Abstract
In plants, light signals caused by the presence of neighbors accelerate stem growth and flowering and induce a more erect position of the leaves, a developmental strategy known as shade-avoidance syndrome. In addition, mutations in the photoreceptors that mediate shade-avoidance responses enhance disease susceptibility in Arabidopsis thaliana. Here, we describe the Arabidopsis constitutive shade-avoidance1 (csa1) mutant, which shows a shade-avoidance phenotype in the absence of shade and enhanced growth of a bacterial pathogen. The csa1 mutant has a T-DNA inserted within the second exon of a Toll/Interleukin1 receptor–nucleotide binding site–leucine-rich repeat (TIR-NBS-LRR) gene, which leads to the production of a truncated mRNA. Arabidopsis plants transformed with the truncated TIR-NBS-LRR gene recapitulate the mutant phenotype, indicating that csa1 is a dominant-negative mutation that interferes with phytochrome signaling. TIR-NBS-LRR proteins have been implicated in defense responses in plants. RPS4, the closest homolog of CSA1, confers resistance to Pseudomonas syringae and complements the csa1 mutant phenotype, indicating that responses to pathogens and neighbors share core-signaling components in Arabidopsis. In Drosophila melanogaster and Caenorhabditis elegans, TIR domain proteins are implicated in both development and immunity. Thus, the dual role of the TIR domain is conserved across kingdoms.
INTRODUCTION
Plants respond to the presence of potential competitors thanks to the perception of the low proportions of red compared with far red of the light transmitted or reflected by green tissues of neighbor plants (Ballaré, 1999; Smith, 2000; Franklin and Whitelam, 2005). Low red:far-red ratios are perceived by phytochromes, which exist in two photointerconvertible forms, an inactive red-absorbing form (Pr) and an active far-red-absorbing form (Pfr) (Quail, 2002). The high red:far-red ratio characteristic of environments with low plant density moves the phytochrome system toward the Pfr form, which inhibits stem growth, hyponasty (upward movement of leaves), and flowering (Ballaré, 1999; Smith, 2000). Conversely, in dense canopies, the red:far-red ratio is reduced, and the photoequilibrium is dominated by inactive phytochrome. This releases stem growth, hyponasty, and flowering, causing tall plants with erect leaves to less likely become shaded by neighbors (Ballaré, 1999; Smith, 2000; Franklin and Whitelam, 2005).
Phytochrome B (phyB) is the member of the phytochrome family that contributes most strongly to shade-avoidance responses (Yanovsky et al., 1995; Franklin et al., 2003). The inactive form of phyB is localized in the cytosol, whereas the active form accumulates in the nucleus (Nagatani, 2004) and affects the expression of many genes (Devlin et al., 2003; Tepperman et al., 2004).
A number of components mediating phyB signaling have been identified through forward or reverse genetic screens, including transcription factors, kinases, phosphatases, and proteins with unknown biochemical functions (Chen et al., 2004). However, the molecular mechanism through which phyB exerts its activity is still unclear.
Here, we describe the Arabidopsis thaliana constitutive shade-avoidance1 (csa1) mutant, which displays shade-avoidance responses in the absence of shade. The csa1 mutant phenotype is caused by a mutation in a Toll/Interleukin1 receptor–nucleotide binding site–leucine-rich repeat (TIR-NBS-LRR)–type gene. TIR-NBS-LRRs are plant disease resistance genes of particular interest due to their homology to Toll-like receptors that trigger innate immune responses in animals (Holt et al., 2003). In plants, TIR-NBS-LRRs confer resistance against specific pathogen strains in accordance to the gene-for-gene model (Flor, 1956). The closest homolog of CSA1, RPS4, confers resistance to Pseudomonas syringae pv tomato strain DC3000 expressing avrRps4 (Gassmann et al., 1999). Interestingly, RPS4 complements the csa1-1 mutant phenotype, indicating that the ability to respond to pathogens and neighbors is mediated, at least in part, through common signaling components in Arabidopsis.
RESULTS AND DISCUSSION
Isolation and Physiological Characterization of csa1
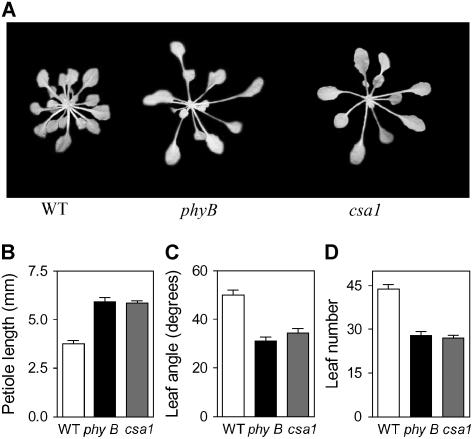
The csa1 mutant was isolated in a screening of activation-tagged lines (Weigel et al., 2000) for seedlings displaying a shade-avoidance phenotype (long stem) under high red:far-red ratios (i.e., in the absence of a shade signal). csa1 also showed long petioles, erect and pale (low chlorophyll content) leaves, and early flowering under short (noninductive) photoperiods (Figure 1), resembling the phyB mutant.
Figure 1.
csa1 Shows the Shade-Avoidance Developmental Pattern under High Red:Far-Red Ratios and Resembles the phyB Mutant.
(A) Representative 30-d-old plants.
(B) Petiole length.
(C) Leaf angle.
(D) Rosette leaf number (reduced leaf number indicates early flowering).
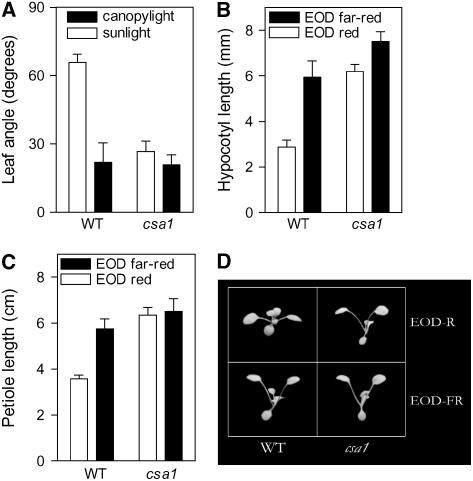
To test whether shade-avoidance responses were impaired in the csa1 mutant, we compared the leaf angle of wild-type and csa1 plants placed for 5 h either under sunlight (red:far-red ratio = 1.1) or under a dense canopy (red:far-red ratio = 0.2) of the grass Cynodon dactylon. While leaf angle dramatically decreased in wild-type plants placed under the grass canopy compared with plants left under sunlight, no significant response was observed for csa1 (Figure 2A).
Figure 2.
csa1 Growth and Development under Natural and Simulated Shade.
(A) The response to natural shade. Leaf angle of seedlings grown for 14 d under fluorescent white light and placed on the morning of the 15th day in the field under either sunlight or canopy shade light for 5 h.
(B) Response to simulated shade. Hypocotyl length of seedlings grown for 3 d under short days (8 h white light/16 h darkness) ended with a 5-min red or far-red light pulse.
(C) and (D) Response to simulated shade. Wild-type and csa1 seedlings grown for 2 weeks under short days (8 h white light/16 h darkness) ended with a 5-min pulse of either red or far-red light.
The shade imposed by the grass canopy simultaneously reduces the red:far-red ratio and total fluence rate of light (Franklin and Whitelam, 2005). To analyze the specific response to changes in light quality, wild-type and csa1 seedlings were grown under white-light photoperiods (8 h) ended with a pulse (5 min) of either red or far red, which establish high or low Pfr levels during the subsequent dark period, respectively (Fankhauser and Casal, 2004). End-of-day (EOD) far red significantly promoted hypocotyl (stem) and petiole growth in wild-type plants, while responses to EOD far red were significantly reduced (Figure 2B) or absent (Figure 2C) in the csa1 mutant.
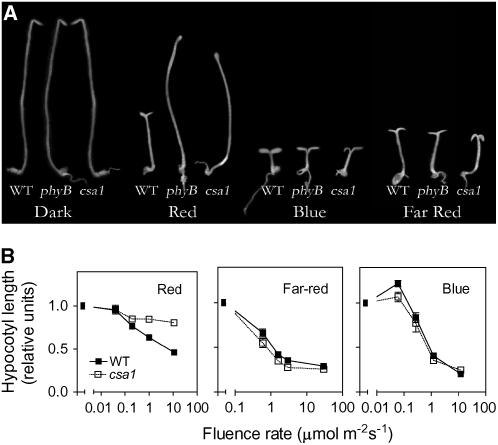
In seedlings that grow on seed reserves in complete darkness, exposure to light signals a major shift in developmental pattern, including the arrest of hypocotyl growth (see wild type in Figure 3A). csa1 showed poor inhibition of hypocotyl growth under red light, the spectral region that most efficiently activates phyB, but it retained normal responses to far-red or blue light compared with darkness (Figures 3A and 3B), which are mediated mainly by another member of the phytochrome family (phyA; Quail, 2002) and cryptochromes (Cashmore et al., 1999). This indicates that csa1 is not generally defective in light regulation of cell growth, but it is specifically impaired in red light signaling.
Figure 3.
The Long-Hypocotyl Phenotype of csa1 Seedlings Is Red Light Specific.
(A) Wild-type, phyB, and csa1 mutant seedlings grown under continuous red, far-red, blue light, or darkness for 3 d.
(B) Hypocotyl-length fluence-rate response curves of wild-type and csa1 mutant seedlings.
csa1 Specifically Impairs phyB Signaling
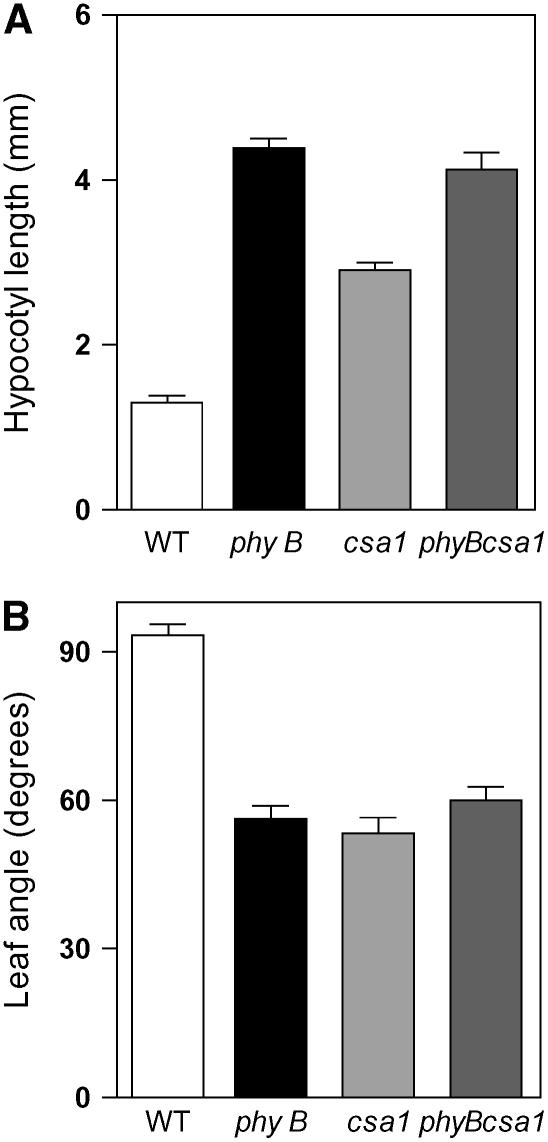
The csa1 mutant strongly resembles the phyB mutant under white light (Figure 1) and also showed reduced responses to changes in the red:far-red ratio (Figure 2) and to red light signals (Figure 3), typically perceived by phyB. To address the relationship between PHYB and CSA1, we compared the behavior of the phyB single mutant with that of the phyB csa1 double mutant. We found that while the csa1 mutation alone significantly promoted hypocotyl elongation and a more erect position of the leaves under short photoperiods of white light with high red:far-red ratios, the csa1 mutation had no additional effect on these traits when present in a phyB mutant background (Figure 4). These observations indicate that the csa1 mutation alters Arabidopsis development by specifically impairing the phyB signaling pathway.
Figure 4.
csa1 Specifically Impairs phyB Signaling.
(A) Hypocotyl length of wild-type, csa1, phyB, and phyB csa1 seedlings grown for 4 d under short days (8 h white light/16 h darkness).
(B) Leaf angle of wild-type, csa1, phyB, and phyB csa1 seedlings grown for 14 d under short days (8 h white light/16 h darkness).
The csa1 Phenotype Is Caused by a T-DNA Insertion in a TIR-NBS-LRR–Type Gene
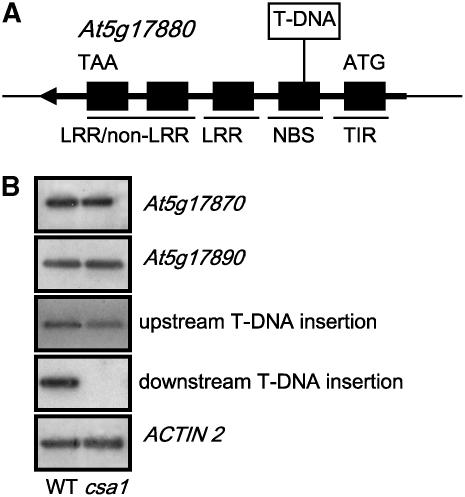
The T-DNA insertion, which includes a multimerized 35S enhancer sequence from the Cauliflower mosaic virus (CaMV), cosegregated with the mutant phenotype and with resistance to the selection marker Basta (data not shown). Cloning of flanking DNA revealed that the T-DNA was inserted within the second exon of At5g17880, a gene encoding a member of the TIR-NBS-LRR gene family (Figure 5A).
Figure 5.
csa1 Results from a T-DNA Insertion in a TIR-NBS-LRR–Type Gene.
(A) Schematic representation of the TIR-NBS-LRR gene At5g17880 and the insertion of the T-DNA within its second exon.
(B) Expression analysis of genes flanking At5g17880 and of the first and fourth exons of At5g17880 by RT-PCR in wild-type and csa1 seedlings.
Since the T-DNA insert in csa1 contains a strong enhancer, the mutant phenotype could be the result of (1) overexpression of a gene flanking At5g17880, (2) lack of function of the protein encoded by the At5g17880 gene, or (3) a dominant-negative effect on TIR-NBS-LRR proteins caused by the expression of a truncated At5g17880 protein.
The expression of neighbor genes At5g17870 and At5g17890 was not significantly different between the wild type and csa1 (Figure 5B). Using primers that amplify either the first or the fourth exon of At5g17880, we could detect mRNA expression of this gene upstream but not downstream of the T-DNA insertion in the mutant, while expression of both exons was detected in the wild type (Figure 5B). This clearly indicates that an mRNA with a truncated At5g17880 open reading frame, and most likely a truncated At5g17880 protein, is being expressed in csa1.
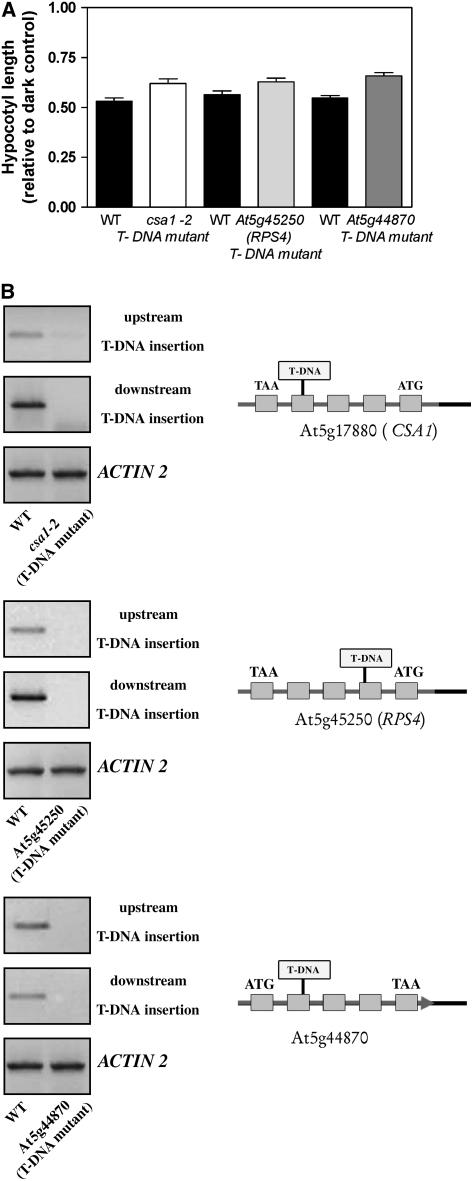
A T-DNA mutant (SALK_023219) carrying an insertion (without enhancer sequences) within the fourth exon of the At5g17880 gene enhanced hypocotyl growth under red light (Figure 6A), providing independent genetic evidence for the role of the CSA1 gene in light signaling. However, this allele that we named csa1-2 did not have the strong csa1 phenotype when grown in white light with a high red:far-red ratio (data not shown) and showed a weaker phenotype than csa1-1 under red light (Figure 6A) in spite of the fact that the CSA1 mRNA was almost undetectable (Figure 6B). Thus, only a small part of the csa1-1 phenotype can be accounted for by lack of At5g17880 function.
Figure 6.
Reduced Sensitivity to Red Light in TIR-NBS-LRR Mutants.
(A) T-DNA insertion alleles in CSA1 (csa1-2) and other TIR-NBS-LRR loci show reduced inhibition of hypocotyl growth under red light.
(B) Expression levels of CSA1, RPS4, and AT5g44870 determined by RT-PCR in wild-type and T-DNA insertion mutants. T-DNA insertion sites for each mutant are shown.
T-DNA Insertions in Other TIR-NBS-LRR Proteins Impair Deetiolation in Red Light
There are ∼90 TIR-NBS-LRR genes in Arabidopsis, and many of them act as early components of the core-sensing mechanism that mediates pathogen perception in plants (Holt et al., 2003). TIR-NBS-LRR genes that are highly related to At5g17880 (Meyers et al., 2003) include RPS4, a gene that has been implicated in defense responses against a bacterial pathogen (Gassmann et al., 1999), and AT5g44870, a gene whose expression has been shown to be regulated by phyA and phyB (Tepperman et al., 2001, 2004). Thus, the lack of a strong phenotype in the csa1-2 mutant could result from redundancy with other family members. In support of this idea, small but significant reductions in the inhibition of hypocotyl elongation by red light were also observed in null mutants carrying T-DNA insertions within the At5g17880 homologs RPS4 and At5g44870 (Figure 6), which share 64% and 63%, respectively, of sequence similarity with CSA1 across the entire protein.
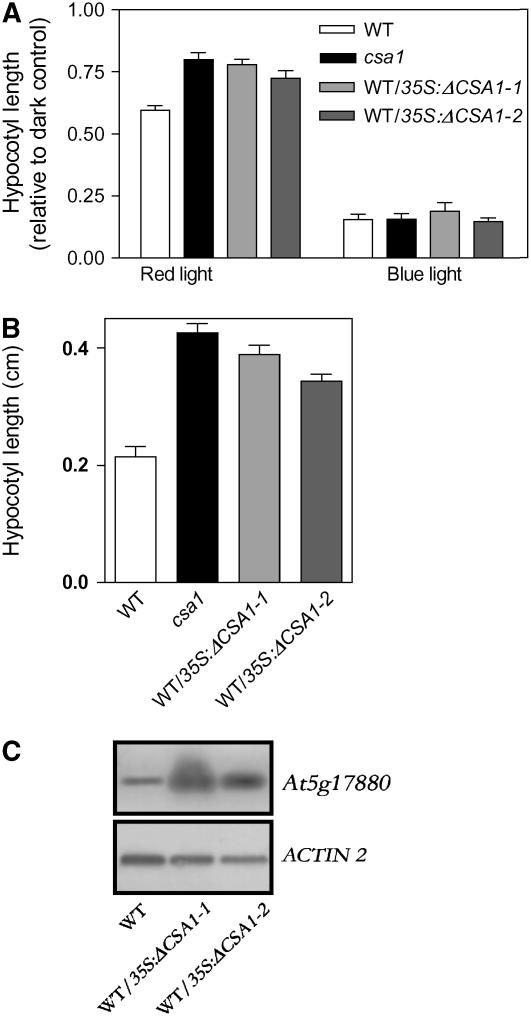
Expression of a Truncated CSA1 Gene Phenocopies the csa1 Mutant
The observation that the csa1-1 phenotype is semidominant (see Supplemental Figure 1 online), together with the lack of a strong phenotype in the csa1-2 allele, support the idea that part of the light signaling defects of csa1-1 results from expression of the truncated TIR-NBS-LRR gene (Figure 5B). To test this idea, we transformed Arabidopsis wild-type plants with a construct containing the truncated At5g17880 gene under the control of the strong and constitutive CaMV 35S promoter. Seedlings from independent transgenic lines expressing the truncated TIR-NBS-LRR gene showed significantly reduced red, but not blue light, inhibition of hypocotyl elongation (Figure 7A). Another phyB-mediated response, the red-light inhibition of hypocotyl negative gravitropism, was also impaired in these plants (see Supplemental Figure 5 online). Finally, these transgenic plants also showed long hypocotyls under white light with a high red:far-red ratio (i.e., in the absence of shade signals) (Figure 7B). Taken together, the above data indicate that a significant part of the csa1 mutant phenotype is the result of a dominant-negative effect caused by the expression of a truncated TIR-NBS-LRR protein of ∼212 amino acids, completely lacking the LRR and part of the NBS domains.
Figure 7.
Expression of 35S:ΔCSA1 in the Wild-Type Background Phenocopies the csa1 Plant.
(A) Hypocotyl length of wild-type and two independent transgenic plants expressing the 35S:Δcsa1 construct, grown for 4 d under red or blue light.
(B) Hypocotyl length of plants grown for 4 d under white light with a high red:far-red ratio (8 h light/16 h darkness).
(C) Expression levels of CSA1 in wild-type plants and two independent transgenic plants expressing the 35S:Δcsa1 construct determined by RT-PCR using primers that amplify the first exon.
The molecular analysis of transcripts encoding TIR-NBS-LRR proteins indicates the existence of alternative spliced forms, some of which partially resemble the truncated CSA1 form (Jordan et al., 2002). However, we were not able to detect alternative spliced forms of CSA1 (E. Petrillo, M.J. Yanovsky, and A. Kornblihtt, unpublished data). Whether the alternative spliced forms of other TIR-NBS-LRRs, which resemble the truncated CSA1 gene, modulate light signaling remains to be determined.
The TIR-NBS-LRR Gene RPS4 Rescues the csa1 Phenotype
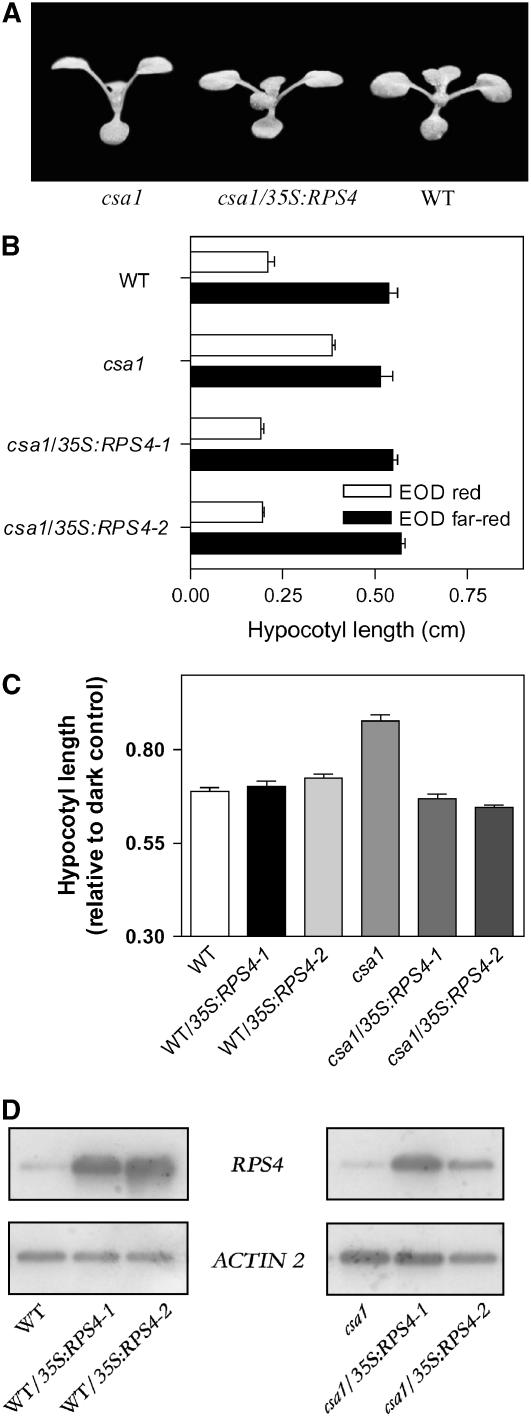
In animals, Toll-like receptors signal through homodimerization of TIR domains as well as heterodimerization with the TIR domain of adapter proteins (Yamamoto et al., 2004). Interestingly, truncated proteins containing only the TIR domain function as dominant-negative forms that block signaling downstream of TIR domain receptors (Dupraz et al., 2000). By analogy with the animal model, the truncated form of CSA1 that retains intact only the TIR domain could form nonproductive heterodimers with other proteins bearing TIR domains (Meyers et al., 2002). RPS4 is the TIR-NBS-LRR protein with the highest degree of sequence similarity to CSA1. If the truncated form of CSA1 interferes with RPS4 function, increasing RPS4 levels should attenuate the mutant phenotype. Indeed, transgenic lines overexpressing RPS4 in a csa1-1 background showed a wild-type phenotype when grown under white light with a high red:far-red ratio (e.g., short petioles, nonerect dark green leaves, and normal flowering time) (Figure 8A; see Supplemental Figure 4 online). Overexpression of RPS4 also restored the ability of csa1-1 plants to show differential responses to EOD red compared with far-red light pulses, similar to those present in wild-type plants (Figure 8B). Furthermore, hypocotyl length of csa1 plants transformed with the RPS4 gene was not different from that of wild-type seedlings under red light (Figure 8C) or darkness (wild type, 1.14 ± 0.06 cm; csa1-1/35S:RPS4-1, 1.22 ± 0.09 cm; and csa1-1/35S:RPS4-2, 1.14 ± 0.03 cm), indicating that overexpression of RPS4 in a csa1 background did not produce constitutively dwarf plants, but plants with normal responses to red light. Indeed, overexpression of RPS4 also rescued another response to red light, the phyB-mediated inhibition of negative gravitropism (see Supplemental Figure 5 online). Finally, RPS4 had no effect on red light inhibition of hypocotyl elongation when overexpressed in a wild-type background (Figure 8C) at levels comparable with those obtained in a csa1-1 background (Figure 8D), suggesting that RPS4 overexpression enhanced red light sensitivity in the csa1-1 mutant attenuating the dominant-negative effect caused by the truncated CSA1, rather than through a nonspecific effect of RPS4 on cell growth. Taken together, our results strongly suggest that csa1 impairs phytochrome signaling by interfering with the action of RPS4 and possibly other related TIR-NBS-LRR proteins.
Figure 8.
Overexpression of RPS4 in csa1 Rescues the csa1-1 Mutant Phenotype.
(A) csa1, csa1/35S:RPS4, and wild-type plants grown under white light.
(B) csa1/35S:RPS4 seedlings behave as wild-type plants in response to EOD red and far-red treatments.
(C) Hypocotyl length of wild-type, two independent wild-type/35S:RPS4 transgenic lines, csa1, and two independent csa1/35S:RPS4 transgenic lines grown for 4 d under red light.
(D) Expression levels of RPS4 determined by RT-PCR.
TIR-NBS-LRRs and Phytochrome Signaling
The negative effect of csa1 on phyB signaling could be due to a reduction in phyB levels, to an effect on phyB subcellular localization, to a reduction in the stability of the Pfr form (i.e., enhanced reversion to the Pr form), or to an effect on a component of the phyB signaling pathway that acts downstream from the photoreceptor. We could not detect effects of csa1 on PHYB levels (Figure 9A) or nuclear localization (see Supplemental Figure 2 online). In addition, if the negative effect of csa1 on phyB signaling were due to an increased dark reversion of the active Pfr form to the inactive Pr form, the difference in hypocotyl length between the csa1 mutant and the wild type should be reduced under very high fluences of red light because the Pfr form will be regenerated at a higher rate. In contrast with the above prediction, the hypocotyl length difference between the csa1 mutant and wild-type plants remained the same even at extremely high fluences of red light (see Supplemental Figure 3 online), suggesting that the defect of the csa1 mutant is not associated with an enhanced Pfr-to-Pr dark reversion.
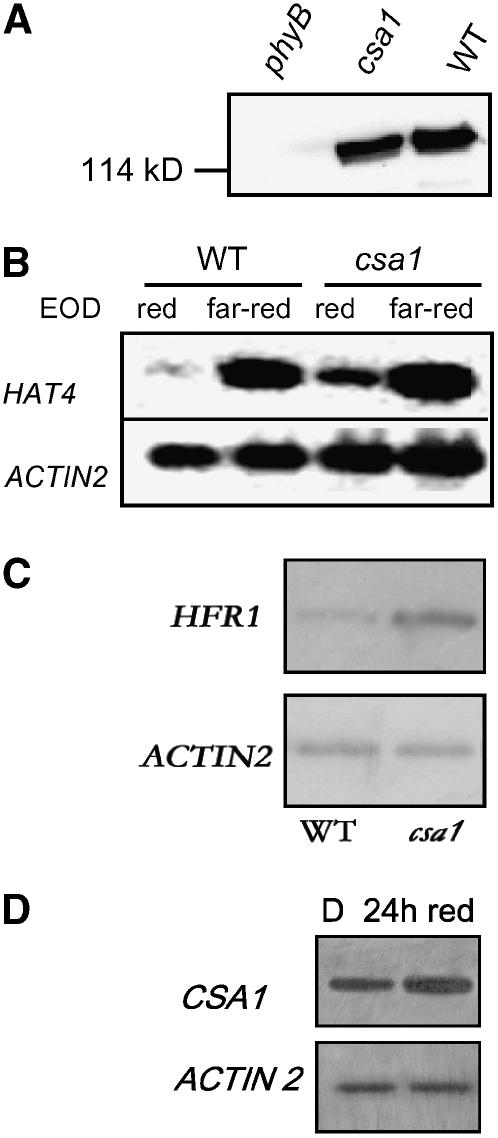
Figure 9.
Molecular Analysis of csa1 and CSA1 Expression.
(A) PHYB levels detected by monoclonal antibodies are normal in csa1 seedlings grown under white light.
(B) HAT4 expression detected by RT-PCR is enhanced in csa1 compared with wild-type seedlings grown under white light and exposed to EOD red light.
(C) HFR1 expression determined by RT-PCR is enhanced in csa1 compared with wild-type seedlings grown under white light.
(D) CSA1 expression detected by RT-PCR is enhanced in etiolated wild-type seedlings exposed for 24 h to continuous red light compared with plants kept in darkness (D).
A dramatic increase in the levels of expression of the homeodomain transcription factor HAT4 gene (also known as ATHB-2) is one of the earliest responses to a reduction in the red:far-red ratio (Carabelli et al., 1996; Devlin et al., 2003). Indeed, HAT4/ATHB-2 is a direct primary target of phytochrome action in the shade-avoidance syndrome because its expression increases rapidly under low red:far-red ratios even in the absence of protein synthesis (Roig-Villanova et al., 2006). In the csa1 mutant, HAT4 expression is already elevated under high red:far-red ratios (Figure 9B). A substantial response to the EOD far-red treatment remains in csa1, but this also occurs in phyB mutants (Carabelli et al., 1996).
Low red:far-red ratios also increase the expression of the basic helix-loop-helix gene HFR1 that antagonizes shade-avoidance responses (Sessa et al., 2005). HFR1 mRNA levels were already increased in the csa1 mutant compared with wild-type plants under high red:far-red ratios, indicating that both positive (HAT4/ATHB2) and negative (HFR1) arms of the shade-avoidance signaling cascade are constitutively elevated in csa1 mutant plants (Figure 9C).
The levels of expression of CSA1 are somewhat elevated in seedlings exposed to 24 h of red light compared with the dark controls (Figure 9D). Red and far-red light have also been shown to promote the expression of the CSA1 homolog At5g44870 after exposing etiolated seedlings to those light signals for several hours (Tepperman et al., 2001, 2004). However, this is not necessarily the main mechanism by which light controls TIR-NBS-LRR activity. In particular, 1 h of irradiation with red light in etiolated seedlings causes a rapid reduction in HAT4 expression without detectable reductions in the expression of At5g17880 or its homologs RPS4 and At5g44870 (Monte et al., 2004; data not shown).
TIR-NBS-LRR proteins like RPS4 are generally predicted to be cytosolic (Gassmann et al., 1999). PhyB is mainly localized in the cytosol in darkness and is translocated into the nucleus under red light. PhyB activity occurs mainly in the nucleus, but the possibility that some phyB activity occurs in the cytoplasm cannot be excluded (Nagatani, 2004). We have not been able to detect physical interactions between CSA1 and phyB in the yeast two-hybrid system (data not shown), strengthening the idea that TIR-NBS-LRR proteins affect phyB signaling downstream from the photoreceptor. An α importin homolog, which mediates the import of specific proteins to the nucleus, has recently been shown to be required for the function of a TIR-NBS-LRR protein in Arabidopsis (Palma et al., 2005). Thus, TIR-NBS-LRR could affect phyB signaling through their effect on another protein that is translocated to the nucleus and interacts with phyB or phyB signaling components.
Plant hormones like ethylene and auxin have been implicated in shade-avoidance responses. However, ethylene appears to mediate shade-avoidance responses to reductions in incident blue light rather than to reductions in the red:far-red ratio (Franklin and Whitelam, 2005). Since responses to blue light are not affected in csa1-1, it is unlikely that the constitutive shade-avoidance phenotype of csa1 is due to alterations in ethylene levels or signaling. In addition, the red1 mutant is involved in the control of auxin homeostasis (Hoecker et al., 2004) and shows elongated hypocotyls and leaf petioles, but it does not flower early as the phyB and csa1 mutants do (Wagner et al., 1997), suggesting that csa1 affects a signaling step upstream of auxin.
csa1-1 is the mutant that most closely resembles the phyB mutant phenotype. The gigantea mutant has elongated hypocotyls under red light but flowers late rather than early (Huq et al., 2000). The elf3 (Hicks et al., 1996), elf4 (Khanna et al., 2003), and srr1 (Staiger et al., 2003) mutants show morphological and flowering phenotypes that resemble that of phyB but they also show either complete disruption of circadian rhythms or a severe alteration in period length, phenotypes that are not obviously observed in phyB (Salomé et al., 2002) or in csa1-1 mutants (data not shown). Although ELF3 interacts physically with phyB, genetic experiments suggest that they do not act on the same pathway in the control of hypocotyl growth and could be part of a complex interplay between light and clock signaling components (Yanovsky and Kay, 2001).
Thus, taken together, the above results strongly suggest that TIR-NBS-LRR proteins like CSA1 and RPS4 mediate or modulate phyB signaling by either acting as early components of the pathway or by modulating the levels or activity of an early step of this signaling cascade.
Light Signaling and Defense Responses Are Linked in Arabidopsis
Recent microarray studies indicate that the expression of many plant disease resistance genes is modulated by the activity of phytochromes (Tepperman et al., 2001; Devlin et al., 2003). Interestingly, the growth of an incompatible strain of P. syringae is actually enhanced in the phyA phyB double mutant (Genoud et al., 2002). Furthermore, the psi2 mutant of Arabidopsis, isolated for its exaggerated responses to light, develops light-dependent necrotic lesions that mimic those induced during the hypersensitive response triggered by plant pathogens during incompatible interactions (Genoud et al., 1998).
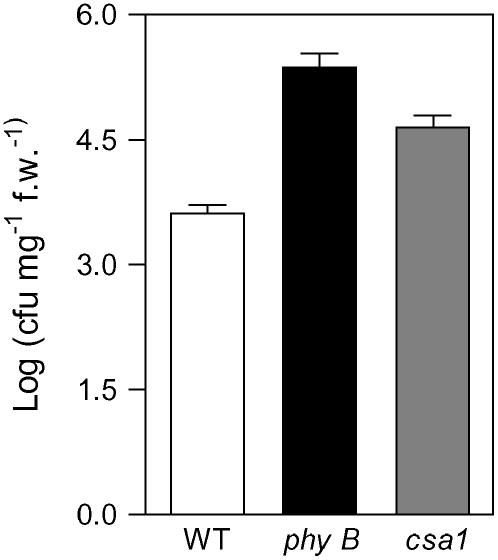
We found that the growth of an incompatible strain of P. syringae was also enhanced in csa1 mutant plants grown under high red:far-red ratios (Figure 10). This observation, together with the fact that the csa1 mutant phenotype can be reduced through expression of the disease resistance gene RPS4, strongly suggests that photomorphogenic and defense responses interact very early in their signaling cascades by sharing core-signaling components.
Figure 10.
Growth of an Avirulent Strain of P. syringae 3 d after Plant Inoculation.
Data are expressed as colony forming units (cfu) per milligram of fresh weight (f.w.).
Thus, our finding constitutes a first step toward understanding the molecular mechanisms underlying the crosstalk observed between photomorphogenic and defense responses. In the future, the identification and characterization of proteins that interact with CSA1, as well as transcriptomic analysis of csa1 mutants, will help us understand more precisely how TIR-NBS-LRRs participate in light signaling. It is worth mentioning, however, that the initial step through which TIR-NBS-LRR proteins mediate defense responses in plants is still poorly understood.
TIR Domain Proteins Play Roles in Immunity and Development in Plants and Animals
A dual role for TIR domain proteins in development and immunity has been reported in the animal kingdom. Although mammalian Toll-like receptors are known exclusively for their roles as mediators of innate immunity (Iwasaki and Medzhitov, 2004), the prototypic receptor Toll was originally identified in Drosophila melanogaster for its role in dorsoventral patterning and only later shown to be involved in the D. melanogaster innate immune system (Imler and Hoffmann, 2002). In Caenorhabditis elegans, TIR domain proteins also play roles in immunity and development (Pujol et al., 2001; Couillault et al., 2004; Liberati et al., 2004). How this dual role arose in evolution is a matter of extensive debate (Johnson et al., 2003). The presence of related proteins with similar functions in plants and animals suggests an ancient evolutionary origin for these receptors. It has been postulated that protists, the common ancestors of plants and animals, may have used similar receptors to sense the presence of competitors (Magor and Magor, 2001). Thus, our observation that TIR domain proteins play a role in developmental responses to neighbor plants may reflect the conservation of an ancient role for this protein family.
METHODS
Genetic Screening
Activation-tagged lines generated in the Columbia (Col) accession of Arabidopsis thaliana (Weigel et al., 2000) were obtained from the ABRC (stock no. CS21995). Seeds were surface sterilized and plated on Murashige and Skoog growth medium without sucrose. The plates were kept at 4°C in the dark for 4 d and then transferred for 1 week to a growth chamber with an 8-h photoperiod of white light provided by fluorescent lamps (Philips TLD 15W/54; 150 μmoles m−2 s−1) at 22°C. Seedlings with long hypocotyls were isolated and crossed to Col wild-type plants. F2 seedlings of a cross between the wild type and csa1 (originally identified as T3) segregated 3:1 basta resistant:basta sensitive, suggesting that there was one locus containing the T-DNA. Three other mutants were identified in this screening, T1, T2, and T4. One was an allele of elf3, and the identity of the other mutants is still unknown because the T-DNA insertions do not cosegregate with the mutant phenotype.
Growth Conditions and Phenotypic Characterization
Seeds were sown in clear plastic boxes on 0.8% agar-water, incubated in darkness at 4°C for 3d, given 2 h of white light at 22°C, and transferred to darkness to induce homogeneous germination and then to different light or dark conditions for 3 d (22°C). Alternatively, the seedlings were transplanted to pots and transferred to short days (8 h white light/16 h darkness). Petiole length and leaf angle between the petiole and a line perpendicular to the soil surface were measured for the second fully expanded leaf of these plants. The number of leaves in the rosette was counted when the tip of the flowering primordia reached 1-cm height. Physiological data are the mean ± se of 4 to 20 replicates from at least two independent experiments.
Light Treatments
White light was provided by fluorescent tubes (Philips TLD 15W/54). EOD far red was provided by incandescent lamps combined with a water filter and an RG9 filter. Continuous far red was provided by incandescent lamps in combination with a water filter, a red acetate filter, and six 2-mm-thick blue acrylic filters (Paolini 2031; La Casa del Acetato). Red light was provided by light-emitting diodes, and blue light was provided by fluorescent white light tubes filtered through a blue acrylic filter. The red:far-red ratio was measured with a Skye meter (SKR 100, remote probe SKR 110; Skye Instruments).
Cloning of the Mutant Locus
The flanking genomic sequence at the T-DNA insertion site was recovered using the thermal asymmetric interlaced–PCR protocol (Liu et al., 1995). The T-DNA insertion was confirmed by PCR with specific primers. Perfect cosegregation (in 220 chromosomes analyzed) between the T-DNA insertion and the mutant phenotype was observed in F2 seedlings derived from a cross between the csa1 mutant and wild-type Col.
Mutant Alleles and Genotyping
Plants of each mutant line were genotyped using two PCR reactions, one for the mutant allele with the T-DNA insertion and one for the wild-type allele. The csa1-1 mutant allele was genotyped by PCR with primers TDNACSA1 (5′-TAATAACGCTGCGGACATCTAC-3′) and WTA (5′-CAACATCTGGAGAGCTAGTG-3′). To amplify the wild-type allele, we used primer WTA in combination with primer WTB (5′-GCTGGTGAAGTCTACTGACA-3′). The other T-DNA mutant alleles used in this study were obtained from the ABRC. The csa1-2 line (SALK_023219) has an insertion in exon 4. The T-DNA insertion in RPS4 (SALK_012799) has an insertion in exon 2. The T-DNA insertion in At5g44870 (SALK_087262) has an insertion in exon 2. For csa1-2, the wild-type allele was amplified with primers 023219LP (5′-CATCGCATCGTTTAAGCGCAC-3′) and 023219RP (5′-TGTTGTTCATTGCCCCAGGAT-3′), and the mutant allele was amplified with primers LBFH (5′-CGCTTGCTGCAACTCTCTCAGG-3′) and 023219RP.
For the T-DNA insertion in RPS4, the wild-type allele was amplified with primers 012799LP (5′-TGGCAGCTTTCTAAGCACCAAT-3′) and 012799RP (5′-GAGTTGGATCGCTTGCCTCAA-3′), and the mutant allele was amplified with primers LBFH and 012799RP. For the T-DNA insertion in At5g44870, the wild-type allele was amplified with primers 087262LP (5′-CATCCTCCACCCAACTAGGGA-3′) and 087262RP (5′-GCAAGTTCCTTGGCGAATGTG-3′) and the mutant allele with primers LBFH and 087262RP.
Recapitulation of the csa1 Mutant Phenotype
The ΔCSA1 construct was generated using primers that amplified the At5g17880 locus from the ATG start codon (5′-GACGGATCCATGACAAGCTCCTCCTCCTG-3′), adding a BamHI site to the site of insertion of the T-DNA within the second exon (5′-GTAGTCGACTTACTTAATTCGGAGCTCGATTC-3′), adding a TAA stop codon and the restriction site SalI. The PCR fragment amplified with the above primers was introduced into the binary vector CHF3 (Staiger et al., 2003) between the 35S promoter and the rbcs terminator. The resulting plasmid was introduced into Agrobacterium tumefaciens strain GV3101 and used to transform Arabidopsis plants of the Col ecotype. Fourteen transgenic lines were generated, and two lines with the strongest phenotype were used for phenotypic characterization.
Complementation of the csa1 Mutant Phenotype with RPS4
RPS4-Col driven by the CaMV 35S promoter was obtained by cloning a 6-kb genomic RPS4 fragment containing 0.5 kb of 5′ upstream and 1.5 kb of 3′ downstream sequence into vector pMD1, a derivative of vector pBI121 (CLONTECH) with a synthetic polylinker replacing the β-glucuronidase reporter gene (courtesy of M. Dixon, Sainsbury Laboratory, Norwich, UK). This construct was introduced into Agrobacterium strain GV3101 and used to transform csa1 mutant plants.
RT-PCR Analysis
Total RNA was extracted with the RNeasy miniprep kit (Qiagen). One microgram was used for the RT reaction with ImProm-II reverse transcriptase (Promega). Amplification of genomic DNA was undetectable in non-retrotranscribed controls. PCR products were detected in DNA gel blots using standard methodology in the exponential range of amplification. Primer sequences will be provided upon request.
Protein Blots
Extracts were prepared according to Martinez-García et al. (1999) and subjected to SDS-PAGE in 1.5-mm-thick, 4.5/7.5% stacking/resolving gels. Proteins were electroblotted to nitrocellulose, and PHYB was detected using a monoclonal antibody kindly provided by Peter Quail (University of California, Berkeley, and USDA Plant Gene Expression Center, Albany, CA).
Bacterial Infection
The bacterial growth assay was performed as described by Tornero and Dangl (2001). The bacterial strain used was Pseudomonas syringae pv tomato DC3000, containing the avirulence gene avrRpt2.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_121794 (At5g17880) and NM_123893 (At5g45250).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The csa1 Mutation Is Semidominant.
Supplemental Figure 2. Cellular Localization of PHYB:GFP in csa1 Mutant Plants.
Supplemental Figure 3. The Long Hypocotyl Phenotype of csa1 Seedlings Is Sustained under High Fluences of Red Light.
Supplemental Figure 4. RPS4 Overexpression Rescues the csa1 Mutant Phenotype.
Supplemental Figure 5. Red Light Inhibition of Hypocotyl Negative Gravitropism.
Supplementary Material
Acknowledgments
This work was supported by grants from Fundación Antorchas and Universidad de Buenos Aires to M.J.Y., Agencia Nacional de Promoción Científica y Tecnológica (BID 1201 OC/AR PICT 11631 and PICT 15098) to J.J.C. and M.J.Y., and National Institutes of Health GM56006 and GM067837 to S.A.K. We thank Romina Sellaro for her assistance with figure preparation, Malena Alvarez for providing bacterial strains, and the ABRC for providing seed stocks.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Marcelo J. Yanovsky (yanovsky@ifeva.edu.ar).
Online version contains Web-only data.
References
- Ballaré, C.L. (1999). Keeping up with the neighbors: Phytochrome sensing and other signaling mechanisms. Trends Plant Sci. 4 97–102. [DOI] [PubMed] [Google Scholar]
- Carabelli, M., Morelli, G., Whitelam, G.C., and Ruberti, I. (1996). Twilight-zone and canopy shade induction of the ATHB2 homeobox gene in green plants. Proc. Natl. Acad. Sci. USA 93 3530–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore, A.R., Jarillo, J.A., Wu, Y.-J., and Liu, D. (1999). Cryptochromes: Blue light receptors for plants and animals. Science 284 760–765. [DOI] [PubMed] [Google Scholar]
- Chen, M., Chory, J., and Fankhauser, C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38 87–117. [DOI] [PubMed] [Google Scholar]
- Couillault, C., Pujol, N., Reboul, J., Sabatier, L., Guichou, J.F., Kohara, Y., and Ewbank, J.J. (2004). TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat. Immunol. 5 488–494. [DOI] [PubMed] [Google Scholar]
- Devlin, P.F., Yanovsky, M.J., and Kay, S.A. (2003). A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol. 133 1617–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupraz, P., Cottet, S., Hamburger, F., Dolci, W., Felley-Bosco, E., and Thorens, B. (2000). Dominant negative MyD88 proteins inhibit interleukin-1beta/interferon-gamma-mediated induction of nuclear factor kB-dependent nitrite production and apoptosis in b cells. J. Biol. Chem. 275 37672–37678. [DOI] [PubMed] [Google Scholar]
- Fankhauser, C., and Casal, J.J. (2004). Phenotypic characterization of a photomorphogenic mutant. Plant J. 39 747–760. [DOI] [PubMed] [Google Scholar]
- Flor, H.H. (1956). The complementary genic systems in flax and flax rust. Adv. Genet. 8 29–54. [Google Scholar]
- Franklin, K.A., Praekelt, U., Stoddart, W.M., Billingham, O.E., Halliday, K.J., and Whitelam, G.C. (2003). Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol. 131 1340–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin, K.A., and Whitelam, G.C. (2005). Phytochromes and shade avoidance responses in plants. Ann. Bot. (Lond.) 96 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann, W., Hinsch, M.E., and Staskawicz, B.J. (1999). The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 20 265–277. [DOI] [PubMed] [Google Scholar]
- Genoud, T., Buchala, A., Chua, N.-H., and Métraux, J.-P. (2002). Phytochrome signaling modulates the SA-perceptive pathway in Arabidopsis. Plant J. 31 87–95. [DOI] [PubMed] [Google Scholar]
- Genoud, T., Millar, A.J., Nishizawa, N., Kay, S.A., Schäfer, E., Nagatani, A., and Chua, N.H. (1998). An Arabidopsis mutant hypersensitive to red and far-red light signals. Plant Cell 10 889–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks, K.A., Millar, A.J., Carre, I.A., Somers, D.E., Straume, M., Meeks-Wagner, D.R., and Kay, S.A. (1996). Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274 790–792. [DOI] [PubMed] [Google Scholar]
- Hoecker, U., Toledo-Ortiz, G., Bender, J., and Quail, P. (2004). The photomorphogenesis-related mutant red1 is defective in CYP83B1, a red light-induced gene encoding a cytochrome P450 required for normal auxin homeostasis. Planta 219 195–200. [DOI] [PubMed] [Google Scholar]
- Holt, B.R., Hubert, D., and Dangl, J. (2003). Resistance gene signaling in plants–Complex similarities to animal innate immunity. Curr. Opin. Immunol. 15 20–25. [DOI] [PubMed] [Google Scholar]
- Huq, E., Tepperman, J.M., and Quail, P.H. (2000). GIGANTEA is a nuclear protein invoved in phytochrome signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 97 9789–9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imler, J., and Hoffmann, J. (2002). Toll receptors in Drosophila: A family of molecules regulating development and immunity. Curr. Top. Microbiol. Immunol. 270 63–79. [DOI] [PubMed] [Google Scholar]
- Iwasaki, A., and Medzhitov, R. (2004). Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 5 987–995. [DOI] [PubMed] [Google Scholar]
- Johnson, G.B., Brunn, G.J., Tang, A.H., and Platt, J.L. (2003). Evolutionary clues to the functions of the Toll-like family as surveillance receptors. Trends Immunol. 24 19–24. [DOI] [PubMed] [Google Scholar]
- Jordan, T., Schornack, S., and Lahaye, T. (2002). Alternative splicing of transcripts encoding Toll-like plant resistance proteins - What's the functional relevance to innate immunity? Trends Plant Sci. 7 392–398. [DOI] [PubMed] [Google Scholar]
- Khanna, R., Kikis, E., and Quail, P. (2003). EARLY FLOWERING 4 functions in phytochrome B-regulated seedling de-etiolation. Plant Physiol. 133 1530–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberati, N.T., Fitzgerald, K.A., Kin, D.H., Feinbaum, R., Golenbock, D.T., and Ausubel, F.M. (2004). Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc. Natl. Acad. Sci. USA 101 6593–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8 457–463. [DOI] [PubMed] [Google Scholar]
- Magor, B.G., and Magor, K.E. (2001). Evolution of effectors and receptors of innate immunity. Dev. Comp. Immunol. 25 651–682. [DOI] [PubMed] [Google Scholar]
- Martinez-García, J.F., Monte, E., and Quail, P.H. (1999). A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 20 251–257. [DOI] [PubMed] [Google Scholar]
- Meyers, B., Morgante, M., and Michelmore, R. (2002). TIR-X and TIR-NBS proteins: Two new families related to disease resistance TIR-NBS-LRR proteins encoded in Arabidopsis and other plant genomes. Plant J. 32 77–92. [DOI] [PubMed] [Google Scholar]
- Meyers, B.C., Kozik, A., Griego, A., Kuang, H., and Michelmore, R.W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte, E., Tepperman, J.M., Al-Sady, B., Kaczorowski, K.A., Alonso, J.M., Ecker, J.R., Li, X., Zhang, Y., and Quail, P.H. (2004). The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl. Acad. Sci. USA 101 16091–16098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani, A. (2004). Light-regulated nuclear localization of phytochromes. Curr. Opin. Plant Biol. 7 708–711. [DOI] [PubMed] [Google Scholar]
- Palma, K., Zhang, Y., and Li, X. (2005). An importin alpha homolog, MOS6, plays an important role in plant innate immunity. Curr. Biol. 15 1129–1135. [DOI] [PubMed] [Google Scholar]
- Pujol, N., Link, E.M., Liu, L.X., Kurz, C.L., Alloing, G., Tan, M.W., Ray, K.P., Solari, R., Johnson, C.D., and Ewbank, J.J. (2001). A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr. Biol. 11 809–821. [DOI] [PubMed] [Google Scholar]
- Quail, P.H. (2002). Phytochrome photosensory signaling networks. Nat. Rev. Mol. Cell Biol. 3 85–93. [DOI] [PubMed] [Google Scholar]
- Roig-Villanova, I., Bou, J., Sorin, C., Devlin, P.F., and Martínez-García, J.F. (2006). Identification of primary target genes of phytochrome signaling. Early transcription control during shade avoidance responses in Arabidopsis. Plant Physiol. 141 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé, P., Michael, T., Kearns, E., Fett-Neto, A., Sharrock, R., and McClung, C.R. (2002). The out of phase 1 mutant defines a role for PHYB in circadian phase control in Arabidopsis. Plant Physiol. 129 1674–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa, G., Carabelli, M., Sassi, M., Ciolfi, A., Possenti, M., Mittempergher, F., Becker, J., Morelli, G., and Ruberti, I. (2005). A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev. 19 2811–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H. (2000). Phytochromes and light signal perception by plants-an emerging synthesis. Nature 407 585–590. [DOI] [PubMed] [Google Scholar]
- Staiger, D., Allenbach, L., Salathia, N., Fiechter, V., Davis, S.J., Millar, A.J., Chory, J., and Fankhauser, C. (2003). The Arabidopsis SRR1 gene mediates phyB signaling and is required for normal circadian clock function. Genes Dev. 17 256–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman, J.M., Hudson, M.E., Khanna, R., Zhu, T., Chang, S.H., Wang, X., and Quail, P.H. (2004). Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. Plant J. 38 725–739. [DOI] [PubMed] [Google Scholar]
- Tepperman, J.M., Zhu, T., Chang, H.-S., Wang, X., and Quail, P.H. (2001). Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. USA 98 9437–9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero, P., and Dangl, J.L. (2001). A high-throughput method for quantifying growth of phytopathogenic bacteria in Arabidopsis thaliana. Plant J. 28 475–481. [DOI] [PubMed] [Google Scholar]
- Wagner, D., Hoecker, U., and Quail, P.H. (1997). RED1 is necessary for phytochrome B-mediated red light-specific signal transduction in Arabidopsis. Plant Cell 9 731–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, M., Takeda, K., and Akira, S. (2004). TIR domain-containing adaptors define the specificity of TLR signaling. Mol. Immunol. 40 861–868. [DOI] [PubMed] [Google Scholar]
- Yanovsky, M.J., Casal, J.J., and Whitelam, G.C. (1995). Phytochrome A, phytochrome B and HY4 are involved in hypocotyl growth responses to natural radiation in Arabidopsis: Weak de-etiolation of the phyA mutant under dense canopies. Plant Cell Environ. 18 788–794. [Google Scholar]
- Yanovsky, M.J., and Kay, S.A. (2001). Signaling networks in the plant circadian system. Curr. Opin. Plant Biol. 4 429–435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.