild-type PXO99 (circles) and raxR− (squares) strains. (B) Diluted PXO99 Xoo cultures were recultured without treatment (circles), with AvrXa21-active HPLC fractions (no. 2, triangles; no. 17, diamonds) or an inactive HPLC fraction (no. 24, squares).
Furthermore, we tested raxST gene expression in the wild-type strain with and without treatment with HPLC-fractionated supernatant to address the question of whether a semipurified form of AvrXa21 could itself induce rax gene expression (Fig. 3B). Reverse-phase HPLC-C18 fractions purified from 10 ml of wild-type Xoo were tested for activity in the leaf-dip bioassay described above. Two bioactive fractions (nos. 2 and 17) and one randomly chosen inactive fraction (no. 24) were added separately to 10 ml of the diluted Xoo cells (105 cfu/ml) for reculture. Fig. 3B shows that treatment with both active fractions strikingly increases the raxST gene expression at low cell population density, whereas treatment with inactive fraction had less effect on raxST gene expression, similar to untreated cells. This autoinduction is consistent with a role of the product of the rax gene pathway, namely AvrXa21, in QS (55).
The characteristics of the Xoo rax genes suggest a QS system with features similar to those typically found in Gram-positive bacteria. For example, oligopeptides are the predominant signaling molecules for genetic competence in Bacillus substilis (58), virulence in Staphylococcus aureus (59), and the production of antimicrobial peptides, inducing bacteriocins and lantibiotics, in lactic acid bacteria (60, 61). As QS molecules, these peptides also activate their own expression and have been given the name autoinducible peptides (AIPs). AIPs are known to be secreted by ABC transporters that are similar to RaxB (44) and are sensed by two-component systems. Significantly, secretion of the putative AvrXa21 peptide by a TOSS would be a previously undescribed occurrence of an AIP in Gram-negative bacteria.
In summary, our data suggest that the AvrXa21 elicitor is a secreted peptide and hint that the biological function of the AvrXa21 elicitor is as a QS signal molecule, the production of which is regulated in a cell-density-dependent manner in the Gram-negative bacteria Xoo by a two-component system. Thus, the rice XA21 PRR effectively eavesdrops on the AvrXa21-mediated molecular conversation among Xoo, using AvrXa21 as an indicator of the presence of a pathogenic bacterial population of significant size and leading to plant responses that effectively halt infection (Fig. 1). Sequence analysis of the genomes of several phytopathogenic bacteria that are similar to Xoo reveals an abundance of the rax classes of molecular components. Xoo, Xcc, Xanthomonas axonopodis pv. citri, and Xylella fastidiosa each possess genes for ≈30 two-component systems together with three to five TOSSs that, like RaxB, are predicted to transport peptides. Furthermore, it is apparent that X. fastidiosa does not use the TTSS for pathogenesis because genome analysis has revealed the absence of these components in this bacterium. Rather, X. fastidiosa contains TOSS genes similar to raxA, raxB, and raxC as well as sequences encoding as many as 10–12 peptides that are candidates for secretion (C. Dardick and P.C.R., unpublished results). These observations highlight that the interactions among phytopathogens, their hosts, and the environment are more sophisticated than are currently recognized.
Our data suggesting that the function of the AvrXa21 pathogen-associated molecule is as a QS signaling molecule may present a critical clue to answering the question, why is AvrXa21 maintained in Xoo regardless of its detection by a host protein? Based on others' results that suggest decreased fitness of rax mutants, AvrXa21 may be essential for Xoo as a signal molecule for cell–cell communication. We hypothesize that Xoo may use QS to coordinate infection and production of virulence factors. QS has global effects on bacterial growth, survival, and interactions with eukaryotes because QS-responsive genes compose up to 10% of the P. aeruginosa transcriptome (62). In that report, a large portion of QS-promoted genes code for membrane proteins and protein export apparatuses involved in secretion of virulence factors. A relationship between QS and virulence also has been described for the human pathogen Vibrio cholerae (63). Interestingly, in another case, a host can produce QS mimic molecules that interfere with the QS-regulated behaviors of the infecting bacteria (64). Given the abundance of other QS peptides and small molecules such as bacteriocins and lantibiotics in the host vicinity, it will not be surprising if these peptides are shown to play a role in the interaction of other bacteria with their hosts, as has been found for a homoserine lactone QS molecule from P. aeruginosa (57). In principle, recognition by plants of TOSS elicitors could be exploited to generate new types of specific and environmentally benign pesticides that induce plant defense responses.
Perspective
The data presented here as well as results of others in recent years establish that there is not a clear dichotomy between PAMP recognition and Avr recognition; rather, plant innate immunity functions as a continuum between the two general types (Table 2, which is published as supporting information on the PNAS web site) (16). PAMPs have been defined as being derived from conserved structures required for pathogen function, whereas Avr factors are thought to be maintained by specific strains as virulence factors. The features of AvrXa21 are a hybrid between these two categories. The AvrXa21 molecule appears to be conserved in Xcc, but sulfation seems to provide specificity to the system, just as flagellin recognition by rice is modulated by glycosylation (65, 66). Further evidence that plant-recognized PAMPs are not entirely conserved comes from a recent article showing that sequence variation of flagellin among Xcc strains modulates FLS2-dependent pathogen recognition by Arabidopsis (16). Also, compromised virulence and survival of avrXa21− strains in the field is consistent with a crucial role for AvrXa21 in Xanthomonad QS. On the other hand, AvrXa21 and XA21 fulfill the standard gene-for-gene model of the dominant resistance gene interacting with the bacterial virulence factor as well (35). Thus, AvrXa21 has features that are similar to both PAMPs and other Avrs.
The fusion of PRR recognition of Avr and PAMP classes proposed in this article has implications for the mechanism of plant innate immunity. Recognition of PAMPs by the animal innate immune system has been proposed to act as guide for the adaptive immune system as to the nature of the infecting pathogen (2). There is not an obvious need for such a function in plants because each receptor can be tied to a specific response through signal transduction. Nonetheless, it is clear that the plant innate immune system recognizes both conserved and less conserved molecules. As recently articulated (67), the fact that plant responses to PAMPs often do not lead to resistance may be attributable to evolutionary selection for plant innate immune perception to require “priming” or “two-hits” so that resources are not unnecessarily wasted in responding to nonpathogenic or small populations of bacteria. In such a model, PAMP perception constitutes an early warning, and, indeed, pretreatment with lipopolysaccharide or flg22 leads to resistance to subsequent treatments with normally pathogenic bacteria (6, 16, 17). However, it may be that the necessity for the factor that primes the plant innate immune system to be a PAMP rather than an Avr molecule is simply a matter of experimental history. Because they are less useful in an agricultural context, plant pathologists have not looked for PRRs and corresponding Avr molecules that do not lead to significant resistance. However, the XA21D receptor, which lacks a transmembrane and kinase domain and confers only partial resistance, may be such a molecule (34). XA21D would likely not have been discovered without experiments showing that its gene family member, Xa21, conferred complete resistance. Conversely, as mentioned above, FLS2-dependent resistance to bacteria that express flagellin also has been observed (17). Physiologically relevant experiments in diverse genetic backgrounds may lead to further evidence that responses mediated by PRRs for both more and less conserved pathogen-associated molecules (i.e., PAMPs and Avrs) are also part of a continuum, with some functioning more often in priming and others more often leading to immediate resistance. Already among described Avr PRRs there is a diversity of strengths, with some leading to superresistance (68). Thus, in possessing receptors for more conserved molecules, it may be that plants are simply taking advantage of the conserved nature of PAMPs as targets for recognition rather than making a functional distinction in terms of responses.
Several examples support the idea that plants gain a selective advantage by making use of a variety of strategies for pathogen recognition beyond the classes of molecules that typically have been discussed (27, 32), both in terms of the molecule detected and the cognate PRRs. AvrXa21 is currently the only known elicitor that is secreted by a TOSS. Until recently, LRRs constituted the only signal recognition domain described for a cloned PRR RLK; however, a recently cloned RLK conferring rice blast resistance, Pi-d2, instead possesses an extracellular lectin-binding domain (37). Moreover, sequence analyses suggest that RLKs and receptor-like proteins with other, diverse extracellular binding domains also are involved in plant innate immunity (33, 42). Similarly, plants appear to use other, entirely different classes of molecules as PRRs as well. Indeed, the first cloned PAMP high-affinity binding site was the soybean β-glucan elicitor binding protein (69). Localized to the extracellular side of the cell membrane, it is predicted to be tethered to the membrane via interactions with a receptor complex (70). Yet another perception strategy is represented by Xa27, which codes for a defense protein the promoter of which serves as the PRR by directly or indirectly interacting with the cognate, nuclear-localized AvrXa27 type III effector (71).
Our recent results suggest that the AvrXa21 elicitor is a TOSS-dependent secreted peptide that represents a previously undescribed family of signaling molecules for QS via a two-component system. Although QS molecules are known to be widely conserved and essential for bacterial communication, this example illustrates that such a QS molecule triggers plant innate immune responses. Our continuing work to identify the AvrXa21 molecule and understand the physical basis of its recognition may allow for identification of similar pathogen-associated molecules from plants and animal pathogens and their cognate host receptors.
Supplementary Material
Acknowledgments
This work was financially supported by National Institutes of Heath Grant GM55962. S.-W.L. was partially supported by a grant from the Korean Science and Engineering Foundation through the Plant Science Research Center, Kyung Hee University, Suwon, Korea. The work of S.-W.H. was partially supported by a grant for the Graduate Study Abroad Scholarship from the Korean Science and Engineering Foundation.
Abbreviations
- LRR
leucine-rich repeat
- PAMP
pathogen-associated molecular pattern
- PRR
pathogen recognition receptor
- RLK
receptor-like kinase
- Xoo
Xanthomonas oryzae pv. oryzae
- NBS
nucleotide-binding site
- TTSS
type three secretion system
- TOSS
type one secretion system
- R
arginine
- D
asparatate
- IRAK
interleukin 1 receptor-associated kinase
- Xcc
X. campestris pv. campestris
- QS
quorum sensing.
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “From Functional Genomics of Model Organisms to Crop Plants for Global Health,” held April 3–5, 2006, at The National Academy of Sciences in Washington, DC. Papers from this Colloquium will be available as a collection on the PNAS web site. The complete program is available on the NAS web site at www.nasonline.org/functional_genomics.
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Girardin SE, Sansonetti PJ, Philpott DJ. Trends Microbiol. 2002;10:193–199. doi: 10.1016/s0966-842x(02)02334-x. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R, Janeway CA., Jr Cell. 1997;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 3.Janeway CA, Jr, Medzhitov R. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 4.Ramos HC, Rumbo M, Sirard JC. Trends Microbiol. 2004;12:509–517. doi: 10.1016/j.tim.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Leulier F, Parquet C, Pili-Floury S, Ryu JH, Caroff M, Lee WJ, Mengin-Lecreulx D, Lemaitre B. Nat Immunol. 2003;4:478–484. doi: 10.1038/ni922. [DOI] [PubMed] [Google Scholar]
- 6.Erbs G, Newman M-A. Mol Plant Pathol. 2003;4:421–425. doi: 10.1046/j.1364-3703.2003.00179.x. [DOI] [PubMed] [Google Scholar]
- 7.Jurk M, Heil F, Vollmer J, Schetter C, Krieg AM, Wagner H, Lipford G, Bauer S. Nat Immunol. 2002;3:499. doi: 10.1038/ni0602-499. [DOI] [PubMed] [Google Scholar]
- 8.Brunner F, Rosahl S, Lee J, Rudd JJ, Geiler C, Kauppinen S, Rasmussen G, Scheel D, Nurnberger T. EMBO J. 2002;21:6681–6688. doi: 10.1093/emboj/cdf667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Werling D, Jungi TW. Vet Immunol Immunopathol. 2003;91:1–12. doi: 10.1016/s0165-2427(02)00228-3. [DOI] [PubMed] [Google Scholar]
- 10.Barton GM, Medzhitov R. Science. 2003;300:1524–1525. doi: 10.1126/science.1085536. [DOI] [PubMed] [Google Scholar]
- 11.Inohara N, Nunez G. Nat Rev Immunol. 2003;3:371–382. doi: 10.1038/nri1086. [DOI] [PubMed] [Google Scholar]
- 12.Nurnberger T, Brunner F, Kemmerling B, Piater L. Immunol Rev. 2004;198:249–266. doi: 10.1111/j.0105-2896.2004.0119.x. [DOI] [PubMed] [Google Scholar]
- 13.Felix G, Duran JD, Volko S, Boller T. Plant J. 1999;18:265–279. doi: 10.1046/j.1365-313x.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- 14.Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G. Plant Cell. 2006;18:465–476. doi: 10.1105/tpc.105.036574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomez-Gomez L, Felix G, Boller T. Plant J. 1999;18:277–284. doi: 10.1046/j.1365-313x.1999.00451.x. [DOI] [PubMed] [Google Scholar]
- 16.Sun W, Dunning FM, Pfund C, Weingarten R, Bent AF. Plant Cell. 2006;18:764–779. doi: 10.1105/tpc.105.037648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 18.Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 19.Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G. Plant Cell. 2004;16:3496–3507. doi: 10.1105/tpc.104.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindeberg M, Stavrinides J, Chang JH, Alfano JR, Collmer A, Dangl JL, Greenberg JT, Mansfield JW, Guttman DS. Mol Plant Microbe Interact. 2005;18:275–282. doi: 10.1094/MPMI-18-0275. [DOI] [PubMed] [Google Scholar]
- 21.van't Slot KAE, Knogge W. Crit Rev Plant Sci. 2002;21:229–271. [Google Scholar]
- 22.Flor HH. Annu Rev Phytopathol. 1971;9:275–296. [Google Scholar]
- 23.Niño-Liu DO, Ronald PC, Bogdanove AJ. Mol Plant Pathol. 2006;7:303–324. doi: 10.1111/j.1364-3703.2006.00344.x. [DOI] [PubMed] [Google Scholar]
- 24.Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala'Aldeen D. Microbiol Mol Biol Rev. 2004;68:692–744. doi: 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staskawicz BJ, Mudgett MB, Dangl JL, Galan JE. Science. 2001;292:2285–2289. doi: 10.1126/science.1062013. [DOI] [PubMed] [Google Scholar]
- 26.Mudgett MB. Annu Rev Plant Biol. 2005;56:509–531. doi: 10.1146/annurev.arplant.56.032604.144218. [DOI] [PubMed] [Google Scholar]
- 27.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 28.Thomma BP, Penninckx IA, Broekaert WF, Cammue BP. Curr Opin Immunol. 2001;13:63–68. doi: 10.1016/s0952-7915(00)00183-7. [DOI] [PubMed] [Google Scholar]
- 29.Mukherjee S, Keitany G, Li Y, Wang Y, Ball HL, Goldsmith EJ, Orth K. Science. 2006;312:1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- 30.Szurek B, Marois E, Bonas U, Van den Ackerveken G. Plant J. 2001;26:523–534. doi: 10.1046/j.0960-7412.2001.01046.x. [DOI] [PubMed] [Google Scholar]
- 31.Dodds PN, Lawrence GJ, Catanzariti AM, Teh T, Wang CI, Ayliffe MA, Kobe B, Ellis JG. Proc Natl Acad Sci USA. 2006;103:8888–8893. doi: 10.1073/pnas.0602577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dangl JL, Jones JDG. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 33.Shiu SH, Bleecker AB. Plant Physiol. 2003;132:530–543. doi: 10.1104/pp.103.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang GL, Ruan DL, Song WY, Sideris S, Chen L, Pi LY, Zhang S, Zhang Z, Fauquet C, Gaut BS, et al. Plant Cell. 1998;10:765–779. doi: 10.1105/tpc.10.5.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, et al. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- 36.Sun X, Cao Y, Yang Z, Xu C, Li X, Wang S, Zhang Q. Plant J. 2004;37:517–527. doi: 10.1046/j.1365-313x.2003.01976.x. [DOI] [PubMed] [Google Scholar]
- 37.Chen X, Shang J, Chen D, Lei C, Zou Y, Zhai W, Liu G, Xu J, Ling Z, Cao G, et al. Plant J. 2006;46:794–804. doi: 10.1111/j.1365-313X.2006.02739.x. [DOI] [PubMed] [Google Scholar]
- 38.Brueggeman R, Rostoks N, Kudrna D, Kilian A, Han F, Chen J, Druka A, Steffenson B, Kleinhofs A. Proc Natl Acad Sci USA. 2002;99:9328–9333. doi: 10.1073/pnas.142284999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu W-H, Wang Y-S, Liu G-Z, Chen X, Tinjuangjun P, Pi L-Y, Song W-Y. Plant J. 2006;45:740–751. doi: 10.1111/j.1365-313X.2005.02638.x. [DOI] [PubMed] [Google Scholar]
- 40.Rooney HC, Van't Klooster JW, van der Hoorn RA, Joosten MH, Jones JD, de Wit PJ. Science. 2005;308:1783–1786. doi: 10.1126/science.1111404. [DOI] [PubMed] [Google Scholar]
- 41.McDowell JM, Woffenden BJ. Trends Biotechnol. 2003;21:178–183. doi: 10.1016/S0167-7799(03)00053-2. [DOI] [PubMed] [Google Scholar]
- 42.Dardick C, Ronald P. PLoS Pathog. 2006;2:14–28. doi: 10.1371/journal.ppat.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen Y, Sharma P, da Silva FG, Ronald P. Mol Microbiol. 2002;44:37–48. doi: 10.1046/j.1365-2958.2002.02862.x. [DOI] [PubMed] [Google Scholar]
- 44.Goes da Silva F, Shen Y, Dardick C, Burdman S, Yadav R, Sharma P, Ronald P. Mol Plant Microbe Interact. 2004;17:593–601. doi: 10.1094/MPMI.2004.17.6.593. [DOI] [PubMed] [Google Scholar]
- 45.Burdman S, Shen Y, Lee S-W, Xue Q, Ronald P. Mol Plant Microbe Interact. 2004;17:602–612. doi: 10.1094/MPMI.2004.17.6.602. [DOI] [PubMed] [Google Scholar]
- 46.Kauffman HE, Reddy APK, Hsieh SPY, Merca SD. Plant Dis Rep. 1973;57:537–541. [Google Scholar]
- 47.Zhu W, MaGbanua MM, White FF. J Bacteriol. 2000;182:1844–1853. doi: 10.1128/jb.182.7.1844-1853.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang GL, Song WY, Ruan DL, Sideris S, Ronald PC. Mol Plant Microbe Interact. 1996;9:850–855. doi: 10.1094/mpmi-9-0850. [DOI] [PubMed] [Google Scholar]
- 49.Bowman KG, Bertozzi CR. Chem Biol. 1999;6:R9–R22. doi: 10.1016/S1074-5521(99)80014-3. [DOI] [PubMed] [Google Scholar]
- 50.Roche P, Debelle F, Maillet F, Lerouge P, Faucher C, Truchet G, Denarie J, Prome J-C. Cell. 1991;67:1131–1143. doi: 10.1016/0092-8674(91)90290-f. [DOI] [PubMed] [Google Scholar]
- 51.Choi SH, Lee SW, Han SS, Lee DK, Noh TH. Kor J Breed. 2003;35:283–288. [Google Scholar]
- 52.Lewis HA, Furlong EB, Laubert B, Eroshkina GA, Batiyenko Y, Adams JM, Bergseid MG, Marsh CD, Peat TS, Sanderson WE, et al. Structure (London) 2001;9:527–537. doi: 10.1016/s0969-2126(01)00613-x. [DOI] [PubMed] [Google Scholar]
- 53.Charles TC, Jin S, Nester EW. Annu Rev Phytopathol. 1992;30:463–484. doi: 10.1146/annurev.py.30.090192.002335. [DOI] [PubMed] [Google Scholar]
- 54.Fuqua WC, Winans SC, Greenberg EP. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waters CM, Bassler BL. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 56.Smith RS, Fedyk ER, Springer TA, Mukaida N, Iglewski BH, Phipps RP. J Immunol. 2001;167:366–374. doi: 10.4049/jimmunol.167.1.366. [DOI] [PubMed] [Google Scholar]
- 57.Vikstrom E, Magnusson K-E, Pivoriunas A. Microbes Infect. 2005;7:1512–1518. doi: 10.1016/j.micinf.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 58.Tortosa P, Dubnau D. Curr Opin Microbiol. 1999;2:588–592. doi: 10.1016/s1369-5274(99)00026-0. [DOI] [PubMed] [Google Scholar]
- 59.Novick RP, Muir TW. Curr Opin Microbiol. 1999;2:40–45. doi: 10.1016/s1369-5274(99)80007-1. [DOI] [PubMed] [Google Scholar]
- 60.Risoen PA, Brurberg MB, Eijsink VG, Nes IF. Mol Microbiol. 2000;37:619–628. doi: 10.1046/j.1365-2958.2000.02029.x. [DOI] [PubMed] [Google Scholar]
- 61.Kleerebezem M, Quadri LE, Kuipers OP, de Vos WM. Mol Microbiol. 1997;24:895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- 62.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. J Bacteriol. 2003;185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. Cell. 2002;110:303–314. doi: 10.1016/s0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- 64.Teplitski M, Chen H, Rajamani S, Gao M, Merighi M, Sayre RT, Robinson JB, Rolfe BG, Bauer WD. Plant Physiol. 2004;134:137–146. doi: 10.1104/pp.103.029918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taguchi F, Shimizu R, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y. Plant Cell Physiol. 2003;44:342–349. doi: 10.1093/pcp/pcg042. [DOI] [PubMed] [Google Scholar]
- 66.Takeuchi K, Taguchi F, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y. J Bacteriol. 2003;185:6658–6665. doi: 10.1128/JB.185.22.6658-6665.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ausubel FM. Nat Immunol. 2005;6:973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- 68.Greenberg JT, Yao N. Cell Microbiol. 2004;6:201–211. doi: 10.1111/j.1462-5822.2004.00361.x. [DOI] [PubMed] [Google Scholar]
- 69.Umemoto N, Kakitani M, Iwamatsu A, Yoshikawa M, Yamaoka N, Ishida I. Proc Natl Acad Sci USA. 1997;94:1029–1034. doi: 10.1073/pnas.94.3.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mithofer A, Fliegmann J, Neuhaus-Url G, Schwarz H, Ebel J. Biol Chem. 2000;381:705–713. doi: 10.1515/BC.2000.091. [DOI] [PubMed] [Google Scholar]
- 71.Gu K, Yang B, Tian D, Wu L, Wang D, Sreekala C, Yang F, Chu Z, Wang GL, White FF, Yin Z. Nature. 2005;435:1122–1125. doi: 10.1038/nature03630. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.