Abstract
S-Adenosylmethionine (SAM) is formed exclusively in the cytosol but plays a major role in plastids; SAM can either act as a methyl donor for the biogenesis of small molecules such as prenyllipids and macromolecules or as a regulator of the synthesis of aspartate-derived amino acids. Because the biosynthesis of SAM is restricted to the cytosol, plastids require a SAM importer. However, this transporter has not yet been identified. Here, we report the molecular and functional characterization of an Arabidopsis thaliana gene designated SAM TRANSPORTER1 (SAMT1), which encodes a plastid metabolite transporter required for the import of SAM from the cytosol. Recombinant SAMT1 produced in yeast cells, when reconstituted into liposomes, mediated the counter-exchange of SAM with SAM and with S-adenosylhomocysteine, the by-product and inhibitor of transmethylation reactions using SAM. Insertional mutation in SAMT1 and virus-induced gene silencing of SAMT1 in Nicotiana benthamiana caused severe growth retardation in mutant plants. Impaired function of SAMT1 led to decreased accumulation of prenyllipids and mainly affected the chlorophyll pathway. Biochemical analysis suggests that the latter effect represents one prominent example of the multiple events triggered by undermethylation, when there is decreased SAM flux into plastids.
INTRODUCTION
S-Adenosylmethionine (SAM) is essential for all organisms because it is the predominant methyl donor in reactions catalyzed by methyltransferases. Additionally, SAM is a source of aminoalkyl and amino groups used for the biogenesis of polyamines and biotin (Hanson and Roje, 2001; Moffatt and Weretilnyk, 2001; Fontecave et al., 2004). In plants, SAM is also converted to 1-aminocyclopropane-1-carboxylic acid, the direct precursor of the plant hormone ethylene (Kevin et al., 2002). SAM also provides the methylene group used in the biogenesis of cyclopropane fatty acids that accumulate in many types of seeds (Bao et al., 2002, 2003).
Earlier biochemical studies revealed that in plants, SAM is synthesized by several soluble isoenzymes (Dogbo and Camara, 1986) located in the cytosol (Wallsgrove et al., 1983). Accordingly, the genome sequence of Arabidopsis thaliana encodes four SAM synthase proteins that all apparently lack organellar targeting sequences; therefore, plastids and mitochondria are strictly dependent upon SAM imported from the cytosol. The import of cytosolic SAM into plastids is crucial for plastid metabolism (Figure 1). SAM is an allosteric activator of plastid Thr synthase (Madison and Thompson, 1976; Aarnes, 1978; Giovanelli et al., 1984; Curien et al., 1998) and, thus, potentially controls the flux of O-phosphohomoserine into the biosynthesis of Thr and Met (Giovanelli et al., 1989; Galili and Hofgen, 2002) (Figure 1). Imported SAM is also used by several plastid methyltransferases, the substrates of which range from small molecules such as prenyllipids (e.g., chlorophylls, plastoquinone, tocopherol, and phylloquinone) (Bouvier et al., 2005; DellaPenna, 2005) and the diterpene antioxidant carnosic acid (Munne-Bosch and Alegre, 2001) to macromolecules such as plastid DNA (Nishiyama et al., 2002) and Rubisco (Black et al., 1987; Grimm et al., 1997; Ying et al., 1999; Trievel et al., 2003). In the latter case, the α-amino group of the N-terminal Met in the processed form of the small subunit and the ɛ-amino group of Lys-4 in the large subunit are methylated (Ying et al., 1999). However, the biological significance of Rubisco methylation is unknown at present.
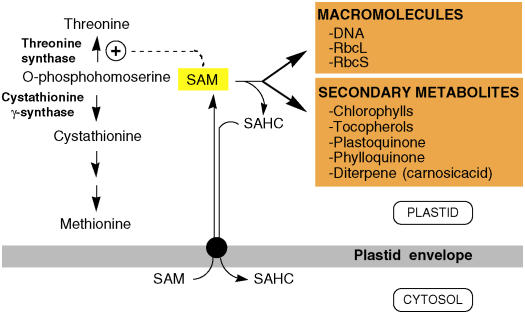
Figure 1.
Role of Plastid SAMT in Plants.
SAM is synthesized exclusively in the cytosol and transported to the stroma through a plastid envelope–localized SAMT (black circle). In plastids, SAM is engaged in the allosteric activation of Thr synthase or used by methyltransferases acting on macromolecules and secondary metabolites as shown. During these methyl transfer reactions, each SAM releases one S-adenosylhomocysteine (SAHC) as a by-product, which is exported in the cytosol. RbcL and RbcS refer to the large and small subunits of ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco).
During the transmethylation reaction, each SAM releases one SAHC, which behaves as a potent inhibitor of SAM-dependent methyltransferases (Poulton, 1981). For instance, in the prenyllipid pathway, the methylation of Mg-protoporphyrin IX (MgProto IX) to Mg-protoporphyrin IX methyl ester (MgProto IX Me) and the methylation of γ-tocopherol to α-tocopherol are inhibited by micromolar concentrations of SAHC (Koch et al., 2003; Shepherd et al., 2003). Therefore, it has been suggested that the efficiency of methyltransferases is dependent upon the efficient removal of SAHC by SAHC hydrolase (Poulton, 1981). The only known route for SAHC catabolism in eukaryotes is mediated by SAHC hydrolase, a cytosolic enzyme that catalyzes the hydrolysis of SAHC into adenosine and homocysteine (Hanson and Roje, 2001). Because of this, it is widely accepted that plastids and mitochondria must import cytosolic SAM and export SAHC to the cytosol.
Biochemical studies using isolated rat mitochondria (Horne et al., 1997) and spinach (Spinacia oleracea) chloroplasts (Ravanel et al., 2004) demonstrated that SAM uptake is mediated by a saturable, carrier-mediated process. In both cases, the transport assay revealed that uptake of external SAM occurred in counter-exchange with either SAM or SAHC, in agreement with earlier findings (Petrotta-Simpson et al., 1975). Despite the importance of SAM to plastid metabolism, the transporter catalyzing its import from the cytosol has not been characterized as a molecular entity. Genes encoding nonorganellar and organellar SAM transporters (SAMTs) have been identified in several evolutionarily diverse organisms. The first class includes structurally unrelated plasma membrane SAMTs from yeast (Rouillon et al., 1999) and the obligate intracellular bacterium Rickettsia prowazekii, the causative agent of epidemic typhus (Tucker et al., 2003). The second class contains mitochondrial SAMTs from yeast (Marobbio et al., 2003) and human mitochondria (Agrimi et al., 2004).
Here, we report the identification and functional characterization of a plastid SAMT from Arabidopsis that we designated SAMT1. SAMT1 is moderately similar to yeast and human SAM mitochondrial transporters. Both Arabidopsis T-DNA SAMT1 knockouts and Nicotiana benthamiana SAMT1-silenced plants have severely growth-retarded phenotypes and an altered plastid prenyllipid metabolism, characteristics that are consistent with an undermethylation status.
RESULTS
Molecular Characterization of SAMT1
A BLASTP search (Altschul et al., 1997) revealed that the closest Arabidopsis homolog (E-value, 9e-28; score, 126) of the yeast mitochondrial SAMT (SAM5p) (Marobbio et al., 2003) is encoded by At4g39460. The corresponding protein, provisionally named SAMT1, showed 31% amino acid identity with SAM5p. Based on this evidence, we amplified and cloned SAMT1 and its homologs from Capsicum annuum (Ca SAMT1) and Nicotiana benthamiana (Nb SAMT1) using single-stranded cDNAs transcribed from total RNA. The gene products of SAMT1, Ca SAMT1, and Nb SAMT1 have approximate molecular masses of 35 kD. The peptide sequence of SAMT1 exhibits 76% identity to the sequences of the putative Ca SAMT1 and Nb SAMT1 homologs, which, in turn, have 28% identity with the yeast SAM5p sequence (see Supplemental Figure 1 online). SAMT1 has 62% identity to the predicted amino acid sequence of At1g34065 when compared pairwise (see Supplemental Figure 1 online). A BLAST search revealed no significant similarity between SAMT1 and R. prowazekii SAMT (Tucker et al., 2003). The N-terminal regions of SAMT1, Ca SAMT1, and Nb SAMT1 displayed sequence motifs that could be identified as putative plastid transit peptides using ChloroP (http://genoplante-info.infobiogen.fr/predotar/) (Emanuelsson et al., 1999) and TargetP (http://www.cbs.dtu.dk/services/TargetP/) (Emanuelsson et al., 2000) (see Supplemental Figure 1 online). Analysis of SAMT1 using the transmembrane prediction program TMpred (http://www.ch.embnet.org/software/TMPRED_form.html) revealed that SAMT1 is a membrane-bound protein with five membrane-spanning domains located at residues 54 to 72, 99 to 118, 133 to 152, 230 to 250, and 285 to 304 (see Supplemental Figure 1 online). These domains were also predicted in yeast SAM5p.
Expression Pattern of SAMT1
The expression profile of SAMT1 was analyzed using the Digital Northern Tool of Genevestigator (Zimmermann et al., 2004, 2005). SAMT1 (At4g39460) is ubiquitously expressed in Arabidopsis but is most highly expressed in the pedicel, the shoot apex, and juvenile leaves (see Supplemental Figure 2 online). To identify genes that show similar expression patterns as SAMT1, we performed cluster analysis (Eisen et al., 1998) of the AtGenExpress developmental series microarray data (http://www.weigelworld.org/resources/microarray/AtGenExpress/) (Schmid et al., 2005). SAMT1 belongs to a highly correlated cluster (r = 0.86) of 68 genes, all of which are expressed predominantly in photosynthetic tissues such as rosette leaves and in the vegetative shoot apex but are strongly repressed in roots and later stages of seed development (see Supplemental Table 1 online). In accordance with the proposed function of SAMT1, the cluster contains genes involved in heme and chlorophyll biosynthesis (At5g14220, putative protophorphyrin oxidase) and methyltransferases [At3g62000, O-methyltransferase family 3 protein; At3g21300, RNA methyltransferase protein; At3g56330, N(2),N(2)-dimethylguanosine tRNA methyltransferase; At2g48120, pale cress protein (PAC)]. The pac mutant, which is defective in the PAC, was previously shown to have altered chloroplast and leaf development, and it was suggested that PAC is involved in chloroplast mRNA maturation (Holding et al., 2000).
Cellular Localization of SAMT1
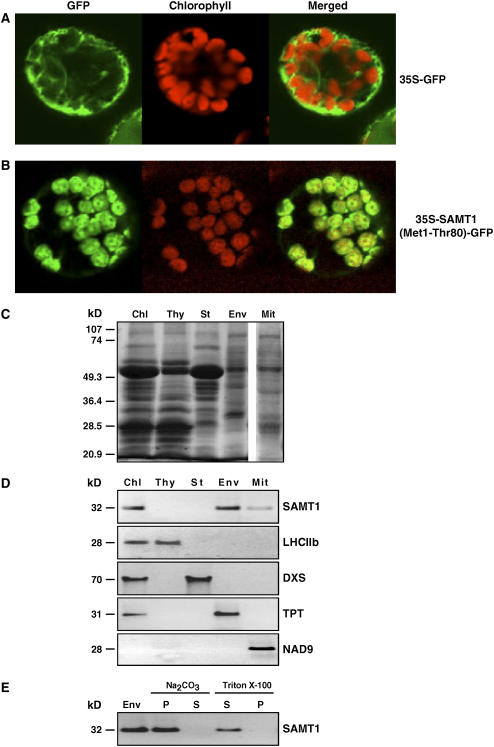
Because the organellar targeting of proteins predicted by algorithms cannot be considered completely reliable, the subcellular localization of SAMT1 was experimentally tested using green fluorescent protein (GFP) fusion proteins and antibodies raised against recombinant SAMT1. The similarity between SAMT1 and yeast mitochondrial SAMT starts at residue 55 (see Supplemental Figure 1 online). The computer-based programs ChloroP (http://www.cbs.dtu.dk/services/ChloroP/) and TargetP (http://www.cbs.dtu.dk/services/TargetP/) predicted a cleavage of SAMT1 between residues 23 and 24. To validate this prediction, a construct was made in which the first 80 N-terminal amino acids (Met-1 to Thr-80) of SAMT1 were fused to GFP. The resulting DNA construct was subcloned downstream of the cauliflower mosaic virus 35S promoter to give 35S-SAMT1(Met1-Thr80)-GFP. A construct without translational fusion between the first 80 N-terminal residues and the GFP (35S-GFP) was used as a control. Both constructs were introduced into N. benthamiana protoplasts using the methods of Goodall et al. (1990). After incubation at 25°C for 20 h, the protoplast preparations were examined by confocal laser scanning microscopy. The fluorescent images revealed that the transiently expressed (Met1-Thr80)-GFP was localized exclusively in tobacco chloroplasts characterized by their red autofluorescence, whereas the control 35S-GFP was detected in the cytosol (Figures 2A and 2B).
Figure 2.
Subcellular and Suborganellar Localization of SAMT1 in the Plant Cell.
The first 80 N-terminal amino acids (Met-1 to Thr-80) of SAMT1 were fused translationally to the green fluorescent protein (GFP). The corresponding DNA construct was fused to the cauliflower mosaic virus 35S promoter to give 35S-SAMT1(Met1-Thr80)-GFP. A construct without translational fusion between the first 80 N-terminal residues and the GFP (35S-GFP) was used as a control. Each construct was introduced into tobacco protoplasts using polyethylene glycol–mediated transformation, and the localization of the fluorescence was examined 20 h after transformation. The merged images of GFP and chlorophyll autofluorescence are shown.
(A) GFP fluorescence in a protoplast expressing the construct without translational fusion between GFP and the first 80 N-terminal amino acids (35S-GFP).
(B) GFP was translationally fused to the first 80 N-terminal amino acids (Met-1 to Thr-80) of SAMT1 to give 35S-SAMT1(Met1-Thr80)-GFP.
(C) SDS-PAGE analysis of total chloroplast (Chl), thylakoid membrane (Thy), stromal (St), chloroplast envelope membrane (Env), and total mitochondrial (Mit) proteins. Fifty micrograms of proteins was separated and stained using Coomassie blue. The mobilities of standard proteins of known molecular mass are indicated at left.
(D) Immunoblot analysis of total chloroplast (Chl), thylakoid membrane (Thy), stromal (St), chloroplast envelope membrane (Env), and total mitochondrial (Mit) proteins. Equal amounts of proteins (50 μg) were separated by SDS-PAGE, blotted, and subjected to protein gel blot analysis using SAMT1-specific antibodies or antibodies against the following marker proteins: light-harvesting chlorophyll a/b complex (LHCIIb; thylakoid membranes), 1-deoxy-d-xylulose 5-phosphate synthase (DXS; chloroplast stroma), triose phosphate translocator (TPT; chloroplast envelope membranes), and NADH dehydrogenase (NAD9) for the respiratory chain NADH dehydrogenase (complex I) of mitochondria. Numbers at left indicate molecular masses.
(E) Suborganellar localization of SAMT1. Chloroplast envelope membranes (Env) were treated with 0.5 M Na2CO3, pH 11.5, or 2% Triton X-100 for 30 min at 0°C. After ultracentrifugation (100,000g), the pellet (P) and the supernatant (S) were used for immunoblot analysis using anti-SAMT1. The molecular mass of SAMT1 is indicated at left.
To further analyze the compartmentation of SAMT1, intact purified Arabidopsis chloroplasts, chloroplast subfractions (chloroplast envelope membranes, thylakoid membranes, and stroma), and mitochondria were isolated and subjected to SDS-PAGE and immunoblot analysis (Figures 2C to 2E) using specific polyclonal antibodies raised against recombinant SAMT1 (see Supplemental Figure 3 online).
On immunoblots of chloroplast proteins, the anti-SAMT1 antibody detected a major band with a molecular mass of 32 kD corresponding to the mature form of SAMT1 in total chloroplast and in chloroplast envelope membrane proteins (Figure 2D). Neither thylakoid membrane nor stroma proteins contained immunoreactive SAMT1 (Figure 2D). The purity of the isolated mitochondria and plastids, and of their subcompartments, was tested using antibodies directed against proteins known to be localized in specific chloroplast or mitochondrial compartments. These included anti-LHCIIb for the light-harvesting chlorophyll a/b complex for the thylakoid membranes (Paulsen et al., 1990), anti-TPT for the chloroplast envelope membrane triose phosphate translocator (Flügge et al., 1989), anti-DXS for the stromal 1-deoxy-d-xylulose 5-phosphate synthase (Bouvier et al., 1998), and anti-NAD9 for the respiratory chain NADH dehydrogenase (complex I) of mitochondria (Weiss et al., 1991) (Figure 2D). Immunoblot analysis of mitochondrial proteins with anti-SAMT1 showed a faint signal, corresponding approximately to the same molecular mass protein as that detected previously in chloroplast protein samples (Figures 2C and 2D). This unidentified protein has a localization pattern that coincides with that of the mitochondrial NADH dehydrogenase (NAD9) (Figures 2C and 2D). This immunoreactivity could be attributable to the fact that SAMT1 and At1g34065 share 62% amino acid sequence (see Supplemental Figure 1 online). In addition, several bioinformatic analyses suggest that At1g34065 may be located in the mitochondria (http://www.suba.bcs.uwa.edu.au/) (Picault et al., 2004; Heazlewood et al., 2005).
After treatment of chloroplast envelope membranes with 0.5 M sodium carbonate, pH 11.5, and ultracentrifugation, SAMT1 remained in the alkali-insoluble membrane pellet (Figure 2E). This treatment releases peripheral proteins while not significantly solubilizing integral proteins (Fujiki et al., 1982). Under similar conditions, SAMT1 was not extracted from the chloroplast envelope membranes using 1 M NaCl (data not shown). As would be predicted for an integral protein, SAMT1 was completely solubilized by treatment with 2% Triton X-100 (Figure 2E). The localization of SAMT1 in the chloroplast envelope membranes has been suggested previously by bioinformatic analysis (Koo and Ohlrogge, 2002) and demonstrated by proteomics (Ferro et al., 2002, 2003), supporting the localization data presented in this study. Collectively, these data demonstrate that SAMT1 is an integral membrane protein that is localized in the chloroplast envelope membranes.
Plastid SAMT1 Mediates Specific Counter-Exchange of SAM with SAM and SAHC
Sequence similarity to known proteins allows for the development of testable hypotheses about the possible function of a novel protein. To test our hypothesis that SAMT1 encodes a SAMT, we expressed the corresponding cDNA in yeast cells under the control of a galactose-inducible promoter. The nucleotides encoding the first 35 N-terminal amino acids of the plastid targeting sequence were omitted from the expression construct because the presence of the plastid targeting sequence frequently leads to reduced expression levels in yeast cells; instead, codons encoding an N-terminal hexa-His tag were inserted at the N terminus (see Methods for details), allowing the detection of the recombinant protein with antibodies directed against the His tag.
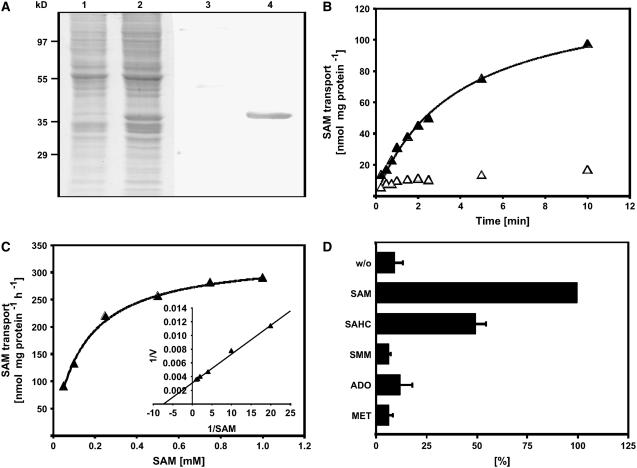
Total membrane fractions were prepared from transgenic yeast harboring the expression construct or an empty vector and analyzed by SDS gel staining and protein gel blot for the expression of SAMT1. Visual inspection of the Coomassie blue–stained SDS-PAGE gel revealed the presence of an additional protein band with an apparent molecular mass of ∼36 kD in the membrane fractions of yeast cells harboring the expression construct (Figure 3A, lane 2) that was not detectable in yeast cells containing the control construct (Figure 3A, lane 1). Proteins from an identical gel were electrotransferred onto a membrane and subsequently immunodecorated using an anti-penta-His monoclonal antibody. A specific signal with an apparent molecular mass of 36 kD was detected in cells harboring the expression construct, but not in control cells (Figure 3A, lanes 3 and 4), indicating that the additional protein band observed in the Coomassie blue–stained gel represents recombinant, His-tagged SAMT1.
Figure 3.
Expression of SAMT1 in Transgenic Cells, and Determination of Apparent Km and Substrate Specificity of Recombinant SAMT1.
(A) Coomassie blue–stained SDS-PAGE gel showing the total membrane fraction of yeast cells transformed with either the empty expression vector (lane 1) or the expression vector harboring the cDNA encoding SAMT1 (lane 2), and protein gel blot of an identical gel (lanes 3 and 4). Control cells and expression lines were processed in parallel, including induction of expression by the addition of galactose. After transfer of proteins from the gel to a membrane, the proteins were immunodecorated using an anti-penta-His antibody. Lanes 1 and 3, control cells; lanes 2 and 4, yeast line expressing SAMT1.
(B) Time kinetics of SAM transport into liposomes reconstituted with SAMT1. Uptake of SAM into liposomes that had been reconstituted with SAMT1 and preloaded with 20 mM SAM was induced by the addition of radiolabeled SAM to the liposome suspension. Aliquots were removed after 20, 30, 40, 60, 80, 120, 140, 300, and 600 s, and the transport reaction was terminated by loading the liposome suspension onto ion-exchange columns. SAM transport was quantified by liquid scintillation counting of the column pass-through. Open symbols and closed symbols refer to liposomes reconstituted with membranes from control cells and cells expressing SAMT1, respectively.
(C) Determination of the apparent Km value of SAMT1. Rates of SAM uptake into liposomes preloaded with 20 mM SAM were quantified independent of various external SAM concentrations. The inset shows a double-reciprocal plot of the data (Lineweaver-Burk plot) that was used to determine the apparent Km value of SAMT1 for SAM transport. Results from one representative experiment of five independent replicates are shown. The apparent Vmax observed in the experiment shown was 357 nmol SAM·mg−1 protein·h−1.
(D) Substrate specificity of SAMT1. The substrate specificity of SAMT1 was determined by quantifying the rate of SAM transport into liposomes that had been preloaded with 20 mM SAM, SAHC, SMM, adenosine, Met, or control substrate (gluconate). SAM transport was quantified as described for (B). The data shown are arithmetic means ± sd from five independent replicates. ADO, adenosine; w/o, control (i.e., without internal substrates).
Membrane fractions from both yeast lines were reconstituted into liposomes that had been preloaded with SAM and assayed for SAM transport activity. Background SAM transport activity of reconstituted membrane proteins from control cells was negligible (Figure 3B, open symbols), whereas liposomes reconstituted with membranes from yeast cells expressing SAMT1 showed high saturable SAM–SAM counter-exchange activity (Figure 3B, closed symbols).
Affinity of SAMT1 for SAM and Substrate Specificity
To investigate the putative function of a transporter protein in vivo, knowledge of its substrate specificity as well as its apparent Km values is required. Using proteoliposomes preloaded with saturating concentrations of SAM, we observed an apparent Km value of 129 ± 16 μM for the uptake of radiolabeled SAM into liposomes (Figure 3C, inset). The apparent Vmax values for SAM–SAM counter-exchange were determined in the five independent experiments conducted to determine the apparent Km for SAM uptake and ranged between 270 and 398 nmol·mg−1 protein·h−1, indicating that the levels of SAMT1 expression and the efficiency of reconstitution were comparable between the independent replicates.
The substrate specificity of SAMT1 was analyzed by measuring the uptake of radiolabeled SAM into proteoliposomes that had been preloaded with saturating concentrations (i.e., 20 mM) of SAM, SAHC, S-methylmethionine (SMM), Met, adenosine, or control substrates, respectively. The SAM–SAM counter-exchange rates were set to 100%, and exchange rates for other substrates were calculated relative to the rates observed for cis/trans SAM transport. Liposomes reconstituted with SAMT1 showed high rates of SAM uptake when preloaded with either SAM (100%) or SAHC (50%), whereas the uptake of SAM into liposomes preloaded with SMM, Met, adenosine, or no counter-exchange substrate was negligible (Figure 3D).
Together, these results clearly demonstrate that SAMT1 represents a high-affinity SAM–SAHC antiporter that has negligible SAM uniporter activity and that does not accept nonspecific counter-exchange substrates such as SMM, Met, or adenosine.
Identification of T-DNA Knockout Mutants
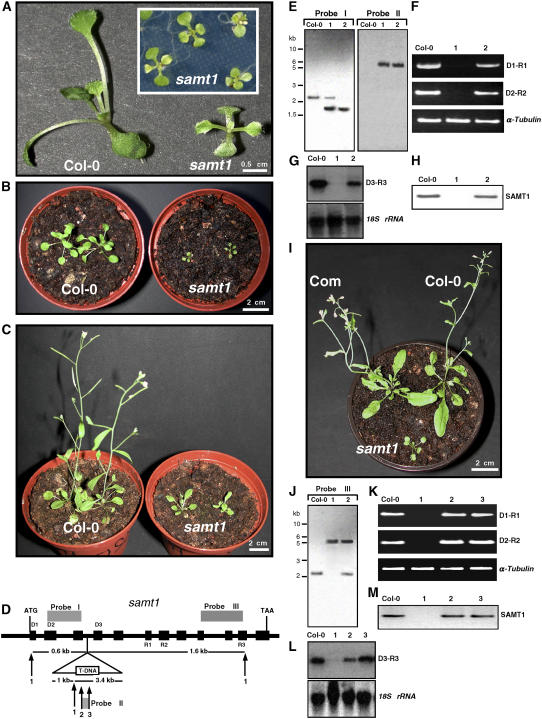
To determine whether SAMT1 has the function of a plastidic SAMT in vivo and whether a mutation in the SAMT1 gene would affect plastid functions, we identified a T-DNA insertion for SAMT1 from the Salk Institute Genomic Analysis Laboratory Collection (Alonso et al., 2003). To this end, the SALK_008248 line (subsequently designated samt1) was selected. The progeny of self-fertilized heterozygous kanamycin-resistant plants grown under nonselective conditions revealed a segregation of pale-green leaf phenotype during the first weeks and a severely growth-retarded phenotype (Figures 4A to 4C). samt1 mutants have very low germination efficiency, with <20% viable seedlings produced. Furthermore, samt1 has a long generation time, at least three times that of wild-type plants (Figures 4B and 4C). The segregation ratio of the encoded kanamycin marker corresponded to 3:1, as expected for a single Mendelian transmission. When heterozygous kanamycin-resistant plants were grown under selective conditions, the kanamycin resistance or sensitivity (Kanr or Kans) and the mutant phenotype segregated in the following ratio: mutant Kanr:wild-type Kanr:wild-type Kans (1:2:1). This finding indicated that the phenotype was genetically linked to the homozygous state of the mutation.
Figure 4.
Phenotypic and Molecular Analyses of Arabidopsis Wild-Type (Col-0) and samt1 Mutant Plants.
(A) Twelve-day-old plants were grown on agar-solidified Murashige and Skoog medium supplemented with 0.5% sucrose.
(B) and (C) Four-week-old (B) and 5-week-old (C) plants were grown for 14 d on Murashige and Skoog agar medium supplemented with 0.5% sucrose before photoautotrophic growth on damp compost.
(D) Gene structure of SAMT1, and position of the T-DNA insertion. Black boxes and thick lines indicate the positions of exons and introns, respectively. The triangle represents the T-DNA (not drawn to scale) inserted in position +641 of the SAMT1 genomic sequence. The positions of the primers used for RT-PCR (D1, D2, D3, R1, R2, and R3) and the probes (I to III) used for RNA and DNA gel blots are indicated. Numbers indicate SacI (1), KpnI (2), and ScaI (3) restriction sites.
(E) DNA gel blot analysis of genomic DNA from SAMT1 (Col-0), heterozygous samt1 (1), and homozygous samt1 (2) plants digested with SacI and probed with probes I and II.
(F) RT-PCR analysis of SAMT1 gene expression in wild-type (Col-0), homozygous samt1 (1), and heterozygous samt1 (2) plants. Primers used are indicated at right. Amplification of α-tubulin mRNA was used as a positive control. PCR was performed for 27 cycles (α-tubulin) and 35 cycles (SAMT1).
(G) RNA gel blot analysis of SAMT1 gene expression in wild-type and samt1 plants. Total RNA (10 μg) was isolated from wild-type (Col-0), homozygous samt1 (1), and heterozygous samt1 (2) plants. Blots were hybridized with SAMT1 probe and with 18S rRNA probe to verify equivalent sample loading.
(H) Protein gel blot analysis of SAMT1 in wild-type (Col-0), homozygous samt1 (1), and heterozygous samt1 (2) plants. Fifty micrograms of chloroplast proteins was separated by SDS-PAGE, blotted, and subjected to immunoblot analysis using SAMT1-specific antibodies.
(I) Complementation of the samt1 mutant. Phenotypes of 5-week-old wild-type (Col-0) and homozygous samt1 and complementation transgenic (samt1/35S-SAMT1) (Com) lines are shown.
(J) DNA gel blot analysis of genomic DNA from wild-type (Col-0), homozygous samt1 (1), and complemented (samt1/35S-SAMT1) (2) plants. Genomic DNA was digested with SacI and hybridized with probe III.
(K) RT-PCR analysis of SAMT1 expression in the wild type (Col-0) and in the homozygous samt1 (1), the heterozygous samt1 (2), and the complemented (samt1/35S-SAMT1) (3) lines. Primers used are indicated at right. Amplification of α-tubulin mRNA was used as a positive control. PCR was performed for 25 cycles (α-tubulin) and 35 cycles (SAMT1).
(L) RNA gel blot analysis of SAMT1. Total RNA (10 μg) used was isolated using the lines shown in (K). Blots were hybridized with the probes used in (G).
(M) Protein gel blot analysis of SAMT1 in wild-type and mutant lines as in (K). An equivalent amount of chloroplast proteins (50 μg) was immunologically probed with anti-SAMT1.
We confirmed by sequencing the T-DNA flanking genomic DNA regions in the samt1 mutant line that the T-DNA insertion is located at position +641 of SAMT1, in the third intron, downstream of the start codon (Figure 4D). This insertion leads to the truncation of SAMT1 after 83 amino acids. One band was observed in the homozygous line, and its size is consistent with the insertion of a single T-DNA (Figure 4E, lane 2, probe II). The absence of SAMT1 mRNA from the samt1 mutant line was verified by RT-PCR, RNA gel blot analysis, and protein gel blot analysis using anti-SAMT1 antibody and purified chloroplasts prepared from leaf protoplasts (Fitzpatrick and Keegstra, 2001). RT-PCR using primers surrounding the targeted insertion region demonstrated that SAMT1 mRNA was absent from the homozygous insertion line but was clearly detectable in the heterozygous line and the wild-type Columbia ecotype (Col-0) (Figure 4F). Arabidopsis α-tubulin was used as a control to assess the quality of the mRNA and the efficiency of the RT-PCR (Figure 4F). The absence of SAMT1 mRNA in the homozygous line was also confirmed by RNA gel blot analysis (Figure 4G). In addition, purified plastids were prepared from the wild-type and heterozygous and homozygous samt1 lines and subjected to protein immunoblot analysis. SAMT1 was not detectable in plastid preparations obtained from the homozygous samt1 line, but it was clearly detectable as a protein with an apparent molecular mass of 32 kD in the heterozygous and wild-type lines (Figure 4H). These data demonstrate that a T-DNA insertion in SAMT1 leads to a null allele of samt1.
To further show that the phenotype of the samt1 mutant was caused by the T-DNA insertion in SAMT1, we attempted genetic complementation of the mutant phenotype by expression of SAMT1 in the mutant background. To this end, a 2.5-kb genomic fragment encoding the amino acid sequence of SAMT1 was placed under transcriptional control of the cauliflower mosaic virus 35S promoter, and the resulting construct (35S-SAMT1) was introduced into the heterozygous (samt1/SAMT1) mutant. From the progeny, we selected complemented lines (samt1/35-SAMT1) that contained both the 35S-SAMT1 transgene and the T-DNA insertion using PCR- and RT-PCR–based genotyping, as described in Methods (see Supplemental Figure 4 online). Five lines (samt1/35S-SAMT1) containing a homozygous samt1 allele and the 35S-SAMT1 transgene were obtained. Figure 4I shows that the phenotype of a representative transgenic line containing samt1 and the SAMT1 transgene (samt1/35S-SAMT1) (Figure 4J) is similar to that of the wild type. In addition, RT-PCR, RNA gel blot, and protein immunoblot analyses revealed the accumulation of SAMT1 mRNA and protein in these homozygous complemented lines (Figures 4K to 4M). Collectively, these data demonstrate that the phenotype of the samt1 mutant is caused by T-DNA insertional inactivation of SAMT1 and that the phenotype conferred by samt1 can be rescued by expression of the wild-type SAMT1 in the mutant background.
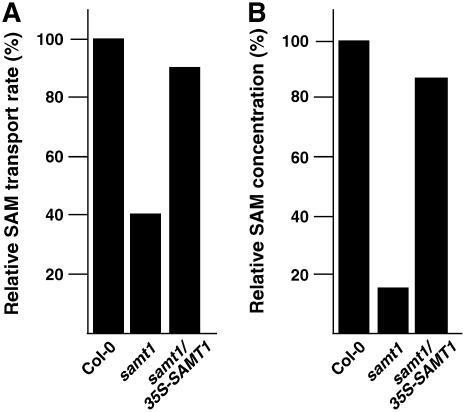
To confirm that the observed phenotype is linked to a SAM transport defect, we evaluated the uptake of labeled [14C]SAM into chloroplasts isolated from the homozygous mutant line and determined the endogenous chloroplast SAM concentration. As shown in Figure 5A, the capacity of chloroplasts isolated from the samt1 lines to import SAM is significantly lower than that of the complemented lines or the wild-type plants. A similar trend was observed concerning the endogenous concentration of chloroplast SAM (Figure 5B). Based on the stromal volume per unit of chlorophyll reported by Winter et al. (1994), the estimated concentrations of SAM in the stroma were equivalent to 1.6 μM in samt1 plants and 9 μM in wild-type plants.
Figure 5.
Plastid SAM Uptake and Endogenous Concentration of SAM.
(A) Plastid uptake was estimated after incubation of isolated chloroplasts with 200 μM [14C]SAM for 1 min. The 100% transport was 375 pmol·mg−1 protein·min−1. Chloroplasts were isolated from wild-type (Col-0), samt1, and complemented (samt1/35S-SAMT1) lines.
(B) Concentration of plastid SAM. The 100% value was 215 pmol/mg protein. Wild-type (Col-0), samt1, and complemented (samt1/35S-SAMT1) lines were analyzed.
Values shown are from two independent experiments that gave similar results.
Effects of SAMT1 Silencing on Plastid Prenyllipids in Arabidopsis and N. benthamiana
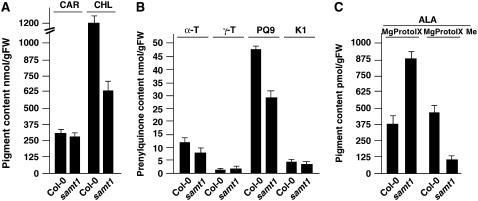
Because SAM is required and channeled toward different subplastial pathways of prenyllipid biosynthesis, we evaluated whether plastid SAMT1 could regulate the supply of SAM engaged in chlorophyll, tocopherol, phylloquinone, and plastoquinone biosynthesis in vivo. The results shown in Figures 6A and 6B indicate that total chlorophyll content and plastoquinone contents were reduced in samt1 compared with wild-type Col-0. The overall level of tocopherol and phylloquinone was slightly lower in samt1 plants; however, the carotenoid content was not significantly modified. A key metabolic step in which plastid SAM is engaged is the conversion of MgProto IX to MgProto IX Me. Hypothesizing that this step might be affected, we incubated detached rosette leaves with 5-aminolevulinate, the chlorophyll precursor. As shown in Figure 6C, the total amount of MgProto IX and the ratio of MgProto IX to MgProto IX Me were higher in samt1 than in wild-type Col-0. These data are consistent with a lower capacity of samt1 to import plastid SAM channeled to the chlorophyll pathway.
Figure 6.
Pigment and Prenyllipid Contents of Wild-Type (Col-0) and samt1 Plants.
(A) Total carotenoid (CAR) and chlorophyll (CHL) contents of wild-type (Col-0) and samt1 plants.
(B) Total α-tocopherol (α-T), γ-tocopherol (γ-T), plastoquinone-9 (PQ9), and phylloquinone (K1) contents of wild-type (Col-0) and samt1 plants.
(C) Rosette leaves were detached and incubated with 5 mM 5-aminolevulinate (ALA) for 12 h in the dark before determination of MgProto IX and MgProto IX Me contents.
Total lipids were extracted from 3-week-old plants and subjected to chromatographic analysis. Values represent means ± sd from three independent analyses. FW, fresh weight.
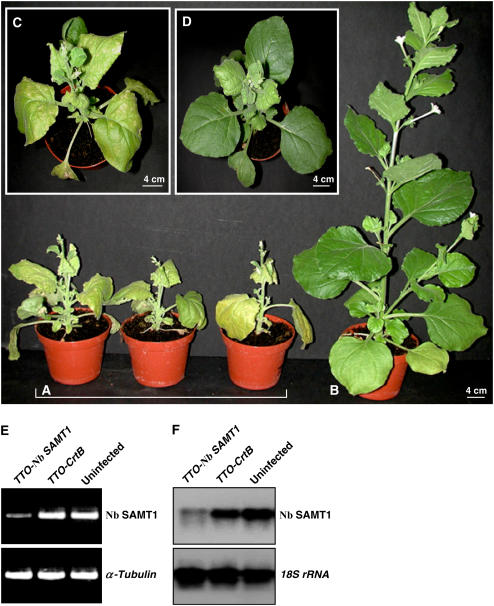
To verify the results obtained with the Arabidopsis insertion mutant using an independent experimental approach, we examined whether identical metabolic changes could be observed in N. benthamiana plants in which the expression of N. benthamiana SAMT1 (Nb SAMT1) had been deliberately silenced using a virus-induced gene-silencing strategy. To this end, we cloned a fragment of Nb SAMT1 into the viral vector TTO and inoculated N. benthamiana plants with infectious TTO-Nb SAMT1 mRNAs. A construct (TTO-CrtB), consisting of TTO harboring a fragment of the bacterial phytoene synthase (CrtB) sequence, was used as a control rather than the empty vector, because expression of the latter sometimes causes adverse effects on plant growth. Three weeks after infiltration, the silencing effect was analyzed. Nb SAMT1–silenced plants were strongly stunted and became pale green (Figures 7A and 7C), whereas control plants infected with TTO-CrtB resembled wild-type plants (Figures 7B and 7D). RT-PCR was performed to evaluate the silencing efficiency of Nb SAMT1 using primers annealing outside of the 5′ and 3′ ends of the fragment used for the TTO-Nb SAMT1 construct. Nb SAMT1 mRNAs were strongly reduced in silenced tissues compared with control tissues infected with TTO-CrtB and wild-type tissues (Figure 7E). A similar trend was observed using mRNA gel blot analysis (Figure 7F).
Figure 7.
Virus-Induced Gene Silencing of N. benthamiana SAMT1 (Nb SAMT1).
(A) N. benthamiana plants infected with TTO-Nb SAMT1 were photographed at 21 d after inoculation.
(B) Uninfected N. benthamiana plants were photographed at 21 d after inoculation.
(C) and (D) N. benthamiana plants infected with TTO-Nb SAMT1 (C) or TTO-CrtB (D) used as a control were photographed at 21 d after inoculation.
(E) RT-PCR analysis of the transcript level of Nb SAMT1. Primers annealing outside of the TTO-Nb SAMT1 construct were used. The expression of α-tubulin was used as a control. Upper uninoculated leaves from three independent plants were used to determine Nb SAMT1 mRNA levels in TTO-Nb SAMT1 (A), TTO-CrtB (D), and uninfected (B) plants. PCR was performed for 25 cycles (α-tubulin) and 32 cycles (Nb SAMT1).
(F) RNA gel blot analysis was performed using total RNA (10 μg) isolated from the same plants described for (E). Blots were hybridized with Nb SAMT1 probe and 18S rRNA probe to verify equivalent sample loading.
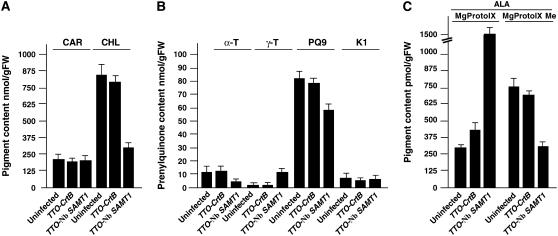
To examine the metabolic changes in plastids from Nb SAMT1–silenced plants, we analyzed the level of plastid pigments and prenyllipids of upper uninoculated leaves 3 weeks after inoculation. The total content of chlorophyll and plastoquinone-9 was reduced significantly in Nb SAMT1–silenced plants (Figures 8A and 8B). This phenomenon was accompanied by an increased accumulation of γ-tocopherol in tissues expressing TTO-Nb SAMT1 (Figure 8B). No such changes were observed in upper uninoculated leaf tissues from control plants infected with TTO-CrtB and in wild-type plants (Figures 8A and 8B). By contrast, the carotenoid content remained essentially unchanged. To evaluate the effect of Nb SAMT1 silencing on the chlorophyll pathway, upper uninoculated leaves were detached from Nb SAMT1–silenced plants, TTO-CrtB–infected plants, and wild-type plants and fed with 5-aminolevulinate, as shown above. Similar to what was observed in the Arabidopsis T-DNA insertion mutant, leaf tissues of Nb SAMT1–silenced plants exhibited a higher content of MgProto IX and higher MgProto IX to MgProto IX Me ratio compared with control tissues of TTO-CrtB and wild-type tissues (Figure 8C). This observation strongly suggests that Nb SAMT1 is required for the import of SAM from the cytosol, as shown above for samt1, and further supports the notion that Nb SAMT1 functions as a transporter in the plastid envelope.
Figure 8.
Pigment and Prenyllipid Contents of Uninfected, TTO-CrtB, and TTO-Nb SAMT1–Silenced Plants.
(A) Total carotenoid (CAR) and chlorophyll (CHL) contents.
(B) Total α-tocopherol (α-T), γ-tocopherol (γ-T), plastoquinone-9 (PQ9), and phylloquinone (K1) contents.
(C) Upper noninoculated leaves were detached and incubated with 5 mM 5-aminolevulinate (ALA) for 12 h in the dark before determination of MgProto IX and MgProto IX Me contents.
Total lipids were extracted from 3-week-old plants and subjected to chromatographic analysis. Values represent means ± sd from three independent analyses. FW, fresh weight.
DISCUSSION
The current literature indicates that in all eukaryotes, SAM is synthesized in the cytosol and translocated into cell organelles via specific carriers. This pathway has been characterized at the molecular level in yeast (Marobbio et al., 2003) and human (Agrimi et al., 2004) mitochondria but has not yet been characterized in plants. Sequence comparison of SAMT1 with these nonplant mitochondrial SAMTs showed only moderate sequence similarity (see Supplemental Figure 1 online). However, SAMT1 displays the characteristic features of members of the mitochondrial carrier family (MCF) of proteins, such as a hydrophobic polypeptide sequence with a tripartite structure composed of five membrane-spanning domains (Picault et al., 2004).
The results presented here indicate that the Arabidopsis SAMT1 gene encodes a plastid-targeted protein. The relatively high degree of sequence similarity of SAMT1 and At1g34065 suggests that these two proteins might be homologs. At1g34065 has been classified as a member of the MCF based on its primary structure (Picault et al., 2004). Although this suggests that At1g34065 may represent a mitochondrial SAMT, further analysis is required. Interestingly, members of the MCF do not seem to be restricted to the mitochondrial inner membrane. Recently, another MCF member was demonstrated to function as a plastidic folate transporter (Bedhomme et al., 2005), and other MCF members have been shown to be involved in adenine nucleotide uniport into plastids (Leroch et al., 2005) and in the uptake of ADP-glucose into maize (Zea mays) endosperm plastids (Sullivan and Kaneko, 1995). Moreover, a yeast peroxisomal ATP transporter also belongs to the MCF (Palmieri et al., 2001), indicating that targeting of MCF members to organelles other than mitochondria is not restricted to the plant kingdom.
SAMT1 could be expressed with high yields in transgenic yeast cells under the control of the galactose-inducible GAL4 promoter, and the activity of the recombinant protein could be reconstituted into liposomes. The uptake of radiolabeled SAM into liposomes reconstituted with SAMT1 was found to be dependent on the preloading of liposomes with suitable counter-exchange substrates such as SAM and SAHC. No significant SAM uniport activity was observed under the conditions used in our study, and counter-exchange of SAM with SMM, Met, or adenosine was negligible. The apparent Km value of recombinant SAMT1 for SAM uptake in the in vitro reconstituted system was ∼130 μM, which is higher than the value of 38 μM determined using intact isolated spinach chloroplasts (Ravanel et al., 2004). This finding might be attributable to species-specific differences between spinach and Arabidopsis, or to the fact that the reconstitution procedure affects protein function. The fact that reconstituted solute transporters display slightly higher Km values than those in their authentic environment is commonly observed. For example, the apparent Km of the plastidic triose phosphate/phosphate translocator reconstituted into liposomes is approximately threefold higher than that determined using intact chloroplasts (Gross et al., 1990; Loddenkotter et al., 1993; Fischer et al., 1997). Also, the apparent Km values observed for plastidic dicarboxylate translocators reconstituted into liposomes are higher than those observed in intact, isolated plastids (Renne et al., 2003). Furthermore, the additional amino acids resulting from the fusion of the hexa-His tag to the N terminus of the transporter protein might influence the kinetic properties of the recombinant protein. Importantly, because of experimental constraints, the fastest uptake kinetics that can be measured in a reconstituted liposome system are in the range of 20 s, significantly slower than the kinetics that can be measured with intact organelles using a silicon-oil filtration-centrifugation system. Hence, the initial rate of the transport kinetics might be underestimated in the liposome system. In this context, it is also interesting that a Km of 75 μM, which is close to the value reported in this study, has been reported in an in vitro system for yeast mitochondria SAMT (Marobbio et al., 2003). It is thus likely that the apparent Km of reconstituted SAMT1 overestimates the true Km value, which is probably lower than that observed in the reconstituted system and closer to the one determined using isolated chloroplasts.
Although SAM is required for various plastid pathways, little is known about plastid SAM homeostasis. Previous studies have shown that decreasing cytosolic SAM synthetase in tobacco using antisense RNA yields chlorotic, senescent, and dwarf plants (Boerjan et al., 1994); however, it has not been established to what extent the levels of plastid SAM were affected. Available data from the Arabidopsis literature indicate that SAMT1 expression is observed in all tissues, with the highest transcript levels in juvenile leaves and shoot apices and the lowest levels in senescent leaves (see Genevestigator [http://www.genevestigator.ethz.ch/]). Interestingly, senescence is considered as a period of SAM limitation (Moffatt and Weretilnyk, 2001). In this context, the accumulation of γ-tocopherol in TTO-Nb SAMT1–silenced plants (Figure 8B) and in senescing tobacco leaves (Falk et al., 2003) could reflect a decreased plastid SAM import.
The biochemical analyses of SAMT1 insertional mutants and of N. benthamiana plants in which Nb SAMT1 expression had been silenced reveal that in the general prenyllipid pathway, the chlorophyll pathway is the most sensitive to SAM limitation. A possible implication of this result is that the SAM-dependent conversion of MgProto IX to MgProto IX Me becomes severely disrupted when the import of SAM into plastids is restricted by SAMT1 activity. Interestingly, tobacco plants expressing antisense SAM-MgProto IX methyltransferase exhibited chlorotic leaves and a reduced-growth phenotype (Alawady and Grimm, 2005), as seen in Nb SAMT1–silenced plants (Figures 7A and 7C). The kinetic constants of enzymes catalyzing the different steps of subplastid prenyllipid biosynthetic pathways are not yet available. Examination of the available literature indicates that plastid methyltransferases involved in the prenylquinone pathway (i.e., SAM-γ-tocopherol methyltransferase with a Km for SAM in the range of 2 to 5 μM) (d'Harlingue and Camara, 1985; Shintani and DellaPenna, 1998; Koch et al., 2003) have 10-fold higher affinity for SAM than Mg-Proto IX methyltransferase, which has a Km of 38 μM (Shepherd et al., 2003). Also, the Km for Rubisco large subunit methyltransferase is in the range of 2 to 8 μM (Zheng et al., 1998). Therefore, one may suggest that when a low concentration of SAM prevails in plastids, plastid methyltransferases having a high affinity for SAM, such as SAM-γ-tocopherol or SAM-Rubisco methyltransferases, are favored compared with SAM-MgProto IX methyltransferase. This is supported by the fact that when spinach chloroplasts were incubated with low concentrations (0.6 μM) of SAM, the methyl groups were preferentially used for RbcS methylation (Black et al., 1987).
However, because SAM is crucial and plastid prenyllipid synthesis was not totally inhibited in samt1, one may ask whether SAMT1 constitutes the sole plastid SAMT or whether alternative pathways exist for SAM import. For a given substrate, one or multiple transport systems might be involved. For example, plastids seemingly possess two independent and structurally unrelated transport systems for folates, one belonging to the MCF (Bedhomme et al., 2005) and the other being related to that of trypanosomes and cyanobacteria (Klaus et al., 2005). In addition, compensatory mechanisms have been observed for plastid transporters (reviewed in Weber et al., 2004, 2005). Under these conditions, it is frequently difficult to determine the contributions of individual transporters to the overall flux of metabolites. From a metabolic perspective, it is interesting that earlier compartmentation studies have shown that several enzymes of the plastid prenyllipid pathway are partially located in the plastid envelope (for review, see Schultz et al., 1985). It has also been shown that the methylation of MgProto IX takes place in the plastid envelope as well as in the thylakoids (Block et al., 2002). The metabolic contribution of the plastid envelope is supported by the fact that in totally bleached leaves, MgProto IX is still produced and used in the retrograde control of nuclear genes (Strand et al., 2003). Finally, it has been shown that the methylation step of demethylplastoquinol to plastoquinone-9 is catalyzed by the genuine 37-kD inner envelope membrane protein (Motohashi et al., 2003). Thus, it could be that the deficiency of plastid SAMT1 is partly compensated by cytosolic SAM that passively diffuses through the plastid outer envelope membrane and that reacts with prenyllipid biosynthetic enzymes located in the inner envelope membrane. The resulting products could be further integrated into the thylakoid membranes through the plastid vesicle-inducing protein (Kroll et al., 2001). The validation of this hypothesis requires further analysis of the topology of plastid prenyllipid methyltransferases. The finding that plastid transporters could influence plant secondary metabolism is not unprecedented. Analysis of the Arabidopsis chlorophyll a/b binding protein underexpression mutant, cue1, offers additional evidence for this idea. CUE1 encodes a plastidic phosphoenolpyruvate/phosphate translocator (At PPT1) (Streatfield et al., 1999). Downregulation of CUE1 resulted in reduced metabolic flux through the shikimate pathway and in a decrease in the level of available plastoquinone (Streatfield et al., 1999).
It remains undetermined whether the downregulation of SAMT1 has an effect on the activity of plastid Thr synthase. In vitro studies revealed that SAM is a potent allosteric regulator of Thr synthase. Inactivation of Thr synthase leads to an imbalance in the regulation of the synthesis of aspartate-derived amino acids (Galili, 1995). Interestingly, even when the total concentration of SAM was increased or decreased in plant cells, plastid Thr synthase was not affected (Shen et al., 2002).
Finally, hierarchical clustering (Eisen et al., 1998) of the Arabidopsis AtGenExpress developmental series gene expression data (Schmid et al., 2005) showed that SAMT1 belongs to a highly correlated cluster consisting of 68 genes that are expressed predominantly in photosynthetic tissues but are strongly repressed in roots and during the later stages of seed development. Of these 68 genes, 48 (71%) are predicted to be targeted to chloroplasts. Because <14% of the proteins encoded by the Arabidopsis genome are predicted to be targeted to chloroplasts, plastid-targeted genes are clearly overrepresented in the SAMT1 cluster, which further emphasizes the role of SAMT1 in plastid function.
METHODS
Materials
Chemicals were purchased from Sigma-Aldrich, and radiochemical [14C]SAM was purchased from GE Healthcare and Amersham Biosciences. Reagents and enzymes for recombinant DNA techniques were obtained from Invitrogen, New England Biolabs, and Promega.
Plant Material
Unless stated otherwise, Arabidopsis thaliana (Col-0), Nicotiana benthamiana, and Capsicum annum were grown on damp compost in a greenhouse under a light regime of 8 h of darkness and 16 h of light. Alternatively, Arabidopsis seeds were grown on 0.8% (w/v) agar-solidified Murashige and Skoog medium supplemented with 0.5% sucrose and kanamycin (50 μg/mL). 5-Aminolevulinate (5 mM) was fed to detached leaves (3-week-old rosette leaves) in the dark for 12 h at room temperature before extraction of porphyrins. For the isolation of Arabidopsis mitochondria, a cell suspension culture was used (May and Leaver, 1993).
Isolation of SAMT1 cDNAs
SAMT1, Ca SAMT1, and Nb SAMT1 cDNAs were cloned from Arabidopsis, C. annuum, and N. benthamiana, respectively, by RT-PCR (Bouvier et al., 1998). The primers used for RT-PCR were derived from Arabidopsis genomic sequences and homologous ESTs from Solanum tuberosum (accession numbers BI178733 and BI179073), Nicotiana tabacum (accession number BP133973), C. annuum (accession number CA518170), and Solanum lycopersicum (accession number BG125859). The cDNAs were sequenced using an automatic DNA sequencer (ABI 3100; Applied Biosystems). Sequence comparisons were performed through the National Center for Biotechnology Information using BLAST programs (Altschul et al., 1997).
Cluster Analysis of Arabidopsis Microarray Gene Expression Data
Microarray gene expression data of the AtGenExpress Developmental Series were downloaded from the AtGenExpress website (http://www.weigelworld.org/resources/microarray/AtGenExpress/), and arithmetic means were calculated for each of the triplicate values of the 79 tested conditions provided by AtGenExpress. The data were loaded into the program Cluster (Eisen et al., 1998) and adjusted by median-centering rows and columns (in this order) for five consecutive rounds each, as described previously (Eisen et al., 1998). After data adjustment, genes and arrays were hierarchically clustered using the Spearman rank correlation coefficient as the metric of similarity and the average linkage-clustering method. The resulting hierarchically clustered gene expression data were visualized using the program TreeView (Eisen et al., 1998), and the cluster containing At4g39460 was identified.
Functional Expression of SAMT1 in Yeast Cells
The DNA encoding the mature part of SAMT1 (amino acid residues 40 to 325 of the precursor protein) was amplified by PCR using a proofreading polymerase (Platinum Pfx polymerase; Invitrogen). As template, the cDNA encoding the precursor of SAMT1 (cloned into pBAD/Thio-TOPO; see below) was used. The forward primer was designed with a KpnI restriction site (5′-CAAAGGCTTTTTGGTACCTGTTA-3′), and the reverse primer contained the XbaI restriction site behind the stop codon (5′-CCTTCTAGATCGCCCTATTCTTC-3′). The PCR product was subcloned into pGEM-T Easy (Promega), and its sequence was verified (Research Technology Support Facility, Michigan State University).
The DNA fragment was cloned in-frame with an N-terminal His tag into pYES-NT, a yeast expression vector (Invitrogen) under the control of the galactose-inducible GAL4 promoter. The resulting construct, pMSU133, was transformed into the Saccharomyces cerevisiae strain INVSc1 (Invitrogen). Transformation of yeast was performed by the lithium chloride method according to the instructions provided by the manufacturer. The transformants were selected on Synthetic Complete minimal agar plates lacking uracil (SC-U).
For heterologous expression of SAMT1, 50 mL of SC-U induction medium containing 2% galactose was inoculated to an OD600 of 0.4 with yeast cells harboring pMSU133 grown overnight in 5 mL of SC-U containing 2% raffinose. The yeast cells were grown for 6 h aerobically at 30°C. Control cultures with an empty vector were processed in parallel. Cells from the induction culture were harvested by centrifugation (3000g, 10 min), resuspended in 500 μL of ice-cold breaking buffer (50 mM sodium phosphate, pH 7.4, 1 mM EDTA, 5% glycerol, 1 mM phenylmethylsulfonyl fluoride, and 0.1 mM 4-[2-aminoethyl]-benzenesulfonylfluoride), and disrupted by vigorous agitation for 2 min with acid-washed glass beads (0.4 to 0.6 mm; Sigma-Aldrich) in a tissue homogenizer (Vibration Mill type MM301; Retsch). Glass beads and cell debris were removed by centrifugation (600g, 1 min, 4°C), and the total membrane fraction was collected from the supernatant by ultracentrifugation (100,000g, 45 min, 4°C). The membrane pellet was resuspended in 50 μL of ice-cold 10 mM HEPES-KOH, pH 7.5, and 0.8 mM MgCl2.
An aliquot of the membrane fraction was analyzed by SDS-PAGE, and the expression of the recombinant protein was detected by SDS-gel blot analysis using a monoclonal anti-penta-His antibody (Qiagen).
Reconstitution of Transport Activities into Liposomes
The 100,000g membrane fraction obtained from 50 mL of galactose-induced yeast cells expressing SAMT1 was reconstituted into liposomes, and the transport activity of SAM was tested in the presence or absence of internal counter-exchange substrates. Liposomes were prepared by sonicating acetone-washed soybean phospholipids (8%, w/v; Sigma-Aldrich) in 100 mM Tricine-KOH, pH 7.8, 30 mM potassium gluconate, and 20 mM internal substrate for 10 min at 4°C (unless stated otherwise). Yeast membranes were incorporated into the liposomes without solubilization by a freeze-thaw step as described previously (Kasahara and Hinkle, 1977; Flügge and Weber, 1994). After thawing at room temperature, the proteoliposomes were sonicated again to form unilamellar vesicles. Unincorporated substrates were removed by passing the liposomes over PD10 gel filtration columns (GE Healthcare) that were equilibrated with 10 mM Tricine-KOH, pH 7.6, 100 mM sodium gluconate, and 50 mM potassium gluconate. The eluted proteoliposomes were used for the uptake experiments within the next 30 min.
The time-dependent uptake of SAM was started by adding [14C]SAM (53 mCi/mmol) to the proteoliposomes at room temperature. The uptake reactions were terminated by passing aliquots over Dowex AG-50W-X8 columns (100 to 200 mesh resin, hydrogen form; Bio-Rad), which had been preequilibrated with 150 mM lithium acetate, pH 7.5 (acetic acid). Radiolabeled substrate that was not incorporated into the liposomes during the uptake assay was removed by binding to the cation-exchange column; radiolabeled substrate contained in the liposomes eluting from the columns was quantified by scintillation counting (Tri-Carb 2100TR; Packard).
Transient Expression of GFP Fusions
The nucleotide fragment encoding the N-terminal 80 amino acids (Met-1 to Thr-80) of SAMT1 was fused in-frame with the codon-optimized GFP S65C mutant gene (Heim et al., 1995), with expression driven by the cauliflower mosaic virus 35S promoter using the pCATsGFP plasmid. Forward 5′-CATGCCATGGGAATGGCTCCTCTTACTCTCTCCGTTGAGTGAAGAGC-3′ and reverse 5′-CATGCCATGGGTAGTCTTAATAGTATCAATTGGGTATAAAGCTGTTTC-3′ primers possessing NcoI restriction sites were used for PCR amplification and insertion into the NcoI site of pCATsGFP. The resulting plasmid, designated 35S-SAMT1(Met1-Thr80)-GFP, and the 35S-GFP used as a control were verified by DNA sequence analysis and introduced into N. benthamiana protoplasts prepared from leaf cuttings. The purified plasmids (20 μg per 2 × 106 protoplasts) were introduced into the protoplasts using polyethylene glycol according to a described procedure (Goodall et al., 1990). Expression of the fusion construct was monitored after 20 h of incubation at 25°C in the dark. The samples were examined using a Zeiss LSM510 confocal laser scanning microscope equipped with an Axiovert 100 inverted microscope. The filter sets used were BP505-545 (excitation, 488 nm; emission, 505 to 545 nm) and LP585 (excitation, 488 nm; emission, 585 nm) to detect GFP and the chlorophyll autofluorescence. The images were processed using ImageJ version 1.33 (National Institutes of Health; http://rsb.info.nih.gov/ij).
Carotenoid, Prenyllipid, and Porphyrin Analysis
Leaf samples were extracted as described previously (Bouvier et al., 2003) except that dichloromethane:methanol (2:1, v/v) was used. After concentration under a nitrogen stream, the lipid extract was analyzed by HPLC using the SpectraSYSTEM (ThermoFinnigan) comprising an SCM1000 solvent degasser, a P1-1000XR gradient pump, an AS3000 autosampler, and a UV6000LP diode array detector. Data acquisition and processing were performed using ChromQuest version 3 software (ThermoFinnigan) (Bouvier et al., 2003). For carotenoids and chlorophylls, a linear gradient of acetonitrile:water (90:10, v/v) to 100% ethyl acetate was used. For plastoquinone and phylloquinone, an isocratic mobile phase composed of methanol:hexane (80:20, v/v) was used. The absorbance of the eluate was monitored at 263 nm. Peak assignments were ascertained based on the spectral changes after reduction of oxidized forms using NaBH4 (Barr and Crane, 1971). Phylloquinone and plastoquinone isolated from tobacco leaves were used as standards for the quantitative determination of the amount of quinones. Tocopherols were separated using an isocratic mobile phase containing acetonitrile:methanol:dichloromethane (60:35:5, v/v), and the eluates were monitored at an excitation wavelength of 290 nm and emission of 330 nm using a scanning fluorescence detector (SpectraSYSTEM FL3000; ThermoFinnigan). For porphyrin analysis, leaf samples were frozen in liquid nitrogen and extracted with acetone:water:NH4OH (9:1:0.1, v/v). HPLC analysis was performed on a C18 reverse phase column as described above. The column was eluted using a linear gradient of acetone:water (60:40, v/v) to acetone for 40 min (La Rocca et al., 2001). Eluates containing MgProto IX and MgProto IX Me were monitored at 415 nm for excitation and 595 for emission. Peak assignments were based on a comparison with authentic MgProto IX (Porphyrin Products).
Generation of SAMT1 Antibodies, and Immunoblot Analysis
Truncated SAMT1 cDNA was amplified by PCR using the forward primer 5′-GTAATGCAGAGTTCACAGCTC-3′ and the reverse primer 5′-TTCTTCTTTGGTTTCTTTAACCG-3′ and cloned into the pBAD/TOPO ThioFusion vector as described previously (Bouvier et al., 2003). Transformed Escherichia coli (TOP10) was grown to an OD600 of 0.5 at 19°C in Luria-Bertani medium containing ampicillin (100 μg/mL) before being induced with 0.02% arabinose and overnight culture. Bacterial cells were pelleted by centrifugation before lysis by sonication in 50 mM Tris-HCl buffer, pH 7.6, containing 1 mM DTT and 2% Triton X-100 (Bouvier et al., 1998). SAMT1 was purified using affinity chromatography at 4°C with ProBond resin (Invitrogen) as described by the manufacturer's protocol, except that 2% Triton X-100 was included in the buffers. The purified protein was used for rabbit immunization.
Isolation and Complementation of the T-DNA Insertion Mutants
A T-DNA insertion line for SAMT1 (At4g39460), seed stock identifier SALK_008248, was identified as samt1 from the Sequence-Indexed Library of Insertion Mutations using the Arabidopsis Gene Mapping Tool (http://signal.salk.edu/cgi-bin/tdnaexpress), and the seed stocks were obtained from the ABRC. The mutant was characterized according to described procedures (Alonso et al., 2003). The genotype of the knockout mutant line was analyzed by PCR and RT-PCR using primers specific for the SAMT1 open reading frame (forward: D1, 5′-ATGGCTCCTCTTACTCTCTCC-3′; D2, 5′-CGAAGAGGGTAATGCAGAGTTC-3′; reverse: R1, 5′-CCTCTGTAGGCACTCGAATC-3′; R2, 5′-GCAATCATTCGAACAGCACTG-3′) and the primer specific for the T-DNA insertion (LBa1, 5′-TGGTTCACGTAGTGGGCCATCG-3′). As a positive control, RT-PCR amplification of α-tubulin mRNA was performed using the forward (5′-GAGATTGTTGATCTGTGCTTG-3′) and reverse (5′-CTCTGCGGAGATGACTGGGGC-3′) primers. The T-DNA insertion was confirmed by DNA gel blot analysis. Five micrograms of genomic DNA was isolated from wild-type and heterozygous and homozygous samt1 mutant plants using the NucleoSpin Plant L kit (Macherey-Nagel), digested with SacI, separated by agarose gel electrophoresis before denaturation, and blotted onto nitrocellulose BA-85 membranes (Schleicher and Schuell). The blots were hybridized with the following 32P-labeled probes: probe I (PCR product [380 bp] amplified using forward 5′-CGAAGAGGGTAATGCAGAGTTC-3′ and reverse 5′-CTGAAGTCTAGTCTTAATAGTATC-3′ primers); probe II (KpnI-ScaI fragment [651 bp] of the T-DNA); and probe III (PCR product [440 bp] amplified using forward 5′-CTTAGTGATCCTGAGAACGCTC-3′ and reverse 5′-GTCAACGATTCCTTGATATTG-3′ primers). Hybridizations were performed at 65°C, and the filters were washed according to standard procedures (Sambrook et al., 1989).
The complementation of the T-DNA mutant was performed using Arabidopsis mutant lines heterozygous for samt1, because of the growth defect of the homozygous line. For complementation of the samt1 mutant, a genomic fragment (2.5 kb) corresponding to the coding region of SAMT1 was amplified by PCR using forward 5′-GCTCTAGAATGGCTCCTCTTACTCTCTCCGTTGATG-3′ and reverse 5′-GCTCTAGATTATTCTTCTTTGGTTTCTTTAACCG-3′ primers containing XbaI sites. The resulting 2.5 kb fragment was sequenced and cloned into the XbaI site of the binary pKYLX71-35S2 vector (Maiti et al., 1993) and introduced into Agrobacterium tumefaciens strain C58C1pMP90. Transformation of heterozygous samt1 was performed according to the floral dip method (Clough and Bent, 1998).
Because the T-DNA mutant and the pKYLX71-35S2 vector used in the complementation experiments both confer resistance to kanamycin, all kanamycin-resistant plants except homozygous samt1 were removed from the agar plates and grown on damp compost at a density of five seedlings per pot. Rosette leaves from the resulting plants were used for genomic DNA extraction using the NucleoSpin Plant kit (Macherey-Nagel). Aliquots of purified DNA from plants growing in the same pot were pooled and used for a first variant of multiplex PCR. To this end, the forward (D1) and the two reverse (TRbcS, 5′-ACAAAATGTTTGCATATCTCTT-3′; and left border LBa1) primers were introduced in the same reaction mixture. The primer TRbcS was designed from the sequence of the pea (Pisum sativum) Rubisco small subunit terminator that is present in the pKYLX71-35S2 vector used to introduce the 35S-SAMT1 transgene. The TRbcs primer hybridizes only to the transgene. The forward (D1) and the reverse (left border LBa1) primers allowed the detection of native samt1. Pools displaying both 2.8- and 1-kb amplicons attributable to D1-TRbcS and D1-Lba1 primer combinations, respectively, were used for a second PCR to select from the pools individual samt1 lines harboring the 35S-SAMT1 transgene. During this second PCR step, purified DNA from each plant of the relevant pool was separately used as a template in combination with D1-TRbcS and D1-LBa1 primers. After this step, isolated samt1 lines harboring the 35S-SAMT1 transgene were used to analyze the absence of native SAMT1 in complemented lines (samt1/35S-SAMT1) by RT-PCR using the forward (D3) and reverse (3′ untranslated region of SAMT1 [3′utr SAMT1], 5′-CAAAGGGGTTACATTGTCACCTTAC-3′) primers. The 3′utr SAMT1 primer is not present in the 35S-SAMT1 transgene. Amplification of α-tubulin mRNA was used as a positive control.
Virus-Induced Gene Silencing
The cDNA fragments (0.5 kb) corresponding N. benthamiana (Nb SAMT1) and Pantoea agglomerans phytoene synthase (CrtB; P22872), used as a control, were amplified by PCR and cloned into the viral vector TTO (Kumagai et al., 1995). For TTO-Nb SAMT1, the forward primer 5′-GCCTCGAGTTGAAGATGCTTCCTGAAAATCTTAGTGC-3′ and the reverse primer 5-TCCCTAGGCCAAAGCACTCTTGGACCGATGCCCTT-3′ containing XhoI and AvrII restriction sites, respectively, were used. For TTO-CrtB, the forward 5′-GCCTCGAGATTACGCCCCGCATGGCGCTCGATCAC-3′ and reverse 5′-TCCCTAGGGCGGCGATCCCAGGCGCTGCCTCCCGC-3′ primers containing XhoI and AvrII restriction sites, respectively, were used. The PCR and N. benthamiana transfection were performed as described previously (Kumagai et al., 1995, 1998). In plants transfected with TTO-Nb SAMT1, the detection of endogenous Nb SAMT1 transcripts was performed by RT-PCR (Bouvier et al., 1998). Total RNA was extracted and used to transcribe a single-strand cDNA using random primers and SuperScript II reverse transcriptase (Invitrogen). Equal amounts of first-strand cDNAs were used as templates for PCR amplification using forward 5′-GGAAATCTTGCTGGAGTCCTACCG-3′ and reverse 5′-CTTTGAGGCTGCATTATCAGGAC-3′ primers annealing outside of the TTO-Nb SAMT1 construct. The N. benthamiana α-tubulin gene was used as an internal positive control for quantification of the relative cDNA amount. To this end, we used the forward 5′-ATGCTTTCATCATATGCCCCTGTG-3′ and reverse 5′-CAGCACCAACTTCCTCGTAATC-3′ primers for amplification. Upper uninoculated leaves from TTO-Nb SAMT1, TTO-CrtB, and wild-type plants were used for analysis.
Plastid and Mitochondria Isolation
Chloroplasts were isolated from Arabidopsis leaf protoplasts (Fitzpatrick and Keegstra, 2001), and mitochondria were isolated from dark-grown Arabidopsis cell suspension cultures (Millar et al., 2001). Chloroplast subfractions (stroma, thylakoid membranes, and envelope membranes) were isolated as described previously (Werner-Washburne et al., 1983) using 5-week-old Arabidopsis (ecotype Col-0) grown with a daylength of 12 h. To investigate whether SAMT1 is integrated into the chloroplast envelope membranes, purified chloroplast envelope membranes equivalent to 500 μg of proteins were extracted with 0.5 M Na2CO3, pH 11.5, or 1 M NaCl for 30 min at 0°C. After centrifugation at 100,000g for 1 h, the pellet containing the membranes and the supernatant were separated. The pellet (membranes) was dissolved in SDS sample buffer (50 mM Tris-HCl, pH 8, 5% SDS, 0.5 M DTT, 30% glycerol, and 0.2% bromophenol blue), whereas the supernatant proteins were precipitated using 20% trichloroacetic acid before solubilization using the SDS sample buffer. Organelles and suborganellar fractions were prepared for electron microscopy as described previously (Bouvier et al., 1998).
Protein Separation and Immunoblot Analysis
The protein content of the extract was measured as described (Smith et al., 1985). Proteins were resolved by SDS-PAGE followed by electroblotting (Bouvier et al., 1998). The polyclonal antibodies to LHCIIb were raised against recombinant pea LHCIIb using a plasmid vector generously provided by H. Paulsen (Paulsen et al., 1990). Antibodies to NAD9 for the respiratory chain NADH dehydrogenase (complex I) of mitochondria were a generous gift of G. Bonnard (Institut de Biologie Moléculaire des Plantes, Centre National de la Recherche Scientifique). Antibodies to the plastid triose phosphate translocator were kindly provided by I. Flügge (Flügge et al., 1989). The antibodies raised against 1-deoxy-d-xylulose 5-phosphate synthase have been described previously (Bouvier et al., 1998). Protein gel blots were visualized using the enhanced chemiluminescence system (Amersham Biosciences).
Uptake of SAM into Plastids
The uptake of SAM into plastids was estimated by silicone layer-filtering centrifugation (Heldt, 1980) using plastids isolated from leaf protoplasts (Fitzpatrick and Keegstra, 2001). Chloroplast suspensions equivalent to 50 μg of chlorophyll were isolated from Col-0 and mutant lines and incubated at 25°C for 1 min in the presence of 50 mM Tris-HCl, pH 7.6, containing 0.33 M sorbitol and 200 μM [14C]SAM before centrifugation and determination of the radioactivity by liquid scintillation counting (Beckman LS-6500). Chlorophyll content was determined as described by Moran and Porath (1980).
Determination of Plastid SAM Concentration
Purified plastids were extracted with 20% trichloroacetic acid, and SAM recovered in the soluble fraction was converted to fluorescent isoindoles subsequently analyzed by HPLC (Capdevila and Wagner, 1998; Moffatt et al., 2002).
RNA Gel Blot Analysis
Total RNA was extracted from different Arabidopsis or N. benthamiana organs and tissues using the NucleoSpin RNA Plant L kit (Macherey-Nagel). RNA gel blot analysis was performed as described previously (Bouvier et al., 2003) using a 32P-labeled SAMT1 probe (PCR product [544 bp] amplified using forward D3 5′-GGAGGTGGAAAAATTGTTTTGAAG-3′ and reverse R3 5′-GTCAACGATTCCTTGATATTG-3′ primers) and a 32P-labeled Nb SAMT1 probe (PCR product [662 bp] amplified using forward 5′-GGAAATCTTGCTGGAGTCCCG-3′ and reverse 5′-CCTTGAGGCTGCATTATCAGGAC-3′ primers). 18S rRNA 32P-labeled probes from Arabidopsis (PCR product [400 bp] amplified using forward 5′-GACGCGGGCTCTGGCTTGCTCTG-3′ and reverse 5′-CTGCAACAACTTAAATATACGC-3′ primers) and N. benthamiana (PCR product [400 bp] amplified using forward 5′-CGATGGTAGGATAGTGGCCTAC-3′ and reverse 5′-CAATTAAGGCCAGGAGCGCATC-3′ primers) were used to verify equivalent sample loading. The prehybridization, hybridization, and washing conditions were as described previously (Sambrook et al., 1989).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AJ627908 (SAMT1), AJ627178 (Ca SAMT1), and AJ619956 (Nb SAMT1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Similarity between Plant, Yeast, and Human Organelle SAMTs.
Supplemental Figure 2. RNA Gel Blot Analysis of SAMT1 in Various Tissues.
Supplemental Figure 3. Expression of Recombinant SAMT1 and Specificity of Anti-SAMT1.
Supplemental Figure 4. Functional Complementation of samt1 with SAMT1.
Supplemental Table 1. Expression Data Showing Genes Clustering with SAMT1.
Supplementary Material
Acknowledgments
We thank Monto Kumagai and the Large Scale Biology Corporation for the TTO vector and Philippe Hugueney (Ecole Normale Supérieure, Lyon, France) and Guido Jach (Max-Planck-Institut für Züchtungsforschung, Cologne, Germany) for the pCATS-GFP vector. We thank Philippe Hammann and Malek Alioua (Institut de Biologie Moléculaire des Plantes [IBMP]) for DNA sequencing and Angèle Geldreich (IBMP) for help with protoplast preparation. We thank Géraldine Bonnard (IBMP) for anti-NAD9 antibody, Ingo Flügge (University of Köln) for anti-TPT antibody, Harald Paulsen (University of Mainz) for pea LHCII cDNA, and Annie Marion-Poll (Institut National de la Recherche Agronomique, Versailles, France) for pKYLX71-35S2 vector. We thank the Salk Institute Genomic Analysis Laboratory for providing the T-DNA insertion mutants. The Inter-Institute Confocal Microscopy Plate-Form was cofinanced by the Centre National de la Recherche Scientifique, the Université Louis Pasteur, the Région Alsace, and the Association pour la Recherche sur le Cancer. This work was supported by European Community Grant QLK3-CT-2000-00809, National Science Foundation Grants MCB-0348074 and MCB-0519740, Department of Energy Grant DE-FG02-04ER15562, and an Alexander-von-Humboldt Foundation Feodor Lynen postdoctoral fellowship to N.L.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Bilal Camara (bilal.camara@ibmp-ulp.u-strasbg.fr).
Online version contains Web-only data.
References
- Aarnes, H. (1978). Regulation of threonine biosynthesis in barley seedlings (Hordeum vulgare L.). Planta 140 185–192. [DOI] [PubMed] [Google Scholar]
- Agrimi, G., Di Noia, M.A., Marobbio, C.M., Fiermonte, G., Lasorsa, F.M., and Palmieri, F. (2004). Identification of the human mitochondrial S-adenosylmethionine transporter: Bacterial expression, reconstitution, functional characterization and tissue distribution. Biochem. J. 379 183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alawady, A.E., and Grimm, B. (2005). Tobacco Mg protoporphyrin IX methyltransferase is involved in inverse activation of Mg porphyrin and protoheme synthesis. Plant J. 41 282–290. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, X., Katz, S., Pollard, M., and Ohlrogge, J. (2002). Carbocyclic fatty acids in plants: Biochemical and molecular genetic characterization of cyclopropane fatty acid synthesis of Sterculia foetida. Proc. Natl. Acad. Sci. USA 99 7172–7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, X., Thelen, J.J., Bonaventure, G., and Ohlrogge, J.B. (2003). Characterization of cyclopropane fatty-acid synthase from Sterculia foetida. J. Biol. Chem. 278 12846–12853. [DOI] [PubMed] [Google Scholar]
- Barr, R., and Crane, F.L. (1971). Quinones in algae and higher plants. Methods Enzymol. 23 372–408. [Google Scholar]
- Bedhomme, M., Hoffmann, M., McCarthy, E.A., Gambonnet, B., Moran, R.G., Rebeille, F., and Ravanel, S. (2005). Folate metabolism in plants: An Arabidopsis homolog of the mammalian mitochondrial folate transporter mediates folate import into chloroplasts. J. Biol. Chem. 280 34823–34831. [DOI] [PubMed] [Google Scholar]
- Black, M.T., Meyer, D., Widger, W.R., and Cramer, W.A. (1987). Light-regulated methylation of chloroplast proteins. J. Biol. Chem. 262 9803–9807. [PubMed] [Google Scholar]
- Block, M.A., Tewari, A.K., Albrieux, C., Marechal, E., and Joyard, J. (2002). The plant S-adenosyl-L-methionine:Mg-protoporphyrin IX methyltransferase is located in both envelope and thylakoid chloroplast membranes. Eur. J. Biochem. 269 240–248. [DOI] [PubMed] [Google Scholar]
- Boerjan, W., Bauw, G., Van Montagu, M., and Inze, D. (1994). Distinct phenotypes generated by overexpression and suppression of S-adenosyl-L-methionine synthetase reveal developmental patterns of gene silencing in tobacco. Plant Cell 6 1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier, F., d'Harlingue, A., Suire, C., Backhaus, R.A., and Camara, B. (1998). Dedicated roles of plastid transketolases during the early onset of isoprenoid biogenesis in pepper fruits. Plant Physiol. 117 1423–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier, F., Rahier, A., and Camara, B. (2005). Biogenesis, molecular regulation and function of plant isoprenoids. Prog. Lipid Res. 44 357–429. [DOI] [PubMed] [Google Scholar]
- Bouvier, F., Suire, C., Mutterer, J., and Camara, B. (2003). Oxidative remodeling of chromoplast carotenoids: Identification of the carotenoid dioxygenase CsCCD and CsZCD genes involved in Crocus secondary metabolite biogenesis. Plant Cell 15 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila, A., and Wagner, C. (1998). Measurement of plasma S-adenosylmethionine and S-adenosylhomocysteine as their fluorescent isoindoles. Anal. Biochem. 264 180–184. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Curien, G., Job, D., Douce, R., and Dumas, R. (1998). Allosteric activation of Arabidopsis threonine synthase by S-adenosylmethionine. Biochemistry 37 13212–13221. [DOI] [PubMed] [Google Scholar]
- DellaPenna, D. (2005). A decade of progress in understanding vitamin E synthesis in plants. J. Plant Physiol. 162 729–737. [DOI] [PubMed] [Google Scholar]
- d'Harlingue, A., and Camara, B. (1985). Plastid enzymes of terpenoid biosynthesis. Purification and characterization of γ-tocopherol methyltransferase from Capsicum chromoplasts. J. Biol. Chem. 260 15200–15203. [PubMed] [Google Scholar]
- Dogbo, O., and Camara, B. (1986). La méthionine adénosyltransférase des végétaux: Mise en évidence de 3 formes. C. R. Acad. Sci. Paris III 303 93–96. [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300 1005–1016. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., and von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk, J., Andersen, G., Kernebeck, B., and Krupinska, K. (2003). Constitutive overexpression of barley 4-hydroxyphenylpyruvate dioxygenase in tobacco results in elevation of the vitamin E content in seeds but not in leaves. FEBS Lett. 540 35–40. [DOI] [PubMed] [Google Scholar]
- Ferro, M., Salvi, D., Brugiere, S., Miras, S., Kowalski, S., Louwagie, M., Garin, J., Joyard, J., and Rolland, N. (2003). Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol. Cell. Proteomics 2 325–345. [DOI] [PubMed] [Google Scholar]
- Ferro, M., Salvi, D., Riviere-Rolland, H., Vermat, T., Seigneurin-Berny, D., Grunwald, D., Garin, J., Joyard, J., and Rolland, N. (2002). Integral membrane proteins of the chloroplast envelope: Identification and subcellular localization of new transporters. Proc. Natl. Acad. Sci. USA 99 11487–11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, K., Kammerer, B., Gutensohn, M., Arbinger, B., Weber, A., Hausler, R.E., and Flugge, U.I. (1997). A new class of plastidic phosphate translocators: A putative link between primary and secondary metabolism by the phosphoenolpyruvate/phosphate antiporter. Plant Cell 9 453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick, L.M., and Keegstra, K. (2001). A method for isolating a high yield of Arabidopsis chloroplasts capable of efficient import of precursor proteins. Plant J. 27 59–65. [DOI] [PubMed] [Google Scholar]
- Flügge, U.I., Fischer, K., Gross, A., Sebald, W., Lottspeich, F., and Eckerskorn, C. (1989). The triose phosphate-3-phosphoglycerate-phosphate translocator from spinach chloroplasts: Nucleotide sequence of a full-length cDNA clone and import of the in vitro synthesized precursor protein into chloroplasts. EMBO J. 8 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flügge, U.I., and Weber, A. (1994). A rapid method for measuring organelle-specific substrate transport in homogenates from plant tissues. Planta 194 181–185. [PubMed] [Google Scholar]
- Fontecave, M., Atta, M., and Mulliez, E. (2004). S-Adenosylmethionine: Nothing goes to waste. Trends Biochem. Sci. 29 243–249. [DOI] [PubMed] [Google Scholar]
- Fujiki, Y., Hubbard, A.L., Fowler, S., and Lazarow, P.B. (1982). Isolation of intracellular membranes by means of sodium carbonate treatment: Application to endoplasmic reticulum. J. Cell Biol. 93 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili, G. (1995). Regulation of lysine and threonine synthesis. Plant Cell 7 899–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili, G., and Hofgen, R. (2002). Metabolic engineering of amino acids and storage proteins in plants. Metab. Eng. 4 3–11. [DOI] [PubMed] [Google Scholar]
- Giovanelli, J., Mudd, S.H., and Dakto, A.H. (1989). Regulatory structure of the biosynthetic pathway for the aspartate family of amino acids in Lemna paucicostata Hegelm. 6746, with special reference to the role of aspartokinase. Plant Physiol. 90 1584–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovanelli, J., Veluthambl, K., Thompsosn, G.A., Mudd, S.H., and Dakto, A.H. (1984). Threonine synthase of Lemna paucicostata H. Plant Physiol. 76 285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall, G.J., Wiebauer, K., and Filipowicz, W. (1990). Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 181 148–161. [DOI] [PubMed] [Google Scholar]
- Grimm, R., Grimm, M., Eckerskorn, C., Pohlmeyer, K., Rohl, T., and Soll, J. (1997). Postimport methylation of the small subunit of ribulose-1,5-bisphosphate carboxylase in chloroplasts. FEBS Lett. 408 350–354. [DOI] [PubMed] [Google Scholar]
- Gross, A., Brückner, G., Heldt, H.W., and Flügge, U.I. (1990). Comparison of the kinetic properties, inhibition and labelling of the phosphate translocators from maize and spinach mesophyll chloroplasts. Planta 180 262–271. [DOI] [PubMed] [Google Scholar]
- Hanson, A.D., and Roje, S. (2001). One-carbon metabolism in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 119–137. [DOI] [PubMed] [Google Scholar]
- Heazlewood, J.L., Tonti-Filippini, J., Verboom, R.E., and Millar, A.H. (2005). Combining experimental and predicted datasets for determination of the subcellular location of proteins in Arabidopsis. Plant Physiol. 139 598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim, R., Cubitt, A.B., and Tsien, R.Y. (1995). Improved green fluorescence. Nature 373 663–664. [DOI] [PubMed] [Google Scholar]
- Heldt, H.W. (1980). Measurement of metabolite movement across the envelope and of the pH in the stroma and the thylakoid space in intact chloroplasts. Methods Enzymol. 69 604–613. [Google Scholar]
- Holding, D.R., Springer, P.S., and Coomber, S.A. (2000). The chloroplast and leaf developmental mutant, pale cress, exhibits light-conditional severity and symptoms characteristic of its ABA deficiency. Ann. Bot. (Lond.) 86 953–962. [Google Scholar]
- Horne, D.W., Holloway, R.S., and Wagner, C. (1997). Transport of S-adenosylmethionine in isolated rat liver mitochondria. Arch. Biochem. Biophys. 343 201–206. [DOI] [PubMed] [Google Scholar]
- Kasahara, M., and Hinkle, P.C. (1977). Reconstitution and purification of the D-glucose transporter from human erythrocytes. J. Biol. Chem. 252 7384–7390. [PubMed] [Google Scholar]
- Kevin, L.I., Wang, C., Li, H., and Ecker, J.R. (2002). Ethylene biosynthesis and signaling networks. Plant Cell 14 (suppl.), S131–S151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus, S.M., Kunji, E.R., Bozzo, G.G., Noiriel, A., de la Garza, R.D., Basset, G.J., Ravanel, S., Rebeille, F., Gregory, J.F., III, and Hanson, A.D. (2005). Higher plant plastids and cyanobacteria have folate carriers related to those of trypanosomatids. J. Biol. Chem. 280 38457–38463. [DOI] [PubMed] [Google Scholar]
- Koch, M., Lemke, R., Heise, K.P., and Mock, H.P. (2003). Characterization of gamma-tocopherol methyltransferases from Capsicum annuum L and Arabidopsis thaliana. Eur. J. Biochem. 270 84–92. [DOI] [PubMed] [Google Scholar]
- Koo, A.J., and Ohlrogge, J.B. (2002). The predicted candidates of Arabidopsis plastid inner envelope membrane proteins and their expression profiles. Plant Physiol. 130 823–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll, D., Meierhoff, K., Bechtold, N., Kinoshita, M., Westphal, S., Vothknecht, U.C., Soll, J., and Westhoff, P. (2001). VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc. Natl. Acad. Sci. USA 98 4238–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai, M.H., Donson, J., Della-Cioppa, G., Harvey, D., Hanley, K., and Grill, L.K. (1995). Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc. Natl. Acad. Sci. USA 92 1679–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai, M.H., Keller, Y., Bouvier, F., Clary, D., and Camara, B. (1998). Functional integration of non-native carotenoids into chloroplasts by viral-derived expression of capsanthin-capsorubin synthase in Nicotiana benthamiana. Plant J. 14 305–315. [DOI] [PubMed] [Google Scholar]
- La Rocca, N., Rascio, N., Oster, U., and Rudiger, W. (2001). Amitrole treatment of etiolated barley seedlings leads to deregulation of tetrapyrrole synthesis and to reduced expression of Lhc and RbcS genes. Planta 213 101–108. [DOI] [PubMed] [Google Scholar]
- Leroch, M., Kirchberger, S., Haferkamp, I., Wahl, M., Neuhaus, H.E., and Tjaden, J. (2005). Identification and characterization of a novel plastidic adenine nucleotide uniporter from Solanum tuberosum. J. Biol. Chem. 280 17992–18000. [DOI] [PubMed] [Google Scholar]
- Loddenkotter, B., Kammerer, B., Fischer, K., and Flügge, U.I. (1993). Expression of the functional mature chloroplast triose phosphate translocator in yeast internal membranes and purification of the histidine-tagged protein by a single metal-affinity chromatography step. Proc. Natl. Acad. Sci. USA 90 2155–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison, J.T., and Thompson, J.F. (1976). Threonine synthetase from higher plants: Stimulation by S-adenosylmethionine and inhibition by cysteine. Biochem. Biophys. Res. Commun. 71 684–691. [DOI] [PubMed] [Google Scholar]
- Maiti, I.B., Murphy, J.F., Shaw, J.G., and Hunt, A.G. (1993). Plants that express a potyvirus proteinase gene are resistant to virus infection. Proc. Natl. Acad. Sci. USA 90 6110–6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marobbio, C.M., Agrimi, G., Lasorsa, F.M., and Palmieri, F. (2003). Identification and functional reconstitution of yeast mitochondrial carrier for S-adenosylmethionine. EMBO J. 22 5975–5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, M.J., and Leaver, C.J. (1993). Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol. 103 621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.H., Sweetlove, L.J., Giege, P., and Leaver, C.J. (2001). Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol. 127 1711–1727. [PMC free article] [PubMed] [Google Scholar]
- Moffatt, B.A., Stevens, Y.Y., Allen, M.S., Snider, J.D., Pereira, L.A., Todorova, M.I., Summers, P.S., Weretilnyk, E.A., Martin-McCaffrey, L., and Wagner, C. (2002). Adenosine kinase deficiency is associated with developmental abnormalities and reduced transmethylation. Plant Physiol. 128 812–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt, B.A., and Weretilnyk, E.A. (2001). Sustaining S-adenosyl-l-methionine-dependent methyltransferase activity in plant cells. Physiol. Plant. 113 435–442. [Google Scholar]
- Moran, R., and Porath, D. (1980). Chlorophyll determination in intact tissues using N,N-dimethylformamide. Plant Physiol. 65 478–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi, R., Ito, T., Kobayashi, M., Taji, T., Nagata, N., Asami, T., Yoshida, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2003). Functional analysis of the 37 kDa inner envelope membrane polypeptide in chloroplast biogenesis using a Ds-tagged Arabidopsis pale-green mutant. Plant J. 34 719–731. [DOI] [PubMed] [Google Scholar]
- Munne-Bosch, S., and Alegre, L. (2001). Subcellular compartmentation of the diterpene carnosic acid and its derivatives in the leaves of rosemary. Plant Physiol. 125 1094–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama, R., Ito, M., Yamaguchi, Y., Koizumi, N., and Sano, H. (2002). A chloroplast-resident DNA methyltransferase is responsible for hypermethylation of chloroplast genes in Chlamydomonas maternal gametes. Proc. Natl. Acad. Sci. USA 99 5925–5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri, L., Rottensteiner, H., Girzalsky, W., Scarcia, P., Palmieri, F., and Erdmann, R. (2001). Identification and functional reconstitution of the yeast peroxisomal adenine nucleotide transporter. EMBO J. 20 5049–5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen, H., Rümler, W., and Rüdiger, W. (1990). Reconstitution of pigment-containing complexes from light-harvesting chlorophyll a/b-binding protein overexpressed in Escherichia coli. Planta 181 204–211. [DOI] [PubMed] [Google Scholar]
- Petrotta-Simpson, T.F., Tamaldge, J.E., and Spence, K.D. (1975). Specificity and genetics of S-adenosylmethionine transport in Saccharomyces cerevisiae. J. Bacteriol. 123 516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picault, N., Hodges, M., Palmieri, L., and Palmieri, F. (2004). The growing family of mitochondrial carriers in Arabidopsis. Trends Plant Sci. 9 138–146. [DOI] [PubMed] [Google Scholar]
- Poulton, J.E. (1981). Transmethylation and demethylation reactions in the metabolism of secondary plant products. In The Biochemistry of Plants, E.E. Conn, ed (New York: Academic Press), pp. 667–723.
- Ravanel, S., Block, M.A., Rippert, P., Jabrin, S., Curien, G., Rebeille, F., and Douce, R. (2004). Methionine metabolism in plants: Chloroplasts are autonomous for de novo methionine synthesis and can import S-adenosylmethionine from the cytosol. J. Biol. Chem. 279 22548–22557. [DOI] [PubMed] [Google Scholar]
- Renne, P., Dressen, U., Hebbeker, U., Hille, D., Flugge, U.I., Westhoff, P., and Weber, A.P. (2003). The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2. Plant J. 35 316–331. [DOI] [PubMed] [Google Scholar]
- Rouillon, A., Surdin-Kerjan, Y., and Thomas, D. (1999). Transport of sulfonium compounds. Characterization of the S-adenosylmethionine and S-methylmethionine permeases from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 274 28096–28105. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Schultz, G., Soll, J., Fiedler, E., and Schulze-Siebert, D. (1985). Synthesis of prenylquinones in chloroplasts. Physiol. Plant. 64 123–129. [Google Scholar]
- Shen, B., Li, C., and Tarczynski, M.C. (2002). High free-methionine and decreased lignin content result from a mutation in the Arabidopsis S-adenosyl-L-methionine synthetase 3 gene. Plant J. 29 371–380. [DOI] [PubMed] [Google Scholar]
- Shepherd, M., Reid, J.D., and Hunter, C.N. (2003). Purification and kinetic characterization of the magnesium protoporphyrin IX methyltransferase from Synechocystis PCC6803. Biochem. J. 371 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani, D., and DellaPenna, D. (1998). Elevating the vitamin E content of plants through metabolic engineering. Science 282 2098–2100. [DOI] [PubMed] [Google Scholar]
- Smith, P.K., Krohn, R.I., Hermanson, G.T., Mallia, A.K., Gartner, F.H., Provenzano, M.D., Fujimoto, E.K., Goeke, N.M., Olson, B.J., Olson, B.J., and Klenk, D.C. (1985). Measurement of protein using bicinchoninic acid. Anal. Biochem. 150 79–85. [DOI] [PubMed] [Google Scholar]
- Strand, A., Asami, T., Alonso, J., Ecker, J.R., and Chory, J. (2003). Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421 79–83. [DOI] [PubMed] [Google Scholar]
- Streatfield, S.J., Weber, A., Kinsman, E.A., Hausler, R.E., Li, J., Post-Beittenmiller, D., Kaiser, W.M., Pyke, K.A., Flugge, U.I., and Chory, J. (1999). The phosphoenolpyruvate/phosphate translocator is required for phenolic metabolism, palisade cell development, and plastid-dependent nuclear gene expression. Plant Cell 11 1609–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, T.D., and Kaneko, Y. (1995). The maize brittle1 gene encodes amyloplast membrane polypeptides. Planta 196 477–484. [DOI] [PubMed] [Google Scholar]
- Trievel, R.C., Flynn, E.M., Houtz, R.L., and Hurley, J.H. (2003). Mechanism of multiple lysine methylation by the SET domain enzyme Rubisco LSMT. Nat. Struct. Biol. 10 545–552. [DOI] [PubMed] [Google Scholar]
- Tucker, A.M., Winkler, H.H., Driskell, L.O., and Wood, D.O. (2003). S-Adenosylmethionine transport in Rickettsia prowazekii. J. Bacteriol. 185 3031–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallsgrove, R.M., Lea, P.J., and Miflin, B.J. (1983). Intracellular localization of aspartate kinase and the enzymes of threonine and methionine biosynthesis in green leaves. Plant Physiol. 71 780–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, A.P., Schneidereit, J., and Voll, L.M. (2004). Using mutants to probe the in vivo function of plastid envelope membrane metabolite transporters. J. Exp. Bot. 55 1231–1244. [DOI] [PubMed] [Google Scholar]
- Weber, A.P., Schwacke, R., and Flugge, U.I. (2005). Solute transporters of the plastid envelope membrane. Annu. Rev. Plant Biol. 56 133–164. [DOI] [PubMed] [Google Scholar]
- Weiss, H., Friedrich, T., Hofhaus, G., and Preis, D. (1991). The respiratory-chain NADH dehydrogenase (complex I) of mitochondria. Eur. J. Biochem. 197 563–576. [DOI] [PubMed] [Google Scholar]
- Werner-Washburne, M., Cline, K., and Keegstra, K. (1983). Analysis of pea chloroplast inner and outer envelope membrane proteins by two-dimensional gel electrophoresis and their comparison with stroma proteins. Plant Physiol. 73 569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter, H., Robinson, D.G., and Heldt, H.W. (1994). Subcellular volumes and metabolite concentrations in spinach leaves. Planta 193 530–535. [Google Scholar]
- Ying, Z., Mulligan, R.M., Janney, N., and Houtz, R.L. (1999). Rubisco small and large subunit N-methyltransferases. Bi- and mono-functional methyltransferases that methylate the small and large subunits of Rubisco. J. Biol. Chem. 274 36750–36756. [DOI] [PubMed] [Google Scholar]
- Zheng, Q., Simel, E.J., Klein, P.E., Royer, M.T., and Houtz, R.L. (1998). Expression, purification, and characterization of recombinant ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit nepsilon-methyltransferase. Protein Expr. Purif. 14 104–112. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., Hennig, L., and Gruissem, W. (2005). Gene-expression analysis and network discovery using Genevestigator. Trends Plant Sci. 10 407–409. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.