Abstract
Sorbitol and sucrose are major products of photosynthesis distributed in apple trees (Malus domestica Borkh. cv. “Greensleeves”) that affect quality in fruit. Transgenic apple plants were silenced or up-regulated for sorbitol-6-phosphate dehydrogenase by using the CaMV35S promoter to define the role of sorbitol distribution in fruit development. Transgenic plants with suppressed sorbitol-6-phosphate dehydrogenase compensated by accumulating sucrose and starch in leaves, and morning and midday net carbon assimilation rates were significantly lower. The sorbitol to sucrose ratio in leaves was reduced by ≈90% and in phloem exudates by ≈75%. The fruit accumulated more glucose and less fructose, starch, and malic acid, with no overall differences in weight and firmness. Sorbitol dehydrogenase activity was reduced in silenced fruit, but activities of neutral invertase, vacuolar invertase, cell wall-bound invertase, fructose kinase, and hexokinase were unaffected. Analyses of transcript levels and activity of enzymes involved in carbohydrate metabolism throughout fruit development revealed significant differences in pathways related to sorbitol transport and breakdown. Together, these results suggest that sorbitol distribution plays a key role in fruit carbon metabolism and affects quality attributes such as sugar–acid balance and starch accumulation.
Keywords: gene silencing, sugar–acid balance, translocation, starch accumulation
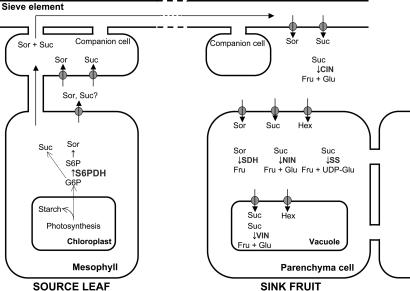
Apples are in the family Rosaceae, which includes temperate species with fleshy fruit. The family produces sorbitol, sucrose, and starch as primary products of photosynthesis (Fig. 1). Sorbitol is synthesized via reduction of glucose-6-phosphate to sorbitol-6-phosphate by aldose-6-phosphate reductase (EC 1.1.1.200), also called sorbitol-6-phosphate dehydrogenase (S6PDH). Sorbitol is the main sugar present in apple leaves and is transported in phloem with sucrose (Fig. 1) (1). Sugars are distributed through a network of sieve elements to sink tissues such as developing fruit, seed, and leaves in a complex process regulated by photosynthetic rate, phloem loading, long-distance translocation and unloading, postphloem transport, and metabolism within sink tissues (2, 3). Transporters of sugar alcohols like sorbitol and other structural components involved in phloem unloading of sorbitol into the apoplast of apple fruit have been identified (4–6).
Fig. 1.
Carbon metabolism in apple. Sorbitol (Sor) and sucrose (Suc) are synthesized in leaf from G6P. Sorbitol is synthesized by S6PDH and sorbitol dehydrogenase. S6PDH is the regulatory step. After translocation to the fruit sink, sorbitol is converted to fructose (Fru) by SDH and sucrose is converted to fructose and glucose (Glu) by invertases (NIN, VIN). Starch synthesis and breakdown (SS) also occurs in fruit. Hex, hexose.
In sink tissues such as fruit, sucrose is metabolized by invertases and sucrose synthase, whereas sorbitol is converted to fructose by sorbitol dehydrogenase (SDH, EC 1.1.1.14) (7, 8). Expression of SDH in developing apple fruit is highest 2–3 weeks after bloom, a period sensitive to carbon availability (9). Additionally, sorbitol and other sugars may regulate expression of SDH mRNA and protein in pear fruit slices (10).
In this article, we describe how sorbitol distribution affects fruit quality using transgenic apple plants with altered levels of S6PDH. Understanding the relationship between sorbitol accumulation and fruit quality is important not only for apple but potentially for other fruits species within the family Rosaceae that synthesize and accumulate sorbitol.
Results
S6PDH Expression and Photosynthate Levels in Leaves.
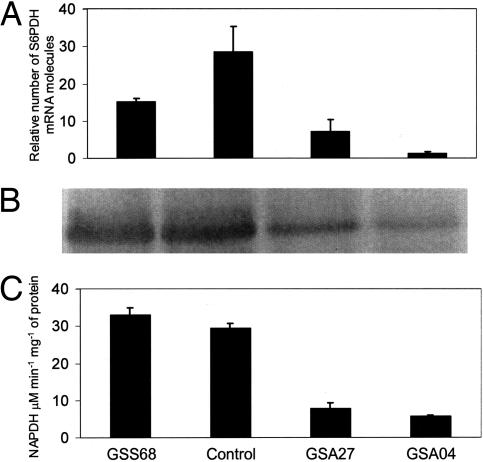
S6PDH transcript levels were examined in transgenic 3-y-old field-grown “Greensleeves” trees on M26 rootstocks, which had set fruit. Three trees each of three clones were selected for further analysis. Clone GSS68 mildly overexpressed S6PDH, and antisense clones GSA27 and GSA04 had different levels of S6PDH suppression. S6PDH transcription was reduced in GSA04 and GSA27, as were protein concentration and activity, but was unexpectedly also reduced in GSS68 (Fig. 2A). In GSA27 and GSA04, the concentration of S6PDH protein was significantly reduced (Fig. 2B); GSS68 had a slight increase in S6PDH (Fig. 2B). S6PDH activity in GSA27 and GSA04 was 20% and 10% of the control, respectively (Fig. 2C).
Fig. 2.
S6PDH RNA expression, protein concentration, and activity in mature leaves of control and transgenic clones. (A) Steady-state transcript levels of S6PDH as revealed by real-time PCR (n = 3). Bars are means and standard errors for each line. (B) Western blot analyses of 20 μg of crude protein extract from each line. (C) S6PDH activity expressed as mmol NADPH·min−1·g−1 of fresh weight (n = 3). Bars are means and standard errors for each line.
Sorbitol accumulation in GSA27 was 30% of the control, and only 22% in GSA04. However, sucrose increased 3.3- and 3.7-fold in GSA27 and GSA04, respectively (Table 1). The ratio of sorbitol to sucrose decreased from 3.4 in the control to 0.3 and 0.2 in GSA27 and GSA04, respectively. In GSS68, leaf sorbitol content was 109% of the control, and the ratio of sorbitol to sucrose increased from 3.4 to 3.8. Starch content increased 1.6-fold for GSA27, 2.1-fold for GSA04, and was unchanged in GSS68 (Table 1).
Table 1.
Nonstructural carbohydrates in five mature, midshoot leaves from each of three control and/or transgenic plants
| Line | Sorbitol, mg·g−1 FW | Sucrose, mg·g−1 FW | Sorbitol-to-sucrose ratio | Starch, mg·g−1 DW |
|---|---|---|---|---|
| GSS68 | 34.98 ± 1.43 | 9.19 ± 1.14 | 3.8 | 39.60 ± 1.47 |
| Control | 32.03 ± 1.60 | 9.46 ± 0.49 | 3.4 | 43.25 ± 3.93 |
| GSA27 | 9.58 ± 0.47* | 31.69 ± 1.15 | 0.3 | 70.43 ± 2.76* |
| GSA04 | 7.08 ± 0.35* | 35.07 ± 4.16 | 0.2 | 89.29 ± 14.76* |
FW, fresh weight; DW, dry weight.
*Significant difference from control at P ≥ 0.05.
Characterization of Shoots and Leaves.
The altered composition of photoassimilates affected vegetative growth (Table 2), leading to distinct phenotypic differences. Shoot lengths were ≈115% of controls in line GSS68 and 90% in line GSA04. Similar trends were seen for leaf area and leaf mass by area, but they were not significant. Leaf net carbon assimilation was significantly lower in both antisense clones at 8:20, 10:15, 12:30, and 14:20 h (Table 6, which is published as supporting information on the PNAS web site). At 16:30 h, only the GSA04 clone was significantly lower. Stem water potential was significantly reduced in both antisense clones at 8:20 h, but only in GSA04 at 10:15 and 12:30 h. Stem water potential and net carbon assimilation for clone GSS68 were similar to the control at all sampling times (Table 6).
Table 2.
Phenotypes of 10 shoots or 50 leaves from each of three individual control or transgenic plants
| Line | Shoot length, cm | Leaf area, cm2 | Leaf mass area−2, g·cm−2 |
|---|---|---|---|
| GSS68 | 103.0 ± 2.22* | 30.74 ± 0.66 | 14.44 ± 0.04* |
| Control | 88.8 ± 3.62 | 28.79 ± 2.60 | 17.01 ± 0.78 |
| GSA27 | 85.2 ± 5.90 | 26.87 ± 1.42 | 18.16 ± 0.37 |
| GSA04 | 80.4 ± 0.87* | 25.48 ± 2.94 | 18.95 ± 0.39 |
*Significant difference from control at P ≥ 0.05.
Analysis of Fruit Pedicel Phloem Exudates.
We further assessed the impact of altered leaf sorbitol concentration by examining phloem exudates from fruit pedicels at 60, 90, and 120 days after full bloom (DAFB). Table 3 shows sorbitol and sucrose extrusion rates during 24 h at 90 DAFB. The ratio of sorbitol to sucrose differed among clones and was similar to the ratio found in leaves. The sorbitol content was 108% of the control in GSS68, but only 60% and 81%, respectively, in the suppressed GSA27 and GSA04. These two lines also extruded ≈16% and 41% less total distributed carbohydrate than the control, respectively (Table 3).
Table 3.
Sorbitol and sucrose in phloem exudates from four fruit stalks per tree from four different trees at 90 DAFB
| Line | Sorbitol, mg·day−1 | Sucrose, mg·day−1 | Sorbitol-to-sucrose ratio | Sorbitol plus sucrose |
|---|---|---|---|---|
| GSS68 | 41.2 ± 4.8 | 12.8 ± 1.1 | 3.2 | 54.0 ± 4.9 |
| Control | 38.3 ± 3.1 | 17.2 ± 1.4 | 2.2 | 55.5 ± 2.6 |
| GSA27 | 15.2 ± 3.7* | 31.2 ± 3.4* | 0.5 | 46.4 ± 5.0 |
| GSA04 | 7.0 ± 1.0* | 25.8 ± 3.4* | 0.3 | 32.8 ± 3.4* |
*Significant difference from control at P ≥ 0.05.
Determination of Nonstructural Carbohydrate Content in Fruit.
Nonstructural carbohydrate concentrations in fruit were determined at harvest (Table 4). In the sorbitol-suppressed lines GSA04 and GSA27, glucose concentrations were 234% and 231% of the control, respectively, but fructose was reduced to 82% in both lines. There was also less sorbitol in these fruits but no significant difference between the sense line and the control. In contrast, sucrose concentrations were similar in all lines. Line GSA04 had less starch than control fruit.
Table 4.
Mean nonstructural carbohydrate and malic acid content of five apple fruits each from three different trees
| Line | Sucrose, mg·g−1 | Glucose, mg·g−1 | Fructose, mg·g−1 | Sorbitol, mg·g−1 | Total, mg·g−1 | Starch, mg·g−1 | Malic acid, mg·g−1 |
|---|---|---|---|---|---|---|---|
| GSS68 | 208.2 ± 15.3 | 40.4 ± 12.4 | 279.6 ± 13.1 | 69.6 ± 5.0 | 597.8 ± 8.7 | 186.7 ± 14.8 | 89.2 ± 4.8 |
| Control | 197.1 ± 20.8 | 62.1 ± 16.4 | 317.0 ± 20.1 | 65.6 ± 7.9 | 641.8 ± 12.7 | 151.3 ± 19.3 | 64.5 ± 5.4 |
| GSA27 | 181.7 ± 3.7 | 145.4 ± 28.9* | 260.3 ± 9.3* | 48.6 ± 2.5* | 636.0 ± 29.5 | 121.1 ± 6.3 | 54.7 ± 1.1 |
| GSA04 | 214.8 ± 14.5 | 143.7 ± 32.2* | 260.7 ± 10.8* | 38.0 ± 5.0* | 657.2 ± 42.6 | 104.8 ± 4.7* | 43.2 ± 2.5* |
*Significant difference from control at P ≥ 0.05.
Determination of Fruit Characteristics.
There were no significant differences among lines in fruit fresh weight. Fruit from all transgenic lines was significantly firmer (Table 5). Soluble solids content was ≈10% higher in GSA04 and GSA27 than in control or GSS68 apples (Table 5). Malic acid was significantly reduced in GSA04 (Table 4), as was titratable acidity (TA) (Table 5). GSA04 fruit juice pH at harvest was higher than the other lines (Table 5). Juice from the sense line GSS68 had twice the TA of GSA04, and lower pH than both antisense lines (Table 5).
Table 5.
Mean quality attributes of cortical tissue from five fruits each from three trees at harvest (120 DAFB)
| Line | Weight, g | Firmness, N | Soluble solids content, % | pH | TA, % |
|---|---|---|---|---|---|
| GSS68 | 167.70 ± 4.18 | 99.37 ± 1.55* | 13.23 ± 0.34 | 3.355 ± 0.015 | 0.82 ± 0.03 |
| Control | 159.17 ± 6.44 | 90.95 ± 2.15 | 13.15 ± 0.36 | 3.457 ± 0.031 | 0.64 ± 0.07 |
| GSA27 | 158.90 ± 4.43 | 99.18 ± 0.51* | 14.70 ± 0.31* | 3.531 ± 0.037 | 0.50 ± 0.05 |
| GSA04 | 165.13 ± 5.50 | 95.60 ± 1.19* | 14.65 ± 0.11* | 3.584 ± 0.017* | 0.42 ± 0.03* |
*Significant difference from control at P ≥ 0.05.
Activities of Key Enzymes Regulating Sugar Metabolism.
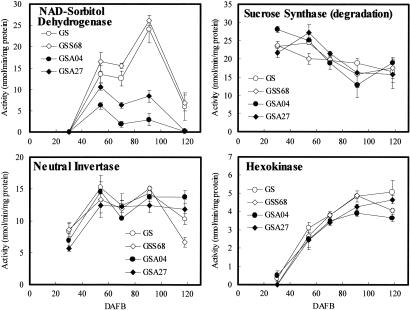
Key enzymes in fruit sugar metabolism were measured to define pathways in fruit affected by sorbitol (Fig. 3). In control fruit, SDH activity was absent at 30 DAFB, peaked at 91 DAFB, and was very low by 118 DAFB. SDH activity of GSS68 was similar to the control, but tended to be higher. Lines GSA04 and GSA27 had reduced SDH activity, especially at 91 DAFB, and it was undetectable at harvest.
Fig. 3.
Activities of sugar metabolism enzymes in control (○) and transgenic apple fruit, GSS68 (◊), GSS04 (●), and GSS27 (♦) during development. Values are means (±SE) of three replicates.
Sucrose synthase (SS) activity decreased during fruit development in all lines (Fig. 3). Neutral invertase (NIN) activity in all lines was lowest at 30 DAFB and then increased and remained roughly constant thereafter (Fig. 3). Vacuolar invertase (VIN) and cell wall-bound invertase (CIN; data not shown) had similar patterns. Fructose kinase (FK) was most active in all lines at 30 DAFB and then decreased gradually (data not shown). Hexokinase (HK) activity in all lines was lowest at 30 DAFB and then increased gradually (Fig. 3).
Expression of Genes for Key Proteins Regulating Sorbitol Metabolism.
We used real-time quantitative TaqMan PCR to analyze individual alleles of sorbitol transport (SOT) and metabolism (SDH) proteins. For each target gene, there was more than one sequence representing allelic forms; we analyzed 21 sequences corresponding to the eight alleles used in this analysis (four SOT and four SDH). PCR primers and TaqMan probes were designed based on sequence information in the Malus × domestica (apple) unigene set at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov). Expression of four alleles each for sorbitol transport and sorbitol dehydrogenase (SOT and SDH) was measured. Expression of SDH2, SDH3, and SDH4 were higher than that of SDH5 (Fig. 4, which is published as supporting information on the PNAS web site). SDH2 expression in control and GSS68 lines was generally higher than in GSA04 and GSA27, except at 54 and 70 DAFB. At 30 DAFB, expression of SDH2 was detected, but no enzyme activity was detected (Figs. 3 and 4). SDH3 expression was higher in control and GSS68 fruit than in GSA04 and GSA27 fruit, except at 30 DAFB.
At 30 DAFB, expression of SOT1, SOT3, and SOT4 was higher in control and GSS68 apples than in GSA04 and GSA27, although expression of SOT2 in GSA04 and GSA27 was higher at 54 DAFB (Fig. 4). We did not detect prominent differences in SOT expression among control, sense, and antisense clones.
Discussion
S6PDH Suppression and Carbon Partitioning in Leaves.
S6PDH expression correlated well with S6PDH protein and enzyme activity, confirming the role of S6PDH as a key limiting step in sorbitol biosynthesis (Fig. 2) (11). S6PDH suppression decreased sorbitol in both source and sink tissues. In mature leaves, sucrose accumulated when sorbitol was suppressed (Table 1). Kanamaru et al. (12) reported that increased sucrose accompanied decreased sorbitol in transgenic apple leaves (Malus domestica Borkh. cv. “Orin”) suppressed for S6PDH. Cheng et al. (13) provide more comprehensive evidence for a compensatory relationship between sorbitol and sucrose in transgenic apple plants with suppressed S6PDH and unchanged photosynthetic assimilation rate. A unique observation here is that line GSS68, which mildly overexpressed S6PDH, accumulated sucrose and starch at levels similar to untransformed controls. Sorbitol-suppressed lines also accumulated more starch (Table 1). Cheng et al. (13) showed that leaf photosynthetic rate in young plants is similar at normal and saturating CO2 concentrations and proposed that plants suppressed for sorbitol synthesis down-regulate fructose 1, 6-bisphosphatase and increase starch accumulation. However, they used young trees, and our trees were >4 years old, growing in the field and bearing fruit. We propose that carbon distribution into starch in leaves of antisense lines may indicate slower translocation of photoassimilate to sinks in antisense trees than in controls. The trees displayed distinct morphological changes (Tables 2 and 6) easily seen in older trees. GSS68 trees were larger than controls, whereas trees of both suppressed lines were more compact. These morphological differences may be due to different carbon assimilation rates (Table 6) and may only be apparent in older, fruit-bearing trees under field conditions. In potato, down-regulation of the sucrose transporter impaired root growth and tuber yield (14). Leggewie et al. (15) found that overexpression of the sucrose transporter altered leaf carbon partitioning and tuber metabolism but not tuber morphology. Altered expression of S6PDH in lines GSA04, GSA27, and GSS68 had little effect on plant morphology in the first 2 years after grafting. Nevertheless, lines with reduced S6PDH were smaller after 3 years, whereas GSS68 trees were more vigorous. Transgenic tobacco and potato plants with reduced expression of the sucrose: proton cotransporter (16, 17) accumulated soluble carbohydrates and produced curled, bleached leaves. In our lines, the leaf mass-to-area ratio increased as the sorbitol-to-sucrose ratio decreased in transgenic clones, and leaves tended to be smaller in the suppressed lines GSA27 and GSA04. The higher leaf mass-to-area ratio in leaves of low sorbitol-producing lines could be explained partly by the increased starch.
Suppression of Sorbitol Partitioning to Fruit.
Altered sorbitol distribution to fruit reduced total carbohydrate in phloem exudates, which were 84% and 59% of the control for GSA27 and GSA04, respectively. Furthermore, the increased starch accumulation and decreased sorbitol during the afternoon indicates that carbon translocation may not proceed at the same rate in antisense and control lines. Line GSS68 translocated less sucrose and had a higher sorbitol-to-sucrose ratio (3.2) than the control (2.2). Analyses of phloem exudates showed that the pattern of distributed carbon resources (sorbitol and/or sucrose) closely resembled that observed in photosynthetic leaves in all lines.
Nonstructural Carbohydrates in Fruit.
Altered S6PDH expression changed carbohydrate concentration in fruit, especially of glucose and malic acid. In GSA04, glucose increased 2.4-fold and starch decreased ≈60% (Table 4). Although the differences were not significant, glucose tended to decrease in line GSS68, whereas starch and malic acid increased (Table 4). Fruit analysis at different developmental stages revealed less starch in line GSA04 as early as 40 DAFB (data not shown). This demonstrates that starch concentration differences were likely because of slower accumulation rather than premature starch degradation. These results contradict BerŸter et al. (7), who proposed that the glucose moiety of sucrose contributes to starch synthesis in fruit. We do not know why the high sucrose in leaves and glucose in fruit of our transgenic lines reduced starch accumulation in fruit. However, we hypothesize that sorbitol contributes to the hexose phosphate pool via fructose and the two related enzymes, sorbitol dehydrogenase and fructokinase. This pathway may be more efficient than the one regulated by sucrose, which uses sucrose synthase and UDP-glucose pyrophosphorylase. Alternatively, lower carbon availability to the fruit, evident from the phloem exudate data, may lead to lower starch accumulation. TA (Table 5) and malic acid concentration (Table 4) in line GSA04 were 33% lower than in control fruit, and ≈52% lower than line GSS68 (Table 4). These data suggest that malic acid synthesis is favored when sorbitol is the predominant sugar transported to the fruit.
Overall Fruit Quality.
Decreased leaf sorbitol to sucrose ratio in antisense lines resulted in lower fresh fruit weight (data not shown). BerŸter (18) showed a relationship between acidity and fresh weight in “Usterapfel” (Malus domestica Borkh.), a mutant with 10-fold increased malic acid. We expected reduced firmness in antisense lines, if anything, because of low starch, low acidity and higher soluble solids (19). However, we found no unfavorable effect of the ratio of sorbitol to sucrose on fruit firmness in any of the lines. Soluble solid content over three years was higher in lines with low sorbitol to sucrose ratio, suggesting that increased sucrose favors soluble sugar rather than starch accumulation in fruit. This may be due to sucrose contributing to glucose formation, which compensated for reduced fructose. Acidity decreased as soluble solids content increased, suggesting that sugar–acid balance was directly correlated to sorbitol concentration or to the sorbitol/sucrose ratio in leaves.
Sugar Metabolism and Fruit Quality.
Of nine carbon-metabolizing enzymes, only SDH had altered activity in transgenic fruit, and this correlated with sorbitol content in phloem exudates (Table 3). Interrupting carbon assimilation by girdling decreased SDH activity in apple fruit (20), and in vitro sorbitol treatment reversed this effect (21). This suggests sorbitol supply may regulate SDH activity in fruit. Our results provide more in vivo evidence to support this hypothesis.
We detected no significant differences in activities of SS (cleavage), VIN (vascular acid invertase), CIN, NIN, FK, HK, SS (synthesis), SPS (sucrose phosphate synthase), and ADP-glucose pyrophosphorylase, all crucial enzymes in sugar metabolism. Activities of SS (cleavage), VIN, CIN, and NIN are also closely related to sink strength, because they are the first enzymes to catabolize translocated sucrose in apple. In general, they contribute to sequential stages of sink initiation, expansion, storage and/or maturation (22). Girdling decreased NIN activity during active starch synthesis (20), which suggests that this enzyme is important for sucrose unloading at that stage. These enzyme activities were not altered in our transgenic fruit, despite changed sucrose content in phloem exudates, so the differences in sucrose may be too small to affect enzyme activities directly.
Sugars can act as signaling molecules (23), so sorbitol may regulate gene expression. SDH activity, protein, and mRNA were increased in sliced tissues of Asian pear fruit not only by sorbitol but also by sucrose, glucose, and mannitol (10). Hexose repressed gene expression of mannitol dehydrogenase in cultured celery cells, and it was suggested that HK and sugar phosphorylation were involved in signaling this repression (24). Here, reduced sorbitol in phloem of antisense lines correlated with suppressed expression of SDH2 and SDH3 in fruit.
No prominent differences in sorbitol transporters were observed in lines with altered sorbitol synthesis, although sorbitol transporters are an important component of sorbitol metabolism (Fig. 4). SOT gene expression was low in tissues of watercored fruit (25), and high sorbitol concentrations are associated with this fruit disorder, suggesting that SOT expression may be regulated by factors other than sugars. We detected no significant changes in gene expression of AGPase, four hexose transporters (HXT1, HXT2, HXT3, and HXT4), NIN1, NIN2, SS1, and SS2 in our transgenic apple fruit (data not shown).
Conclusions
The allocation and distribution of carbon resources from leaves plays a key role in how these resources are used in sink tissues like fruit, where they can affect downstream traits like fruit quality. We successfully reduced carbon allocation to sorbitol through the transgenic suppression of sorbitol synthesis in leaves, increasing the allocation to sucrose and affecting the composition of distributed carbon in fruit, which gave insight into sorbitol's role in apple fruit metabolism. Changing sorbitol availability in leaves altered patterns of glucose, fructose, starch, and malic acid accumulation in apple fruit. Both glucose and malic acid content were affected by the relative amounts of sucrose and sorbitol. The balance and interactions between sweetness and acidity are more important than either taken alone (26), so sorbitol metabolism is key to understanding compositional changes in apple. Sorbitol metabolism in apple and other sorbitol-accumulating members of the Rosaceae family could be a good target for breeding and genetics to manipulate fruit quality traits.
Experimental Procedures.
Plant Transformation and Selection of Transgenic Apple Plants.
The binaries pDU93.0305 with S6PDH cDNA (27) from apple in a sense orientation and pDU93.0330 with S6PDH cDNA in an antisense orientation were constructed as in Tao et al. (11) and introduced into Agrobacterium strain EHA101 as in Wen-jun and Forde (28). Apple transformation was similar to James et al. (29) and involved Agrobacterium-mediated transformation of leaf discs from the apple cultivar “Greensleeves” (30). Selection was achieved with the antibiotic kanamycin. β-glucuronidase (GUS)-specific activity was measured by using fluorometry (31). Selected GUS-positive shoots were rooted in the presence of kanamycin. These plants were then acclimatized in the greenhouse, planted in the field, and allowed to grow on their own roots.
Plant Materials, Experimental Design and Data Collection.
Selected transgenic lines were grafted on M26 rootstock and planted in a randomized block design with 3 × 3-m spacing. Trees were irrigated by using under-canopy microsprinklers and trained to a central leader system. The orchard was managed with standard horticultural practices regarding pruning, fertilization, and weed control. Fruit were thinned to optimize and standardize fruit size.
Five apples were collected from each of three replicate trees of each genotype when firmness of control fruit was between 90 and 95 Newtons. Fruit was transported on ice to the laboratory for analyses. Apples were weighed and evaluated for carbohydrate content, firmness, soluble solids, TA, and pH. Immediately after weighing, some cortical tissue was frozen and kept at −80°C until needed for carbohydrate analysis. Firmness was measured at two peeled points along the equatorial region of each fruit by using a penetrometer with an 11-mm probe. Juice from cortical tissue was used to measure soluble solids (°Brix) by using a refractometer. TA and pH were measured with an automatic titration system.
Ten mature midshoot leaves were chosen at random from each tree and cut at the petiole base. Leaf area was estimated with a Li-3100 leaf area meter (Li-Cor, Lincoln, NE). Leaf samples for dry matter estimates were dried at 60°C to constant weight. Leaf samples for carbohydrate analysis were frozen in liquid nitrogen and kept at −80°C until needed. Carbon-assimilation rates and stomatal conductance of leaves were measured with a portable computerized open-system IRGA (LI-6400; LI-COR under ambient temperature and relative humidity. A cool light source (6400–02 LED) under software control was used. Carbon assimilation rates were measured on three different plants for each transgenic or control line. Stem water potential was measured with a pressure chamber (32) at midday 4 days later. Three to five leaves were randomly selected from each replicate tree of each genotype. Shoot length of current season growth was measured at the end of the growing season.
Phloem Exudate Collection.
Phloem exudates were collected from randomly selected healthy growing fruit stalks as described by Klages et al. (33). Fruit was removed, and the stalk was recut in water and inserted into a 1.5-ml microfuge tube. The tube contained 1.4 ml of 10 mM EDTA (pH 7) 20 μg·ml−1 chloramphenicol in 0.7% agarose. Exudates were collected every 6 h over a 24-hour period. Zero EDTA controls were used at each sampling time. Exudates were extracted overnight in methanol, dried under a stream of nitrogen, and resuspended in sterile water. Soluble carbohydrate was determined by HPLC as described by Perez et al. (34).
Carbohydrates and Organic Acid Analyses.
Fruit and leaf tissues (250 mg) were extracted twice in 10 ml of 80% hot ethanol, with 10 mg of inositol as an internal standard. The extract was dried, resuspended in 1 ml of water, and loaded onto Sep-pack cartridges (34). Soluble sugars were eluted with 4 ml of water. Soluble sugars (sucrose, fructose, glucose, sorbitol, and inositol) and acids were separated by using an Ion-300 (300 mm × 7.8 mm × 10 μm) and a IonGuard GC-801 guard column on an HPLC (Model 1050; Hewlett-Packard, Palo Alto, CA), and detected by using refractive index and UV monitored at 195 nm and 245 nm. Extracts were also analyzed by HPLC-MS: a PE Series 200 Quaternary HPLC with PE (Sciex, Thornhill, ON, Canada) API 2000 mass spectrometer used in the APCI negative-ion mode. The HPLC was equipped with Phenonemex Luna NH2 column (250 mm × 4.6 mm × 5 μm) using a isocratic program of 22% water and 78% acetonitrile at a flow rate of 2.75 ml·min−1.
Starch was determined from the pellet of dry samples extracted twice in hot ethanol as above. The pellet was dried and resuspended in 10 ml of sodium acetate buffer (pH 4.8). The solution was boiled for 10 min to solubilize starch, digested with 50 units of amyloglucosidase, and quantified by HPLC-RI/UV.
Real-Time Quantitative TaqMan PCR.
PCR primers and compatible TaqMan probes were designed by using Primer Express (Applied Biosystems, Foster City, CA). To prevent coamplification of contaminating genomic DNA, primers were designed to cover exon–exon junctions where possible (35).
Total cellular RNA was isolated by the hot borate method (36). Genomic DNA (gDNA) contamination was digested by using RNase-free DNase I (Invitrogen, Carlsbad, CA) for 15 min at 37°C, followed by inactivation at 95°C for 5 min and chilling on ice. Absence of gDNA contamination was confirmed by using a universal 18S TaqMan PCR system on digested total RNA. cDNA was synthesized by using 50 units of SuperScript III reverse transcriptase, 600 ng of random hexadeoxyribonucleotide (pd(N)6) primers, 10 units of RNaseOut (RNase inhibitor), and 1 mM dNTPs (all from Invitrogen) in a final volume of 40 μl. Reverse transcription proceeded for 120 min at 50°C, and then 60 μl of water was added, and the reaction was stopped by heating for 5 min to 95°C and cooling on ice.
Each PCR contained 20× Assay-on-Demand primer, probes for the respective TaqMan system, and commercially available PCR master mix (TaqMan Universal PCR Mastermix, Applied Biosystems) containing 10 mM Tris·HCl (pH 8.3), 50 mM KCl, 5 mM MgCl2, 2.5 mM deoxynucleotide triphosphates, 0.625 unit of AmpliTaq Gold DNA polymerase, 0.25 unit of AmpErase UNG, and 5 μl of diluted cDNA sample in a final volume of 12 μl. Samples were amplified in 96-well plates in an automated fluorometer (ABI PRISM 7700 Sequence Detection System, Applied Biosystems). Amplification conditions were: 2 min at 50°C, 10 min at 95°C, 40 cycles of 15 s at 95°C, and 60 s at 60°C. Fluorescent signals were collected during the annealing temperature, and CT values were extracted with a threshold of 0.04 and baseline values of 3–10.
Housekeeping Gene Validation Experiment.
To determine the most stably transcribed housekeeping gene, a validation experiment was run on each cDNA sample from all tissue types. Three housekeeping genes were used for this experiment: a TaqMan PCR system recognizing plant 18S rRNA (ssrRNA), apple glyceraldehyde 3-phosphate dehydrogenase, and apple ribosomal protein S19. 18S rRNA was transcribed most stably and thus had the least standard deviation across all tissues.
Relative Quantification of Gene Transcription.
Final quantification was done by using the comparative CT method (User Bulletin #2; Applied Biosystems) and reported as relative transcription or the n-fold difference relative to a calibrator cDNA (i.e., lowest target gene transcription). The housekeeping gene 18S rRNA was used to normalize the CT values of the target genes (ΔCT). The ΔCT was calibrated against the weakest signal within each target gene. The linear amount of target molecules relative to the calibrator was calculated by 2−ΔΔCt. All gene transcription was expressed as an n-fold difference relative to the calibrator.
Enzyme Extraction.
To extract S6PDH, mature leaves were collected and frozen in liquid nitrogen. Proteins were extracted as in Lo Bianco and Rieger (37). Tissue (0.5 g) was ground to a fine powder with 0.5 g polyvinyl-pyrrolidone (PVPP) in liquid nitrogen by using a mortar and pestle and then homogenized in an extraction buffer consisting of 50 mM Hepes/NaOH (pH 7.5), 10 mM MgCl2, 1 mM EDTA, 2.5 mM DTT, 1% Tween 20, and 5% (wt/vol) glycerol. The slurry was filtered through Miracloth. After centrifugation at 4,000 × g for 10 min, the supernatant was used for enzyme assays and Western blot analyses.
Five fruits were collected from three trees of each line at 30, 54, 70, 91, and 118 DAFB. They were peeled, frozen in liquid nitrogen, and kept at −80°C until analyses. Tissue for SDH analysis was extracted and assayed with fresh samples after harvest according to Yamaki and Ishikawa (38) with modifications. Cortical tissue was homogenized in 200 mM potassium phosphate buffer (pH 7.8) with 1 mM EDTA, 10 mM sodium ascorbate, 1 mM DTT, 0.1% Tween 20, 5% PVPP, and 2 mM PMSF (Buffer A). The homogenate was squeezed through four layers of Miracloth and centrifuged at 17,000 × g for 30 min. The supernatant was adjusted to 70% ammonium sulfate and centrifuged at 17,000 × g for 30 min. The precipitate was resuspended in Buffer A and applied to a PD10 column (GE Healthcare, Buckinghamshire, U.K.) preequilibrated with 100 mM Tris·HCl (pH 8.0). The eluate was assayed for SDH activity. SS, SPS, VIN, NIN, FK, HK, and ADP-glucose pyrophosphorylase were extracted as above, except that MgCl2 was added to Buffer A, and the column was preequilibrated by 100 mM Tris·HCl (pH 8.0) containing 1 mM MgCl2, 1 mM DTT, and 0.5 mM EDTA. CIN was extracted according to Tanase and Yamaki (39). Cortical tissue was homogenized in 200 mM potassium phosphate buffer (pH 7.8) containing 10 mM 2-mercaptoethanol and centrifuged at 3,000 × g for 30 min. The pellet was washed with 20 mM Tris·HCl (pH 8.0) containing 10 mM 2-mercaptoethanol (Buffer B) and centrifuged at 3,000 × g for 30 min. This process was repeated three times. The pellet was dialyzed against Buffer B, and the dialyzate was assayed for CIN. All procedures were done at 4°C or on ice. Protein concentration was estimated as in Bradford (40).
Enzyme Assay.
S6PDH activity was determined as in Negm and Loescher (41). Fifty microliters of protein extract were added to 950 μl of assay buffer with 0.1 M Tris·HCl (pH 9.0), 0.1 mM NADPH, and 20 mM glucose-6-phosphate (G6P). Net rate was calculated by subtracting the rate for a control with water instead of G6P. The absorbance change at 340 nm over 2 min was recorded, and NADPH oxidation was calculated with an absorption coefficient of 6.22 mM−1.
SDH activity was assayed as in Yamaki and Ishikawa (38), with modifications. The reaction mixture contained 100 mM Tris·HCl buffer (pH 9.5), 1 mM NAD+, 300 mM sorbitol, and the enzyme solution. Enzyme activity was measured as change in absorbance at 340 nm at 25°C.
VIN and CIN activities were assayed according to Tanase and Yamaki (39) in a mixture containing 30 mM potassium acetate (pH 4.5), 200 mM sucrose, and enzyme solution. The reaction was run for 1 h at 30°C and stopped by boiling in water for 3 min before adding 0.75 M Tris·HCl buffer (pH 8.5). The amount of glucose produced from sucrose was determined by the enzyme-coupling method (42). The same process was used for NIN activity, except that 30 mM Hepes-KOH (pH 7.0) was substituted for 30 mM potassium acetate (pH 4.5).
SS and SPS activities were assayed according to Tanase and Yamaki (39), with modification. For SPS, the reaction mixture contained 15 mM Hepes-KOH (pH 8.5), 15 mM fructose-6-phosphate, 2 mM UDP-glucose, 5 mM MgCl2, 50 mM NaF, 1 mM sodium orthovanadate, and enzyme solution. For SS, the reaction mixture contained 15 mM Hepes-KOH buffer (pH 8.5), 15 mM fructose, 2 mM UDP-glucose, 5 mM MgCl2, and enzyme solution. The reaction was incubated for 30 min at 30°C, and stopped by addition of 2.5 N NaOH. Sucrose production was determined by Roe's method (43). For sucrose cleavage activity of SS, the reaction contained 30 mM Hepes-KOH (pH 7.0), 200 mM sucrose, 5 mM UDP, and enzyme solution and was stopped by boiling for 3 min. Fructose production was determined by enzyme coupling (44).
FK and HK activities were assayed according to Kanayama et al. (45) with modification. For HK, the reaction contained 30 mM Hepes-NaOH (pH 7.5), 1 mM MgCl2, 0.6 mM EDTA, 9 mM KCl, 1 mM NAD+, 1 mM ATP, 2 units of G6P dehydrogenase, 30 mM glucose, and enzyme solution. For FK, 2 units of phosphoglucoseisomerase and 30 mM fructose were added. Enzyme activities were determined by changes in absorbance at 340 nm at 25°C.
Protein Analysis.
Twenty micrograms of protein extract was separated by SDS/PAGE according to Laemmli (46) and then transferred to a nitrocellulose membrane as described by Towbin et al. (47). S6PDH was detected by using rabbit anti-S6PDH serum prepared against apple S6PDH. Primary antibodies were visualized by using anti-rabbit serum with alkaline phosphatase.
Statistical Analysis.
All parameters were compared separately in each year by using one-way ANOVA with SAS software (SAS Institute, Cary, NC).
Supplementary Material
Acknowledgments
We thank Dr. Mary Lou Mendum for critical reading of this manuscript. This work was supported by the Washington Tree Fruit Research Commission.
Abbreviations
- DAFB
days after full bloom
- CIN
cell wall-bound invertase
- FK
fructose kinase
- G6P
glucose-6-phosphate
- HK
hexokinase
- NIN
neutral invertase
- SDH
sorbitol dehydrogenase
- S6PDH
sorbitol-6-phosphate dehydrogenase
- SOT
sorbitol transporter
- SPS
sucrose phosphate synthase
- SS
sucrose synthase
- TA
titratable acidity
- VIN
vacuolar acid invertase.
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “From Functional Genomics of Model Organisms to Crop Plants for Global Health,” held April 3–5, 2006, at the National Academy of Sciences in Washington, DC. The complete program is available on the NAS web site at www.nasonline.org/functional_genomics.
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Bieleski RL, Redgwell RJ. Aus J Plant Physiol. 1985;12:657–668. [Google Scholar]
- 2.Patrick JW. Ann Rev Plant Physiol Plant Mol Biol. 1997;48:191–222. doi: 10.1146/annurev.arplant.48.1.191. [DOI] [PubMed] [Google Scholar]
- 3.Patrick JW, Offler CE. J Exp Bot. 2004;52:551–564. [PubMed] [Google Scholar]
- 4.Noiraud N, Maurousset L, Lemoine R. Plant Cell. 2001;13:695–705. doi: 10.1105/tpc.13.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao Z, Maurousset L, Lemoine R, Yoo S-D, van Nocker S, Loescher W. Plant Physiol. 2003;131:1566–1575. doi: 10.1104/pp.102.016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L-Y, Peng Y-B, Pelleschi-Travier P, Fan Y, Lu Y-F, Lu Y-M, Gao X-P, Shen Y-Y, Delrot S, Zhang D-P. Plant Physiol. 2004;135:574–586. doi: 10.1104/pp.103.036632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.BerŸter J, Feusi MES, Rueedi P. J Plant Physiol. 1997;151:269–276. [Google Scholar]
- 8.Kanayama Y, Yamaki S. Plant Cell Physiol. 1993;34:819–823. doi: 10.1093/oxfordjournals.pcp.a029269. [DOI] [PubMed] [Google Scholar]
- 9.Nosarszewski M, Clements AM, Downie AB, Archbold DD. Physiol Plant. 2004;121:391–398. [Google Scholar]
- 10.Iida M, Bantog NA, Yamada K, Shiratake K, Yamaki S. J Am Soc Hort Sci. 2004;129:870–875. [Google Scholar]
- 11.Tao R, Uratsu SL, Dandekar AM. Plant Cell Physiol. 1995;36:525–532. doi: 10.1093/oxfordjournals.pcp.a078789. [DOI] [PubMed] [Google Scholar]
- 12.Kanamaru N, Ito Y, Komori S, Saito M, Kato H, Takahashi S, Omura M, Soejima J, Shitatake K, Yamada K, Yamaki S. Plant Sci. 2004;167:55–61. [Google Scholar]
- 13.Cheng L, Zhou R, Reidel EJ, Sharkey TD, Dandekar AM. Planta. 2005;220:767–776. doi: 10.1007/s00425-004-1384-5. [DOI] [PubMed] [Google Scholar]
- 14.Riesmeier JW, Fluegge U-I, Schulz B, Heineke D, Heldt H-W, Willmitzer L, Frommer WB. Proc Natl Acad Sci USA. 1993;90:6160–6164. doi: 10.1073/pnas.90.13.6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leggewie G, Kolbe A, Lemoine R, Roessner U, Lytovchenko A, Zuther E, Kehr J, Frommer WB, Riesmeier JW, Willmitzer L, Fernie AR. Planta. 2003;217:158–167. doi: 10.1007/s00425-003-0975-x. [DOI] [PubMed] [Google Scholar]
- 16.Riesmeier JW, Willmitzer L, Frommer WB. EMBO J. 1994;13:1–7. doi: 10.1002/j.1460-2075.1994.tb06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buerkle L, Hibberd JM, Quick WP, Kuehn C, Hirner B, Frommer WB. Plant Physiol. 1998;118:59–68. doi: 10.1104/pp.118.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.BerŸter J. Acta Hort. 1998;466:23–28. [Google Scholar]
- 19.Knee M. In: Biochemistry of Fruit Ripening. Seymour G, Taylor J, Tucker G, editors. London: Chapman & Hall; 1993. pp. 325–346. [Google Scholar]
- 20.BerŸter J, Feusi MES. J Plant Physiol. 1997;151:277–285. [Google Scholar]
- 21.Archbold DD. Physiol Plant. 1999;105:391–395. [Google Scholar]
- 22.Koch K. Curr Opin Plant Biol. 2004;7:235–246. doi: 10.1016/j.pbi.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 23.Rolland F, Baena-Gonzalez E, Sheen J. Ann Rev Plant Biol. 2006;57:675–709. doi: 10.1146/annurev.arplant.57.032905.105441. [DOI] [PubMed] [Google Scholar]
- 24.Prata RTN, Williamson JD, Conkling MA, Pharr DM. Plant Physiol. 1997;114:307–314. doi: 10.1104/pp.114.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao Z, Jayanty S, Beaudry R, Loescher W. J Am Soc Hort Sci. 2005;130:261–268. [Google Scholar]
- 26.Hampson CR, Quamme HA, Hall JW, MacDonald RA, King MC, Cliff MA. Euphytica. 2000;111:79–90. [Google Scholar]
- 27.Kanayama Y, Mori H, Imaseki H, Yamaki S. Plant Physiol. 1992;100:1607–1608. doi: 10.1104/pp.100.3.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wen-Jun S, Forde BG. Nucleic Acids Res. 1989;17:8385. doi: 10.1093/nar/17.20.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.James DJ, Passey AJ, Bevan MW. Plant Cell Rep. 1989;7:658–661. doi: 10.1007/BF00272054. [DOI] [PubMed] [Google Scholar]
- 30.Dandekar AM, Teo G, Uratsu SL, Tricoli D. In: Methods in Molecular Biology. Wang L, editor. Totowa, NJ: Humana; 2006. pp. 253–262. [DOI] [PubMed] [Google Scholar]
- 31.Jefferson RA, Kavanaugh TA, Bevan MW. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCutchan H, Shackel KA. J Am Soc Hort Sci. 1992;117:607–611. [Google Scholar]
- 33.Klages K, Donnison H, Wunsche J, Boldingh H. Aus J Plant Physiol. 2001;28:131–139. [Google Scholar]
- 34.Perez AG, Olias R, Espada J, Olias JM, Sanz C. J Agric Food Chem. 1997;45:3545–3549. [Google Scholar]
- 35.Leutenegger CM, Mislin CN, Sigrist B, Ehrengruber MU, Hofmann-Lehmann R, Lutz H. Vet Immunol Immunopathol. 1999;71:291–305. doi: 10.1016/S0165-2427(99)00100-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wan CH, Wilkins TA. Anal Biochem. 1994;223:7–12. doi: 10.1006/abio.1994.1538. [DOI] [PubMed] [Google Scholar]
- 37.Lo Bianco R, Rieger M. HortScience. 2001;36:574. [Google Scholar]
- 38.Yamaki S, Ishikawa K. J Am Soc Hort Sci. 1986;111:134–137. [Google Scholar]
- 39.Tanase K, Yamaki S. J Japan Soc Hort Sci. 2000;69:671–676. [Google Scholar]
- 40.Bradford M. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 41.Negm FB, Loescher WH. Plant Physiol. 1981;67:139–142. doi: 10.1104/pp.67.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamaki S. Plant Cell Physiol. 1980;69:117–121. [Google Scholar]
- 43.Roe JH. J Biol Chem. 1934;107:15–22. [Google Scholar]
- 44.Morell M, Copeland L. Plant Physiol. 1985;78:149–154. doi: 10.1104/pp.78.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanayama Y, Dai N, Granot D, Petreikov M, Schaffer A, Bennett AB. Plant Physiol. 1997;113:1379–1384. doi: 10.1104/pp.113.4.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laemmli MC. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 47.Towbin H, Stachelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.