Abstract
Intracellular active transport is driven by ATP-hydrolyzing motor proteins that move along cytoskeletal filaments. In particular, the microtubule-associated dynein motor is involved in the transport of organelles and vesicles, the maintenance of the Golgi, and mitosis. However, unlike kinesin and myosin, the mechanism by which dynein converts chemical energy into mechanical force remains largely a mystery, due primarily to the lack of a high-resolution molecular structure. Using homology modeling and normal mode analysis, we propose a complete atomic structure and a mechanism for force generation by the motor protein dynein. In agreement with very recent electron microscopy (EM) reconstructions showing dynein as a ring-shaped heptamer, our model consists of six ATPases of the AAA (ATPases associated with various cellular activities) superfamily and a C-terminal domain, which is experimentally known to control motor function. Our model shows a coiled coil spanning the diameter of the motor that accounts for previously unidentified structures in EM studies and provides a potential mechanism for long-range communication between the AAA domains. Furthermore, normal mode analysis reveals that the subunits of the motor that contain the nucleotide binding sites exhibit minimal movement, whereas the rest of the motor is very mobile. Our analysis suggests the likely domain rearrangements of the motor unit that generate its power stroke. This study provides insights into the structure and function of dynein that can guide further experimental investigations into energy transduction in dynein.
Keywords: AAA, homology modeling, molecular motors, electron microscopy, ATP hydrolysis
Motor proteins use energy derived from ATP hydrolysis to move along cytoskeletal filaments (1). In particular, the microtubule-associated molecular motor dynein is involved in the transport of organelles and vesicles, the maintenance of the Golgi, and mitosis (2, 3). Mutations in this protein have been implicated in neurodegenerative diseases (4) and polycystic kidney disease (5). Unlike the motor proteins kinesin and myosin, the mechanism by which dynein converts chemical energy into mechanical force remains largely a mystery. The primary reason for this is the lack of a high-resolution molecular structure. Cytoplasmic dynein is a large multisubunit complex (1.2 MDa) composed of two heavy chains (≈0.5 MDa each) (6), making its structural characterization extremely challenging. In this study, we use homology modeling to propose a structure for the motor unit of the dynein heavy chain, which is the site for energy transduction and force generation (see Fig. 1a). Next, we perform normal mode analysis to determine the large-scale motions of the protein. Together, these results suggest a mechanism for both force generation and regulatory control in dynein.
Fig. 1.
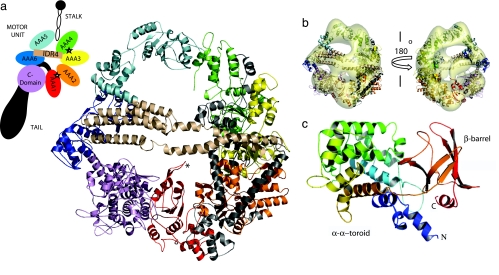
Model of the cytoplasmic dynein heavy chain. (a) Dynein consists of a motor unit (heavy chain) which generates force, a stalk that binds to microtubule, and a tail which oligomerizes with other dyneins. The atomic model of the motor unit contains six domains of the AAA enzyme family and a C domain. Domains are colored and identified according to schema. Active ATP-binding sites are indicated by stars. (b) Model structure from homology fitted to a 25-Å EM map of negatively stained cytoplasmic dynein (8) (EM map courtesy of M. Koonce). Motor unit consists of “smooth” (AAA1-AAA4) and “rough” (AAA5-C domain) sides. Also shown are the linkers between AAA1 and AAA2 (light gray), between AAA2 and AAA3 (black), between AAA3 and AAA4 (light gray), and between AAA5 and AAA6 (wheat). There are densities on the face of the ring. The long arching density putatively is formed by the coiled coil sequentially intermediate between AAA5 and AAA6, whereas the smaller density may be the remnant of the tail fragment. (c) Model of the C domain consists of an α–α toroid modeled from complement component C3d (PDB ID code 1GHQ) and a β-barrel modeled from leukemia-associated rhogef domain (PDB ID code 1TXD).
Sequence analysis of dynein's motor unit indicates that it consists of six concatenated AAA (ATPases associated with diverse cellular activities) subunits, an extended stalk that contains a microtubule binding domain, and a C-terminal domain that is twice the size of an AAA subunit (Fig. 1) (7, 8). Mocz and Gibbons (8) used homology modeling to construct the first structural model of dynein. Their model consisted of the six AAA modules (AAA1–AAA6) arranged as a symmetric hexamer (8). However, recent 3D EM reconstructions of negatively stained cytoplasmic dynein from Dictyostelium discoideum show the motor as a seven-lobe asymmetric ring (Fig. 1b) (9), with the additional subunit presumably formed by the C domain. This observation led Samso and Koonce (9) to propose a revised structure in which the C domain was modeled as an extra AAA subunit. Although the detailed structure of the C domain remains unknown, there is increasing evidence that it plays a key role in regulating the hydrolysis cycle that drives force generation (10). For example, recent enzymatic analysis using recombinant and proteolytic rat motor domain fragments suggests that the C domain regulates ATPase activity (10). Interestingly, 3D EM reconstructions of dynein also revealed additional structures above and below the planar ring that are unaccounted for in either previous model (Fig. 1b) (9). Corroborating these observations are 2D EM images of axonemal dynein showing a stain-filled central cavity that results from a structure that is neither an AAA module nor the C domain (11, 12). We show below that this previously unidentified region is likely a coiled-coil structure that spans the diameter of the motor ring and is formed by the long interdomain region found between subunits AAA5 and AAA6. We postulate that this extended chain mediates long-range communication across the face of the motor, in particular between domains AAA1–AAA4 and domains AAA5–AAA6.
Currently, several models are proposed for dynein's power stroke. In these models, ATP hydrolysis induces movement of either the stalk, the tail or an flexible linker between the motor unit and the tail (8, 9, 11, 13, 14). However, none of these models directly predict how forces are transduced from the primary hydrolytic site to the rest of the rest of the domains. From our structural model and normal mode analysis, we propose a detailed model of the conformational rearrangements in the motor unit that lead to the dynein power stroke and a possible mechanism for interdomain communication.
Results
Modeling the Dynein Motor Unit.
Using homology modeling, we systematically constructed a complete structural model of the motor unit of cytoplasmic dynein from D. discoideum. The model includes the six AAA subunits, the linker regions that connect the subunits, and the C domain. All AAA enzymes consist of two structurally conserved units: an α/β Rossman fold subdomain and an α-helical globular subdomain (Fig. 2a). Despite a <20% sequence identity between proteins in the AAA superfamily, up to 50% of equivalent Cα positions are within 2 Å rmsd (15). To produce improved folds for the six AAA subunits of the dynein motor unit, we used 3DJury (Methods) to search for candidate homologs of these subunits and the linker regions that connect them. 3DJury produces a consensus structure template based on the results of multiple independent structure prediction algorithms. The structural templates obtained in this way have consensus scores well above the confidence threshold of 50, which offers a prediction accuracy of 90% [see supporting information (SI) Table 1, for complete bioinformatics results]. Using these initial alignments, we then built atomic models of the AAA domains and their adjacent linkers using the homology modeling suite in Insight II (Methods). To evaluate the accuracy of the models for each subunit, we compared the local environments of the residues in the predicted structures to the population averaged residue environments determined from known structures (SI Table 2). The profiles score show the current model has better fold than earlier proposed models (8).
Fig. 2.
Model of the first AAA domain. (a) Canonical AAA fold (α/β-subdomain and all-α subdomain) with a synapomorphic pre-Sensor-1 β-hairpin insertion. (Inset) Putative site of primary hydrolysis with ATP docked. Walker A (i.e., P-loop) is red, Walker B is blue, Sensor 1 is yellow, and Sensor 2 is orange. (b) Sequence alignment between the Dictyostelium discoideum AAA1 and template structure RuvB domain (PDB ID code 1HQC).
To determine the residues that form dynein's primary catalytic core, which is located between the first and second AAA subunits, we docked an ATP molecule to the glycine-rich P-loop (1969-GPAGTGKT-1976), which is the putative binding site for the nucleotide phosphate tail (Fig. 2a). Within 5 Å of the docked nucleotide, we found conserved residues in the Walker A and Walker B motifs that bind the β and γ NTP phosphates in all P-loop NTPases. These conserved residues found in dynein include K1975 in Walker A; D2021, E2022, and R2025 in Walker B; and R2145 in Sensor 2 (Fig. 2a). These results are consistent with recent biochemical studies showing the dynein mutant K1975T trapped in a strong-binding state and devoid of motile activity (16).
A notable structural feature found in some AAA enzymes including dynein is a β-hairpin insertion (denoted pre-Sensor-1 β-hairpin) in the α/β-subdomain (Fig. 2a and SI Fig. 5). The pre-Sensor-1 β-hairpin insertion defines a phylogenetic superclade that includes the Holliday junction migration motor protein RuvB that was found to be a consensus template for subunits AAA1, AAA3, and AAA5. In the current model, domains AAA1, AAA3, and AAA5 are predicted to have β-hairpin insertions in positions 2056-KENSKEVELLGGKNISLH-2063, 2767-WRTSDHTWI-2776, and 3743-RLGDQDVDF-3751, respectively. In RuvB, the β-hairpin is essential in interacting with other protein complexes; in particular, mutations in the β-hairpin of RuvB diminished its functional interaction with RuvA, preventing branch migration of the Holliday junctions (17, 18). In dynein, the β-hairpin insertions may perform similar functions. For example, the flexibility of the AAA1 β-hairpin (as shown in the normal mode analysis below) and its proximity to the N-terminal tail suggest that the hairpin may be crucial for coordinating AAA1 and tail movement.
To construct a structural model for the C domain, we followed a protocol similar to the one used to construct models of the AAA subunits. We found that the C domain's first 290 residues consist entirely of α-helices, whereas the remaining 128-residue stretch includes five β-strands and terminates with a helix (Fig. 1c). We then determined a family of candidate proteins that represent good structural templates for the two stretches of the C domain. Interestingly, the candidate templates for the first 290 residues were structures of the complement component C3d [Protein Data Bank (PDB) ID code 1GHQ], which attaches to foreign antigens during immune response (19). Using the C3d fragment as template, the first 290 residues acquired a dome-shaped α–α toroidal fold (19). The remaining five β-strands and last helix were built from the plecktrin homology (PH) domain of the Leukemia-associated RhoGEF (PDB ID code 1TXD) (20), which folds into a flattened seven-stranded β-barrel capped with a C-terminal helix. To obtain the complete structure of the C domain, the two subdomains were docked together using rigid body docking (see Materials and Methods). The α-helical stretch shows higher homology with its template structure than the remaining β-strands suggesting that the function of the C domain is performed by the more conserved α-helical stretch.
Interestingly, the interdomain region between subunits AAA5 and AAA6 (denoted as IDR4) is 231 residues long, comparable with the size of an AAA unit (whereas the length of the other interdomain regions IDR1, IDR2, and IDR3 are 79, 103, and 92, respectively) (Fig. 1). If IDR4 possesses a globular fold, then it would manifest as an additional lobe in the reconstructed EM densities (Fig. 1b) (9), and the motor would appear as an octamer. On the other hand, one of the densities on the face of the motor unit forms a long arch that spans the ring formed by the AAA subunits (Fig. 1b), and is suggestive of a coiled coil. The IDR4 is sufficiently long to span the ≈8-nm facial density of the motor unit. Moreover, coil prediction algorithms assign a coiled-coil structure in the AAA5-IDR4 sequence, although the length of the predicted coil varies for dyneins from different species (SI Fig. 6). The search for structural homologs also resulted in several coiled-coil structures. On the basis of these results, we built IDR4 as a coiled coil using the cytoplasmic domain of serine chemotaxis receptor (PDB ID code 1QU7) (21) as a template.
The smaller lump on the face opposite the arching density (see Fig. 1b and SI Fig. 7) could be the remnant of the dimerizing tail used in the EM studies. Another EM study (22) where the dynein tail has been labeled with antibody-Fab tag showed that the tag is not rigid and can be found at various position around the planar ring. The study suggests that the tail domain docks into the center of the ring and that the tail sequence immediately adjacent to the docking point is flexible. Thus, only the point of attachment near of the tail will exhibit a density because the flexible part will be averaged out, making the smaller facial density (Fig. 1b) the more viable candidate for docking of the N-terminal tail.
Motor Unit Organization.
EM reconstruction of the dynein motor at 25 Å resolution from negatively stained D. discoideum dynein shows a complex of seven densities arranged in an asymmetric ring with “rough” and “smooth” edges (Fig. 1b) (9). The smooth side consists of the more conserved AAA1–AAA4 domains, resembling the symmetric oligomers of other large AAA complexes with known crystal structures. The rough side of the motor consists of AAA5, AAA6, and the C domain. Because the sequences of these domains are less conserved, they are not expected to follow the configuration of homomeric AAA complexes, and, therefore, break the symmetry of the dynein motor domain. This broken symmetry might play an important role in effectively propagating force during the power stroke phase of the hydrolysis cycle (see below).
To preserve functionally relevant interactions between domains AAA1–AAA4 and to construct a regular tetramer for this portion of the motor, we superimposed the models of these subunits onto the σ54 RNA polymerase activator NtrC1 (PDB ID code 1NY6) (23). This protein has a known homogenous heptamer structure consisting of AAA subunits with active catalytic sites. Next, we used the vector-quantization method implemented in SITUS (24) to fit the AAA1–AAA4 tetramer to the EM density (Fig. 1b). To obtain a preliminary orientation of the remaining domains, the atomistic models of subunits AAA5, AAA6, and the C domain were fit separately to their corresponding electron density lobes. We also imposed the constraint that AAA5, IDR4, and AAA6 form a continuous peptide. Thus, AAA5 was oriented such that its C terminus faced the coiled coil. Similarly, AAA6 was oriented such that its N terminus faced IDR4 (SI Text).
Finally, to arrive at the complete model, we docked IDR4 and the rest of the interdomain regions to the seven domains using a rigid-body docking protocol and shape complimentarity as criteria. When the complete atomic model was refit to the EM density of the entire motor unit, SITUS (25) ab initio identified the correct orientation of the domains with a correlation of 0.74 (P < 10−316; Fig. 1b; see SI Text for details).
Normal Mode Analysis of Global Motions.
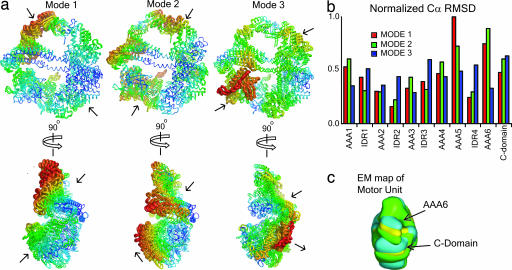
In the final model of the motor unit, the interdomain regions and AAA units form a compact backbone. The most closely packed part of the motor consists of its smooth side where the nucleotide-binding P loops are found. We hypothesized that this compact structure is essential for efficiently transducing forces generated at the ATP hydrolysis site to the extended stalk that contains the microtubule binding domain and is located between subunits AAA4 and AAA5. To investigate this possibility, we performed normal mode analysis to establish the motor unit's dominant modes of motion (Fig. 3 and SI Fig. 8). Normal mode analysis has been shown to accurately identify structural sites that function as pivots and, therefore, can be used to infer global motions of large molecular complexes (26). Normal mode analysis also can be used to explore the intrinsic flexibility of molecular structures. Fig. 3a illustrates atomic displacements associated with the three lowest frequency vibrational modes. The frequencies of modes 2 and 3 are 1.28 and 1.56 times larger than that of mode 1. From Fig. 3 (and the animations provided in SI Movies 1–6), it is evident that the most mobile domains are AAA5, AAA6, and the C domain, whereas AAA1–AAA4 form a more compact structure. These observations are made quantitative in Fig. 3b, which lists the rmsd of the Cα atoms of each subunit for the first three normal modes.
Fig. 3.
Lowest frequency normal modes of dynein motor unit. (a) Superposition of two structures displaced in opposite directions along the normal mode. The size of the backbone is rendered proportional to fluctuations of Cα atoms. Arrows indicate the directions of dominant vibrations. AAA5, AAA6, and C domain exhibit the most prominent variation in domain architecture in the three normal modes. (See SI Movies 1–6.) (b) Mean rmsd of Cα in a domain, normalized by the largest displacement and weighted by inverse frequency. (c) Superposition of reconstructed 3D structures of the motor unit in three distinct stalk conformations from EM studies by Samso and Koonce (9). In the three stalk positions, the side formed by AAA5, AAA6, and C domain exhibit the largest variation (9).
In mode 1, the AAA5 subdomain exhibits an upward motion, whereas AAA6 partially rotates about the IDR4 linker (Fig. 3a). On the other hand, AAA1 to AAA4 and their linkers exhibit minimal displacement. Interestingly, AAA5 is positioned at the base of the stalk that interacts with the microtubule. The fact that the dominant motion of the lowest frequency normal mode occurs at the base of the stalk suggests that the stalk tilts during the motor's power stroke. Mode 2 is characterized by a “squeeze” applied to subunit AAA5 and the C domain coupled with an outward motion by AAA6 (Fig. 3a). Similar to mode 1, in mode 2, AAA1–AAA4 and their linkers exhibit minimal movement. EM 3D reconstructions of the motor unit with stalks positioned at 0°, 25°, and 45° relative to vertical show greatest variation in electron densities corresponding to subunits AAA5 and AAA6 (9) (Fig. 3c). The direction and magnitude of the domain displacements determined for modes 1 and 2 are consistent with these observations (Fig. 3c). For example, the motion predicted to occur in modes 1 and 2 is consistent with the reorientation of subunit AAA6's density observed for different stalk positions (Fig. 3c).
The side views shown in Fig. 3 illustrate that the dominant global motions of the molecule are not coplanar with the motor ring. These results predict that any movement of the stalk or tail will also contain a nonplanar component. A nonplanar bending of the stalk is in agreement with static in situ tomography studies, where stalks are shown to lie obliquely with respect to the dynein ring (27). Moreover, this observation suggests that previous 2D electron micrograph images in which the molecule is forced to lie flat on the substrate might not accurately capture the conformational changes that occur during force generation (27).
In mode 3, the C domain pushes on AAA1. It is not until mode 3 that the linker IDR4 intermediate to AAA5 and AAA6 moves significantly, suggesting that IDR4 is a rigid structure. Similar to modes 1 and 2, mode 3 produces an open-close movement at the main catalytic site, which is found at the interface between AAA1 and AAA2 (Fig. 3 and the animations in SI Movies 1–6). In all three modes, the motion in the catalytic site is smaller compared with the dominant motion in that mode. This observation suggests that, during the mechano-chemical events leading to force production, only minor conformational changes occur in the catalytic site upon binding or release of ATP or ADP. However, this small conformational change is amplified by the rest of the motor domains (as shown below).
The first three normal modes show large movements of the AAA1 pre-Sensor-1 β-hairpin (Fig. 3a) despite minimal displacements of the canonical AAA1 subdomains. Because the β-hairpin projects from the rear of the AAA1 α-β subunit (Figs. 1 and 3), small fluctuations in AAA1 are amplified by the hairpin. This motion makes the hairpin a viable candidate for coupling events in the ATP pocket of subunit AAA1 (the primary hydrolytic site) to the C domain, which has recently been shown to regulate dynein ATPase activity (10). Pre-Sensor-1 β-hairpins in other AAA modules of the motor unit may also mediate interdomain interactions. In other AAA complexes that contain pre-Sensor-1 β-hairpins, this structure plays a significant functional role. For example, in NtrC1, an AAA hexamer that activates bacterial σ54-RNA polymerase holoenzyme, the β-hairpin is used to bind σ54 (28).
Discussion and Conclusion
The predicted structural model of the cytoplasmic dynein motor unit consists of six AAA domains and a C-terminal domain arranged in an asymmetric heptameric ring. The conserved AAA1-AAA4 domains form a tetramer that is organized similarly to other AAA homomer complexes. The less well conserved AAA5, AAA6, and C-terminal domains constitute the rough side of the motor. This asymmetric organization of the motor complex is consistent with the postulated evolutionary origin of the molecule in which the primordial homodimer pairs AAA1–AAA2 and AAA3–AAA4 combined to form a tetramer, with the subunits AAA5 and AAA6 representing later additions to the motor (14).
The β-hairpin insertions inherited by AAA1, AAA3, and AAA5 are unique and functionally important features of AAA enzyme complexes in the PS1BH clade (21). In particular, mutations in the β-hairpin of RuvB abolish its physical and functional interactions with the RuvA DNA recognition protein (18). Because of its close proximity to the primary hydrolytic site as well as the C-terminal and tail domains, the β-hairpin likely plays a role in coordinating the conformational changes involved in the force generating mechano-chemical cycle. This hypothesis can be tested by using site-directed mutations in the sequences corresponding to the β-hairpins.
Another intriguing feature of the homology model is the IDR4 linker that connects subunits AAA5 and AAA6. The model predicts that this structure accounts for the observed density that spans the motor ring. In addition to contributing to the overall rigidity of the motor, IDR4 provides a route for force propagation from the rigid smooth edge of the motor where the nucleotide-binding sites are located to the flexible rough edge. Specifically, IDR4 extends from AAA5, which is at the base of the microtubule-binding stalk, to AAA3, whose nucleotide binding pocket regulates the motor's processivity (14, 16). The IDR4 structure provides a clue to the important question of how distant functional sites communicate with each other to generate a coordinated mechano-chemical cycle.
Studies show that the ATP binding pocket in AAA3 is catalytically active (16, 29). How might this hydrolytic activity in AAA3 affect the microtubule-binding affinity of the stalk? AAA3 domain has two subdomains (Fig. 5), an α/β subdomain and an α subdomain, and the nucleotide binding P-loop is sandwiched between these two subdomains. Manipulations in the P-loop of AAA3 likely induce the open and close movement between these subdomains. Current models assume that perturbations in the AAA3 P loop are propagated to AAA4, which is at the base of the microtubule-binding stalk. Alternatively from our model, the open and close movement in the AAA3 subdomains may be propagated through the coiled coil to the AAA5, which is at the base of the stalk. The orientation of the stalk's globular tip likely dictates the stalks microtubule-binding affinity.
Using 2D images of axonemal dynein, Burgess et al. suggest that a 10-nm segment of the tail forms a linker that curves around the face of the ring (12). In another recent EM study, Meng et al. (22) labeled the cytoplasmic dynein tail with an antibody-Fab tag and used EM and single-particle image analysis. They found the tag at various locations around the planar ring. Collectively, the studies suggest that the tail could dock into the smaller facial lump (Fig. 1b and SI Fig. 7) because the tail's density will be averaged out in the 3D EM reconstruction. In this model, both the tail and IDR4 may interact with the AAA domains, and the less flexible IC4 adds structural stability to the motor unit.
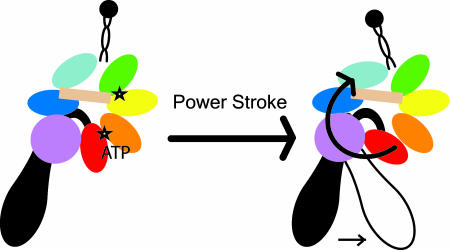
Our analysis of the three lowest frequency normal modes indicates that large scale motions of the motor primarily involve movements in subunits AAA5, AAA6, and the C domain, whereas subunits AAA1–AAA4 function as a rigid structure. This finding is consistent with recent observations from EM reconstructed structures (Fig. 3c) (9, 30). We speculate that the subunits AAA1–AAA4 provide the motor with a stationary backbone against which forces generated in the primary catalytic site can act. This generates conformational changes that propagate sequentially through the C domain, AAA6 and AAA5 and terminate with a movement of the microtubule-binding stalk (Fig. 4).
Fig. 4.
Model of power stroke. Binding ATP or release of ADP+Pi in the hydrolytic sites (indicated by stars) induces conformational change that is primarily propagated through the C domain, AAA6, and AAA5 (see Fig. 1a for domain labels). These domain reorientations cause the stalk or tail to flex about the junction that connects them to the motor unit, thus generating the power stroke.
There are three current models for dynein's power stroke. In the first model, ATP causes a rotation of both the stalk and the tail about the junctions that connect them (8, 9, 13). The second model assumes that a conformational change of the tail swings the motor unit and the stalk together (14). Lastly, the third model assumes that a flexible structural linker between the motor unit and tail bends upon coordinated conformational rearrangements of the AAA domains (10). From our structural model and normal mode analysis, model 2 is unlikely because of the large motions in AAA5 to which the stalk is docked. We propose the possible conformational rearrangements of the domains movements within the motor unit that is the basis of either model 1 or model 2 (Fig. 4). Binding or release of ATP or ADP induces conformational change in the catalytic domain between AAA1 and AAA2. Because of the rigid structure formed by subunits AAA1-AAA4, the disturbance is propagated in a clockwise direction through the C domain, AAA6, and AAA5, causing the microtubule-binding stalk to flex. The change in the angular position of the stalk possibly alters the microtubule binding affinity of the stalk's globular tip. These conformational changes may also play regulatory role, consistent with the findings in enzymatic studies of dynein domain fragments suggesting that the stalk autoinhibits ATP/ADP release in AAA1 and AAA3, and that the C domain also affects the ATPase activity (10).
Using the same 380-kDa dynein fragment from which the EM density is derived, Kon et al. labeled AAA2 and C domain with BFP and the tail with GFP. They found that the dynein adopts at least two conformational states, and the tail undergoes ATP-induced motions relative to the motor domain during transitions between the states (31). On the other hand, an EM study where the tail fragment has been labeled with a FAB-antibody tag, the tail did not have a preferred orientation (22). This discrepancy in results may be due to a difference in experimental conditions, in particular a preparative difference in the nucleotide states. The resolution of this discrepancy warrants further studies. In particular, we propose FRET studies using constructs in which the tail, the motor domain, and the putative coiled coil are labeled with fluorescent proteins.
Materials and Methods
Model Building.
The sequence of the motor domain of cytosolic dynein heavy chain of slime mold D. discodeum (GenBank accession no. P34036) was submitted for threading to 3DJury (http://bioinfo.pl/meta) (32, 33). The templates used in building the AAA1, AAA2, and AAA4 were the Holliday junction migration motor protein RuvB from Thermus thermophilus HB8 (PDB ID code 1HQC; ref. 18), clamp loader gamma complex of Escherichia coli DNA polymerase III (PDB ID code 1JR3; ref. 34), and eukaryotic clamp loader (PDB ID code 1SXJ; ref. 35), respectively. AAA3 and AAA5 were modeled from the same model used by AAA1. Similar to AAA2, AAA6 was built from the clamp loader. The C domain is modeled from complement C3d (PDB ID code 1GHQ; ref. 19) and pH domain of leukemia-associated rhogef (PDB ID code 1TXD; ref. 20). We constructed the atomic models using the Homology suite of INSIGHTII (Accelrys, San Diego, CA). To arrange the domains in a heptamer, we used QRANGE (24) and COLORES (25) (both in SITUS; ref. 24) to fit the individual domain models to a EM map of the dynein motor domain of D. discoideum (9). Finally, we eliminated the clashes in the model structure by running equilibrium simulations using a simplified protein model followed by all-atom reconstruction (36, 37). The rmsd between the models before and after reconstruction is 2.5 Å.
Normal Mode Analysis.
Coordinates of the constructed model were submitted to ELNemo server (http://igs-server.cnrs-mrs.fr/elnemo/index.html) (38) to compute the first 10 normal modes.
A detailed description of the model-building and refinement and normal mode analysis can be found in SI Text.
Supplementary Material
Acknowledgments
We thank M. Koonce for generously providing the EM maps, B. Temple for help and discussions on molecular modeling, and K. Wilcox and S. Barton for a careful reading of the manuscript. This work was supported in part by the Muscular Dystrophy Association Grant MDA3720 (to N.V.D.), American Heart Association Grant 0665361U (to N.V.D.), and National Institutes of Health Grant R01 GM078994-01 (to T.C.E.).
Abbreviations
- PDB
Protein Data Bank.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: Atomic coordinates of the model have been deposited to the Protein Data Bank, www.pdb.org (PDB ID code 2GF8).
This article contains supporting information online at www.pnas.org/cgi/content/full/0602867103/DC1.
References
- 1.Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K. Nature. 2003;421:715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- 2.Holzbaur EL, Vallee RB. Annu Rev Cell Biol. 1994;10:339–372. doi: 10.1146/annurev.cb.10.110194.002011. [DOI] [PubMed] [Google Scholar]
- 3.King SM. Biochim Biophys Acta. 2000;1496:60–75. doi: 10.1016/s0167-4889(00)00009-4. [DOI] [PubMed] [Google Scholar]
- 4.LaMonte BH, Wallace KE, Holloway BA, Shelly SS, Ascano J, Tokito M, Van Winkle T, Howland DS, Holzbaur EL. Neuron. 2002;34:715–727. doi: 10.1016/s0896-6273(02)00696-7. [DOI] [PubMed] [Google Scholar]
- 5.Qin H, Rosenbaum JL, Barr MM. Curr Biol. 2001;11:457–461. doi: 10.1016/s0960-9822(01)00122-1. [DOI] [PubMed] [Google Scholar]
- 6.Hirokawa N. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 7.Neuwald AF, Aravind L, Spouge JL, Koonin EV. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 8.Mocz G, Gibbons IR. Structure (London) 2001;9:93–103. doi: 10.1016/s0969-2126(00)00557-8. [DOI] [PubMed] [Google Scholar]
- 9.Samso M, Koonce MP. J Mol Biol. 2004;340:1059–1072. doi: 10.1016/j.jmb.2004.05.063. [DOI] [PubMed] [Google Scholar]
- 10.Hook P, Mikami A, Shafer B, Chait BT, Rosenfeld SS, Vallee RB. J Biol Chem. 2005;280:33045–33054. doi: 10.1074/jbc.M504693200. [DOI] [PubMed] [Google Scholar]
- 11.Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K. Nature. 2003;421:715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- 12.Burgess SA, Walker ML, Sakakibara H, Oiwa K, Knight PJ. J Struct Biol. 2004;146:205–216. doi: 10.1016/j.jsb.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Vale RD. J Cell Biol. 2000;150:F13–F19. doi: 10.1083/jcb.150.1.f13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asai DJ, Koonce MF. Trends Cell Biology. 2001;11:196–202. doi: 10.1016/s0962-8924(01)01970-5. [DOI] [PubMed] [Google Scholar]
- 15.Smith GR, Contreras-Moreira B, Zhang X, Bates PA. J Struct Biol. 2004;146:189–204. doi: 10.1016/j.jsb.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 16.Kon T, Nishiura M, Ohkura R, Toyoshima YY, Sutoh K. Biochemistry. 2004;43:11266–11274. doi: 10.1021/bi048985a. [DOI] [PubMed] [Google Scholar]
- 17.Han YW, Iwasaki H, Miyata T, Mayanagi K, Yamada K, Morikawa K, Shinagawa H. J Biol Chem. 2001;276:35024–35028. doi: 10.1074/jbc.M103611200. [DOI] [PubMed] [Google Scholar]
- 18.Yamada K, Kunishima N, Mayanagi K, Ohnishi T, Nishino T, Iwasaki H, Shinagawa H, Morikawa K. Proc Natl Acad Sci USA. 2001;98:1442–1447. doi: 10.1073/pnas.031470598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szakonyi G, Guthridge JM, Li DW, Young K, Holers VM, Chen XJS. Science. 2001;292:1725–1728. doi: 10.1126/science.1059118. [DOI] [PubMed] [Google Scholar]
- 20.Kristelly R, Gao G, Tesmer JJG. J Biol Chem. 2004;279:47352–47362. doi: 10.1074/jbc.M406056200. [DOI] [PubMed] [Google Scholar]
- 21.Iyer LM, Leipe DD, Koonin EV, Aravind L. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 22.Meng X, Samso M, Koonce MP. J Mol Biol. 2006;357:701–706. doi: 10.1016/j.jmb.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 23.Lee SY, De la Torre A, Yan DL, Kustu S, Nixon BT, Wemmer DE. Genes Dev. 2003;17:2552–2563. doi: 10.1101/gad.1125603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wriggers W, Milligan RA, McCammon JA. J Mol Graphics Model. 1998;16:283. [Google Scholar]
- 25.Wriggers W, Chacon P. Structure (London) 2001;9:779–788. doi: 10.1016/s0969-2126(01)00648-7. [DOI] [PubMed] [Google Scholar]
- 26.Bahar I, Rader AJ. Curr Opin Struct Biol. 2005;15:586–592. doi: 10.1016/j.sbi.2005.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lupetti P, Lanzavecchia S, Mercati D, Cantele F, Dallai R, Mencarelli C. Cell Motil Cytoskeleton. 2005;62:69–83. doi: 10.1002/cm.20084. [DOI] [PubMed] [Google Scholar]
- 28.Rappas M, Schumacher J, Beuron F, Niwa H, Bordes P, Wigneshweraraj S, Keetch CA, Robinson CV, Buck M, Zhang XD. Science. 2005;307:1972–1975. doi: 10.1126/science.1105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi Y, Edamatsu M, Toyoshima YY. Proc Natl Acad Sci USA. 2004;101:12865–12869. doi: 10.1073/pnas.0403429101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burgess SA, Walker ML, Thirumurugan K, Trinick J, Knight PJ. J Struct Biol. 2004;147:247–258. doi: 10.1016/j.jsb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 31.Kon T, Mogami T, Ohkura R, Nishiura M, Sutoh K. Nat Struct Mol Biol. 2005;12:513–519. doi: 10.1038/nsmb930. [DOI] [PubMed] [Google Scholar]
- 32.Ginalski K, Elofsson A, Fischer D, Rychlewski L. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- 33.Ginalski K, Rychlewski L. Nucleic Acids Res. 2003;31:3291–3292. doi: 10.1093/nar/gkg503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeruzalmi D, O'Donnell M, Kuriyan J. Cell. 2001;106:429–441. doi: 10.1016/s0092-8674(01)00463-9. [DOI] [PubMed] [Google Scholar]
- 35.Bowman GD, O'Donnell M, Kuriyan J. Nature. 2004;429:724–730. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- 36.Ding F, Dokholyan NV. Trends Biotechnol. 2005;23:450–455. doi: 10.1016/j.tibtech.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 37.Ding F, Prutzman KC, Campbell SL, Dokholyan NV. Structure (London) 2006;14:5–14. doi: 10.1016/j.str.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 38.Suhre K, Sanejouand YH. Nucleic Acids Res. 2004;32:W610–W614. doi: 10.1093/nar/gkh368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.