Abstract
Sulfur is an essential macronutrient required for plant growth. To identify key transcription factors regulating the sulfur assimilatory pathway, we screened Arabidopsis thaliana mutants using a fluorescent reporter gene construct consisting of the sulfur limitation-responsive promoter of the SULTR1;2 sulfate transporter and green fluorescent protein as a background indicator for monitoring plant sulfur responses. The isolated mutant, sulfur limitation1 (slim1), was unable to induce SULTR1;2 transcripts under low-sulfur (–S) conditions. Mutations causing the sulfur limitation responseless phenotypes of slim1 were identified in an EIL family transcription factor, ETHYLENE-INSENSITIVE3-LIKE3 (EIL3), whose functional identity with SLIM1 was confirmed by genetic complementation. Sulfate uptake and plant growth on –S were significantly reduced by slim1 mutations but recovered by overexpression of SLIM1. SLIM1 functioned as a central transcriptional regulator, which controlled both the activation of sulfate acquisition and degradation of glucosinolates under –S conditions. Metabolite analysis indicated stable accumulation of glucosinolates in slim1 mutants, even under –S conditions, particularly in the molecular species with methylsulfinylalkyl side chains beneficial to human health. Overexpression of SLIM1 and its rice (Oryza sativa) homologs, but no other EIL genes of Arabidopsis, restored the sulfur limitation responseless phenotypes of slim1 mutants, suggesting uniqueness of the SLIM1/EIL3 subgroup members as sulfur response regulators.
INTRODUCTION
Modern agriculture requires adequate fertilization of sulfur to achieve maximum crop yield and performances. Plants use sulfate, the oxidized form of sulfur existing in the soil, as a sulfur source (Crawford et al., 2000; Leustek et al., 2000; Saito, 2004). By contrast, animals, including humans, require sulfur-containing amino acids and proteins as dietary sulfur sources because of their inability to assimilate sulfate into Cys and Met. Significance of plant sulfate assimilatory pathway is manifested by its ability to fill this metabolic gap in the global sulfur cycle in nature (Crawford et al., 2000). In addition to its basic nutritional importance, sulfur is present in numbers of plant metabolites representing important biological activities as redox controllers, vitamins, coenzymes, flavors, and defense chemicals (Crawford et al., 2000; Leustek et al., 2000; Saito, 2004; Grubb and Abel, 2006; Halkier and Gershenzon, 2006).
Activation of sulfate transport systems is critical for plant growth under a low-sulfur (–S) environment. When the soil environment is inadequately fertilized with sulfate, plants will sustain their growth by increasing the capacities of sulfate uptake systems in roots (Clarkson et al., 1983; Deane-Drummond, 1987; Smith et al., 1995, 1997). In Arabidopsis thaliana, high-affinity sulfate transporters that facilitate the initial uptake of sulfate serve this purpose (Takahashi et al., 2000; Vidmar et al., 2000; Shibagaki et al., 2002; Yoshimoto et al., 2002). Transporters mediating vascular transport of sulfate (Takahashi et al., 1997; Yoshimoto et al., 2003; Kataoka et al., 2004a) and release of vacuolar sulfate can also contribute for efficient use of sulfate pools under –S conditions (Kataoka et al., 2004b). In addition, catabolic recycling of secondary sulfur metabolites and storage compounds may become necessary for adaptation to a –S environment (Hirai et al., 1995, 2005; Kutz et al., 2002). However, such metabolic trade-offs substantially deteriorate the qualities of crop plant species. For cruciferous plants in particular, degradation and reduced production of glucosinolates, the major sulfur-containing secondary metabolites, are critical for both yield and qualities because they act as defense chemicals for plants themselves (Grubb and Abel, 2006; Halkier and Gershenzon, 2006) and are additionally beneficial to humans as cancer-preventive phytochemicals in diets (Talalay and Fahey, 2001).
Sulfur assimilation and glucosinolate production are the two major metabolic processes affected by sulfur nutrition. Recent microarray studies suggested that activation of sulfate acquisition and repression of glucosinolate production may occur in parallel in response to sulfur limitation (Hirai et al., 2003, 2004, 2005; Maruyama-Nakashita et al., 2003, 2005; Nikiforova et al., 2003). Apparently, the entire network of sulfur metabolism is coordinately regulated under the –S environment. Transcription factors and signaling proteins having stimulatory effects on indole glucosinolate biosynthesis have been reported from Arabidopsis (Celenza et al., 2005; Levy et al., 2005; Skirycz et al., 2006). However, regulatory proteins for the sulfate acquisition process that may totally affect the input of sulfur to metabolic pathways have not yet been identified. This study reports identification and functional characterization of a central transcription factor that corresponds to this assimilatory regulation in Arabidopsis. We demonstrate the function of this key transcription factor that essentially contributes to metabolic regulations necessary for adaptation to the –S environment.
RESULTS
Identification of Arabidopsis slim1 Mutants by Fluorescence Imaging
We took a genetic approach to identify the key regulatory proteins controlling the upstream signaling cascades of sulfur metabolism in Arabidopsis. A fluorescent reporter–aided screening was designed to monitor the abundance of SULTR1;2 sulfate transporter in Arabidopsis roots, visualized as an in vivo fluorescence of a jellyfish green fluorescent protein (GFP). SULTR1;2 is a high-affinity sulfate transporter in Arabidopsis, localized in the root hair, epidermis, and cortex of roots, and makes a major contribution to the uptake of sulfate from the environment (Shibagaki et al., 2002; Yoshimoto et al., 2002). The SULTR1;2 mRNA accumulates under –S conditions; therefore, a fusion gene construct, PSULTR1;2-GFP, consisting of a 2160-bp promoter region of SULTR1;2 sulfate transporter gene and the GFP coding sequence exhibits green fluorescence when plants are inadequately supplied with sulfate (Figure 1, parental line; Maruyama-Nakashita et al., 2004b).
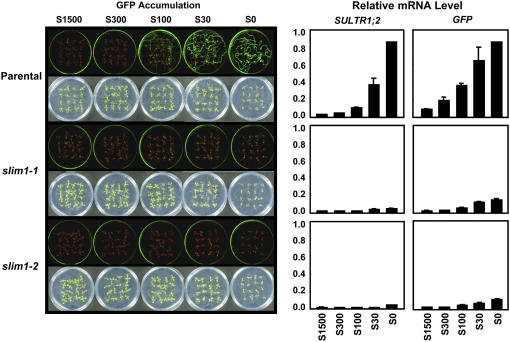
Figure 1.
The –S-Responseless Phenotypes of Arabidopsis slim1 Mutants.
Parental plants and slim1-1 and slim1-2 mutants were grown for 11 d with 1500, 300, 100, or 30 μM of sulfate (S1500, S300, S100, and S30) or with no sulfate (S0). Fluorescence of GFP from the indicator construct (PSULTR1;2-GFP) was visualized under an image analyzer as described in Methods. Bright-field and fluorescent images are presented in the left panels. SULTR1;2 and GFP mRNA contents of roots are shown in the right panels. The mRNA contents were quantified by real-time PCR and normalized using ubiquitin as an internal standard. Values are presented as means ± se (n = 3).
Using this transgenic Arabidopsis as a parental line (Columbia-0 background), ethyl methanesulfonate–mutagenized M2 seeds were generated, and seedlings were screened for the changes in GFP fluorescence on –S culture conditions. The M2 seedlings showing reduced levels of GFP fluorescence were isolated as mutant candidates. Those showing significant reduction of GFP signals were reselected in M3 plants, and F2 populations were generated from the crosses with the parental plants. A family of allelic mutants with cosegregating recessive mutations was chosen for further analysis and named slim1 after their sulfur limitation responseless phenotypes (Figure 1). The slim1-1 and slim1-2 mutants almost completely lacked the fluorescent signals of GFP that should be normally observed in the parental plants on –S media (Figure 1). Commensurate with the abnormality of GFP reporter expression, the induction of SULTR1;2 mRNA under –S was abolished in both slim1-1 and slim1-2 mutants (Figure 1).
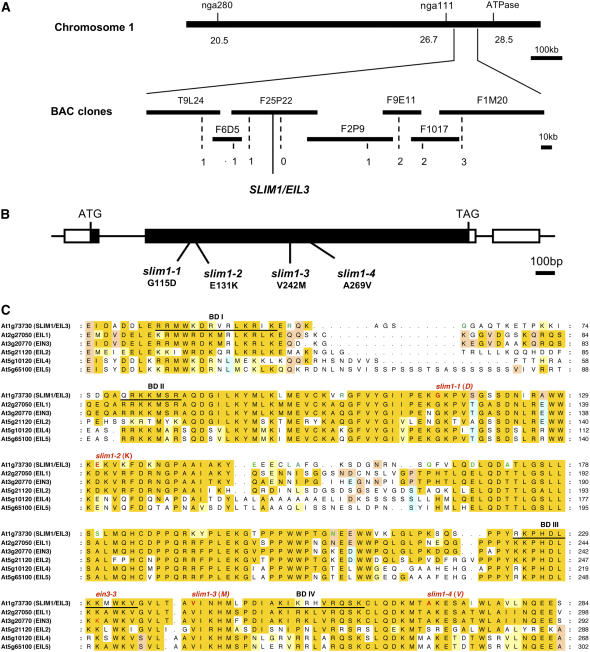
SLIM1 Encodes EIL3
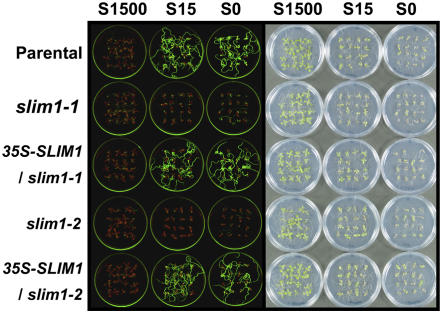
We cloned the SLIM1 gene by a genetic map-based strategy (Figure 2A). Single base substitutions were identified in slim1-1 and slim1-2, causing missense mutations, G115D and E131K, respectively, in the coding region of the ETHYLENE-INSENSITIVE3-LIKE3 (EIL3) gene (At1g73730) (Figures 2B and 2C), a putative EIL family transcription factor whose function has not been verified (Guo and Ecker, 2004). Further analysis identified two additional alleles, slim1-3 (V242M) and slim1-4 (A269V) (Figures 2B and 2C) showing phenotypes identical to slim1-1 and slim1-2. The slim1-3 and slim1-4 mutations were located close to the DNA binding domains BD III (225 to 236) and BD IV (251 to 261) of EIL proteins (Chao et al., 1997; Yamasaki et al., 2005). We further confirmed that overexpression of SLIM1 by cauliflower mosaic virus (CaMV) 35S promoter restores the sulfur limitation responseless phenotypes of slim1-1 and slim1-2 mutants (Figure 3). The expression of GFP from the indicator construct (PSULTR1;2-GFP) was recovered in both mutants by overexpression of SLIM1, establishing the function of SLIM1 in –S-responsive regulation of SULTR1;2 (Figure 3). Expression of SULTR1;2 mRNA was also recovered in both mutants by SLIM1 overexpression (data not shown). SLIM1 mRNA was expressed both in roots and shoots; however, it was not modulated by the changes of sulfur conditions (see Supplemental Figure 1 online). Tissue-specific expression of SLIM1 was examined in transgenic Arabidopsis plants expressing GFP under the control of the SLIM1 promoter (PSLIM1-GFP). GFP signals were predominantly found in the vascular tissues of roots and hypocotyls (see Supplemental Figure 2A online). In roots, GFP was expressed in pericycle and xylem parenchyma cells (see Supplemental Figure 2B online). As expected from its function as a transcription factor, GFP-SLIM1 fusion protein was localized exclusively in nuclei (35S-SLIM1-GFP; see Supplemental Figures 2C and 2D online). Nuclear localization of GFP-SLIM1 was not affected by sulfur availability (data not shown).
Figure 2.
Identification of SLIM1.
(A) Physical map of SLIM1 gene location. Numbers of recombination events are indicated below the BAC clones.
(B) Positions of slim1-1, slim1-2, slim1-3, and slim1-4 alleles. Exons are indicated by thick bars. Black and white bars indicate coding and untranslated regions, respectively.
(C) Alignment of SLIM1 and EIL family proteins in Arabidopsis. Alignment of full protein sequences was performed by the ClustalW program at the DNA Data Bank of Japan (DDBJ) (http://www.ddbj.nig.ac.jp/search/clustalw-j.html), and the N-terminal conserved regions are shown in this figure. Red characters indicate the positions of slim1-1, slim1-2, slim1-3, slim1-4, and ein3-3. Predicted DNA binding domains (BD I to BD IV) are underlined. Amino acid residues conserved in >50% of all 35 EIL family proteins (Figure 8A; see Supplemental Figure 4 online) are highlighted in yellow, and those having similarities with these conserved residues are highlighted in pale yellow. Amino acid residues conserved in >30% but <50% of all EIL family proteins are highlighted in light blue and light orange (two colors were used to distinguish occurrences of different residues at the same position). Green characters are the amino acid residues specific to the SLIM1 family members; they are conserved in >80% (four proteins) of all SLIM1 subfamily proteins (Figure 8A) but are not found in other EIL family proteins at the corresponding positions.
Figure 3.
Complementation of slim1 Mutants by SLIM1 Overexpression.
SLIM1 coding sequence was overexpressed in slim1-1 and slim1-2 mutants under CaMV 35S promoter (35S-SLIM1/slim1-1 and 35S-SLIM1/slim1-2). Plants were grown on S1500, S15, and S0 media, and fluorescence of GFP was monitored as in Figure 1.
SLIM1 Is Required for Plant Growth on the –S Environment
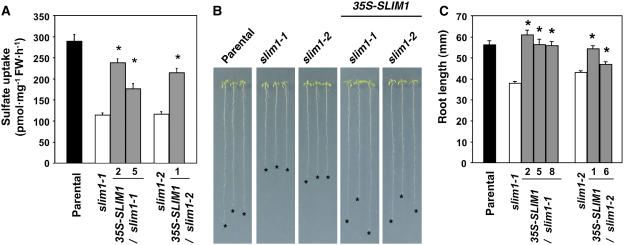
SLIM1 played significant roles in activation of sulfate uptake and proper growth under –S environment (Figure 4). For the sulfate uptake analysis, plants were grown on S15 media containing 15 μM sulfate, and uptake of [35S]sulfate was measured under the same sulfur condition (Figure 4A). The results indicated that high-affinity sulfate uptake activity is reduced ∼60% by slim1-1 and slim1-2 mutations (Figure 4A). By contrast, overexpression of SLIM1 restored these defects in the slim1 mutants (Figure 4A). These results suggested that SLIM1 is required for the –S-responsive induction of the high-affinity sulfate transport system facilitated by SULTR1;2 sulfate transporter in Arabidopsis roots. We further demonstrated the significance of SLIM1 in proper management of plant growth under the –S environment (Figure 4B). Plants were germinated and grown on S0 media with no external addition of sulfate to induce severe –S stress. Both slim1-1 and slim1-2 alleles showed 30% reduction of root growth compared with the parental line (Figures 4B and 4C). The observed growth defects were recovered by overexpression of SLIM1 (Figures 4B and 4C).
Figure 4.
SLIM1 Controls Sulfate Uptake and Plant Growth on –S.
(A) High-affinity sulfate uptake is controlled by SLIM1. Values are presented as means ± se (n = 8). Asterisks indicate statistically significant differences (P < 0.05) between slim1 mutants and SLIM1 overexpressor transgenic lines (2 and 5 for slim1-1, and 1 for slim1-2 backgrounds, respectively). FW, fresh weight.
(B) Root elongation under –S. Plants were vertically grown for 11 d on S0 agar medium. Asterisks indicate the positions of root tips.
(C) Root lengths of the plants in (B). Values are presented as means ± se (n = 25). Asterisks indicate statistically significant differences (P < 0.05) between slim1 mutants and SLIM1 overexpressor transgenic lines (2, 5, and 8 for slim1-1, and 1 and 6 for slim1-2 backgrounds, respectively).
SLIM1 Regulates –S-Responsive Genes
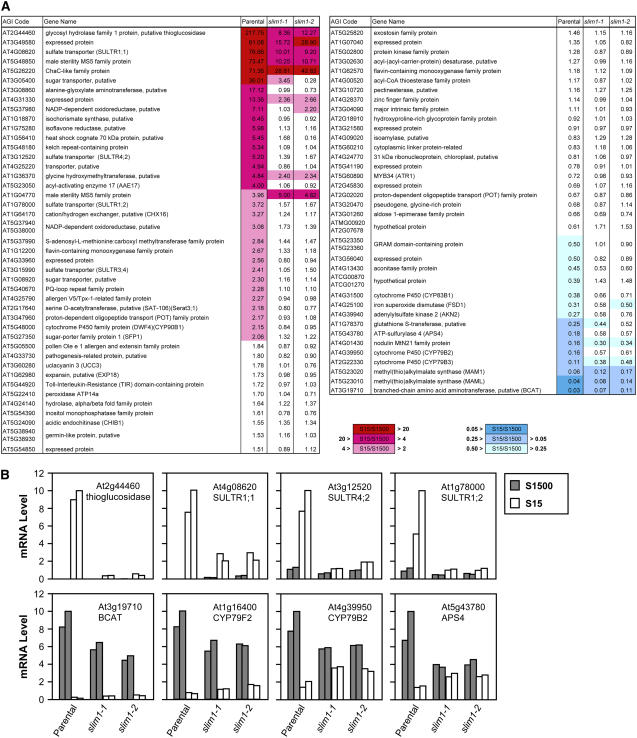
To characterize downstream genes whose transcripts are affected by slim1 mutation, parental line and slim1 mutants were grown on S15 (15 μM sulfate) or S1500 (1500 μM sulfate) media for 10 d, and their root RNAs were hybridized with Affymetrix ATH-1 GeneChip arrays (array data deposited in www.ncbi.nlm.nih.gov/geo under accession no. GSE4455). RNA was prepared from two separate cultures (a and b in Supplemental Table 1 online) to have biological duplicates for each plant line under S15 and S1500 conditions, respectively. Detailed procedures of data analysis are described in Methods. Briefly, after per-chip normalizations, each gene was normalized by that gene's expression level in the control S1500 sample. The –S-responsive genes were selected from the parental line data by Student's t test (P value cutoff: 0.01). The normalized values of S15 samples (S15/S1500 ratios) of –S-responsive genes were compared between the parental line and slim1 mutants to select SLIM1-dependent genes showing statistically significant differences under one-way analysis of variance test with Benjamini and Hochberg multiple testing correction (false discovery rate: 0.05) and Tukey post hoc test. Supplemental Table 1 online represents the list of –S-responsive SLIM1-dependent genes selected under these statistical tests. Figure 5A is a simplified presentation of these genes, visualizing differences of –S responses between the parental line and slim1 mutants.
Figure 5.
Transcriptome Profiles of slim1 Mutants.
(A) List of –S-responsive SLIM1-dependent genes. Genes were extracted through statistical tests of –S responsiveness and SLIM1 dependencies as described in Methods. The normalized values of S15 samples (S15:S1500 ratios) are shown as geometric means of duplicate data. The rates of –S-responsive upregulation and downregulation of the transcripts are highlighted by red and blue color indexes, respectively.
(B) Quantification of transcript levels of SLIM1-dependent genes. Real-time PCR was performed using the gene-specific primers (see Supplemental Table 6 online). Gray and white bars indicate duplicate root RNAs prepared from 10-d-old seedlings on S1500 and S15 conditions, respectively. The mRNA contents were normalized using ubiquitin as an internal standard.
The results indicated that several isoforms of sulfate transporters (SULTR1;1, SULTR1;2, SULTR3;4, and SULTR4;2) were upregulated by sulfur limitation more significantly in the parental line than in both slim1 mutants (Figure 5A; see Supplemental Table 1 online). SULTR1;1, SULTR1;2, and SULTR4;2 play important roles in sulfate acquisition and transport under –S environment (Takahashi et al., 2000; Vidmar et al., 2000; Shibagaki et al., 2002; Yoshimoto et al., 2002; Kataoka et al., 2004b). A putative thioglucosidase (At2g44460) showing clear –S response and SLIM1 dependency is suggested to be involved in hydrolytic degradation of glucosinolates for catabolic sulfur recycling. Around the pathways of Cys synthesis, an –S-inducible isoform of Ser acetyltransferase (Serat3;1) (Kawashima et al., 2005) showed a similar expression profile. In addition, the wild-type response of other –S-upregulated genes with unknown functions was generally abolished in slim1 mutants; the exceptions were MS5 family protein (At1g04770) and putative pectinesterase (At3g10720). The changes of transcript levels of putative thioglucosidase (At2g44460), SULTR1;1, SULTR1;2, and SULTR4;2 were confirmed by quantitative real-time RT-PCR, showing reproducible expression profiles of –S responsiveness and SLIM1 dependencies (Figure 5B).
Another remarkable shift of transcriptome by slim1 mutation was observed in glucosinolate synthesis. They were downregulated by sulfur limitation more significantly in the parental line than in the slim1 mutants (Figure 5A; see Supplemental Table 1 online). Methyl(thio)alkylmalate synthases (MAM1 and MAML) and a putative branched-chain amino acid aminotransferase (At3g19710) for Met chain elongation (Field et al., 2004; Textor et al., 2004), cytochrome P450s for indole glucosinolate biosynthesis (CYP79B2, CYP79B3, and CYP83B1) (Hull et al., 2000; Mikkelsen et al., 2000; Bak et al., 2001; Hansen et al., 2001a), and Myb34 (ATR1), a regulator of indole glucosinolate biosynthetic genes (Celenza et al., 2005), showed these patterns. Adenylylsulfate kinase (AKN2), which provides 3′-phosphoadenosine-5′-phosphosulfate for glucosinolate biosynthesis, and a sulfate assimilation enzyme, ATP sulfurylase (APS4), were also found in this category. On real-time RT-PCR quantification, the transcripts of a putative branched-chain amino acid aminotransferase (At3g19710), CYP79B2, and APS4 were lowered by slim1 mutations under S1500 conditions (Figure 5B). By contrast, they were slightly more abundant in the slim1 mutants than in the parental line under S15 conditions (Figure 5B). In addition, cytochrome P450 CYP79F2, which catalyzes aldoxime formation in the pathway of chain-elongated Met-derived glucosinolate biosynthesis (Hansen et al., 2001b; Reintanz et al., 2001; Chen et al., 2003), was regulated under the same scheme (Figure 5B).
A reduction step catalyzed by 5′-adenylylsulfate reductase (APR) is important for sulfur assimilation, and transcripts for the three APR isoforms accumulate under –S conditions (Gutierrez-Marcos et al., 1996; Setya et al., 1996; Vauclare et al., 2002). This microarray data indicated –S-responsive accumulation of APR2 and APR3 transcripts in the parental line, though they were equally responsive to sulfur limitation in the slim1 mutants (data not shown). The results likely indicate that APR is regulated independent of SLIM1.
Metabolite Accumulation in slim1 Mutants
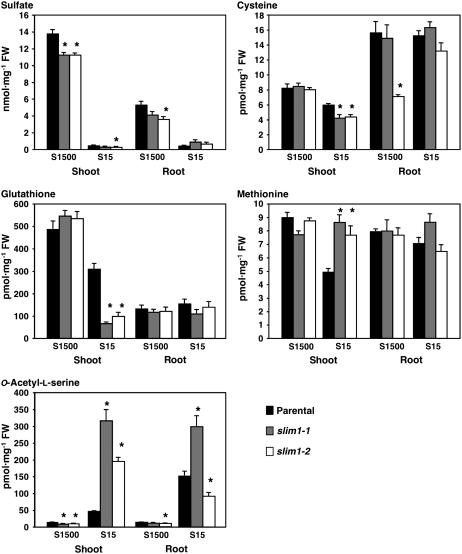
Metabolite analysis emphasized appreciable contribution of SLIM1-mediated regulation to sulfur assimilation and metabolism. Sulfate, Cys, glutathione (GSH), Met, and O-acetylserine (OAS) were measured in parental line and slim1 mutants grown on S15 (15 μM sulfate) or S1500 (1500 μM sulfate) media (Figure 6). The most significant changes were observed in OAS and GSH contents in shoots. Compared with the parental line, the slim1 mutants on S15 media showed overaccumulation of OAS and significant decrease of GSH in shoots (Figure 6). In roots, OAS content was lower in slim1-2 than in the parental line under S15 conditions (Figure 6), but a severer –S condition (S0) induced higher accumulation of OAS in both slim1 mutants (data not shown). Under the same –S condition, slim1 mutations caused a slight decrease and increase of sulfate pools in shoots and roots, respectively. In addition, Cys content was lowered but Met content was increased in shoots by slim1 mutations under S15 conditions.
Figure 6.
Sulfate and Primary Sulfur Metabolite Content of slim1 Mutants.
Plants were grown for 10 d on S15 (15 μM sulfate) or S1500 (1500 μM sulfate) conditions. Means ± se were calculated from triplicate experiments. Asterisks indicate statistically significant differences (P < 0.05) between slim1 mutants and parental line.
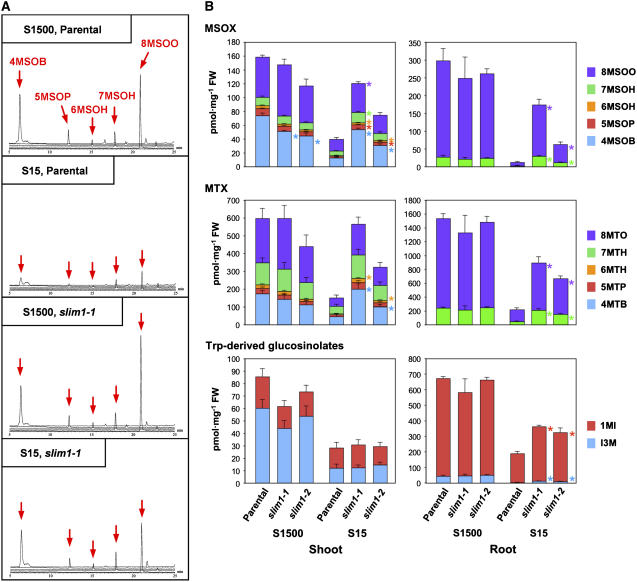
The effects of SLIM1-mediated regulation on secondary sulfur metabolism were evaluated by measuring the glucosinolate contents (Figure 7). As indicated by the mass chromatograms, methylsulfinylalkyl glucosinolate contents were significantly decreased by sulfur limitation in the parental line (Figure 7A). By contrast, substantial amounts of these Met-derived glucosinolates were accumulated in slim1 mutants even under S15 conditions (Figure 7A). These patterns were generally the same in roots for the molecular species with longer alkyl side chains (Figure 7B). Methylthioalkyl glucosinolate contents were modulated exactly the same as in the case of methylsulfinylalkyl glucosinolates; a significant decrease by sulfur limitation was observed in both shoots and roots of the parental line, but these changes were moderated in slim1-1 and slim1-2 mutants (Figure 7B, MTX). Indole glucosinolate contents were also regulated by SLIM1 in roots as in the case of Met-derived species; however, in shoots, they decreased by sulfur limitation in both slim1 mutants and the parental line (Figure 7B).
Figure 7.
Glucosinolate Content of slim1 Mutants.
(A) Profiles of methylsulfinylalkyl glucosinolates analyzed by liquid chromatography–mass spectrometry. Panels show the data of shoot samples of slim1-1 mutant and parental plant. Plants were grown for 10 d under S15 (15 μM sulfate) or S1500 (1500 μM sulfate) conditions. Mass chromatograms of [M-H]− ions for 4-methylsulfinylbutyl glucosinolate (4MSOB; mass-to-charge ratio [m/z] = 436), 5-methylsulfinylpentyl glucosinolate (5MSOP; m/z = 450), 6-methylsulfinylhexyl glucosinolate (6MSOH; m/z = 464), 7-methylsulfinylheptyl glucosinolate (7MSOH; m/z = 478), and 8-methylsulfinyloctyl glucosinolate (8MSOO; m/z = 492) were overlaid in same scales. Positions of 4MSOB, 5MSOP, 6MSOH, 7MSOH, and 8MSOO are indicated by red arrows.
(B) Methylsulfinylalkyl (MSOX), methylthioalkyl (MTX), and Trp-derived indole glucosinolate contents of slim1 mutants. Plants were grown and analyzed as in (A). Means ± se were calculated from triplicate experiments. Statistically significant differences (P < 0.05) between slim1 mutants and the parental line are shown by asterisks at the right of the columns for each compound of MSOX, MTX, and indole glucosinolates. I3M, indole-3-ylmethyl glucosinolate; 1MI, 1-methoxyindol-3-ylmethyl glucosinolate; 4MSOB, 4-methylsulfinylbutyl glucosinolate; 5MSOP, 5-methylsulfinylpentyl glucosinolate; 6MSOH, 6-methylsulfinylhexyl glucosinolate; 7MSOH, 7-methylsulfinylheptyl glucosinolate; 8MSOO, 8-methylsulfinyloctyl glucosinolate; 4MTB, 4-methylthiobutyl glucosinolate; 5MTP, 5-methylthiopentyl glucosinolate; 6MTH, 6-methylthiohexyl glucosinolate; 7MTH, 7-methylthioheptyl glucosinolate; 8MTO, 8-methylthiooctyl glucosinolate.
SLIM1 Proteins Are Functionally Distinguishable from Other EILs
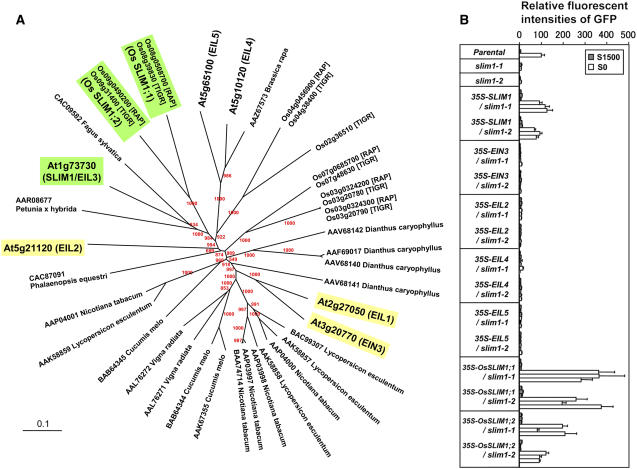
In the Arabidopsis genome, six genes are annotated to encode the EIL family proteins (EIN3 and EIL1 to EIL5) (Figures 2B and 8A; Guo and Ecker, 2004). EIN3 is a transcription factor controlling the expression of ethylene-responsive genes, and EIL1 and EIL2 are the closest functional homologs of EIN3 (Chao et al., 1997; Solano et al., 1998; Guo and Ecker, 2004). On the contrary, the functions of EIL3 (SLIM1), EIL4, and EIL5 have not yet been verified (Guo and Ecker, 2004). To assess whether SLIM1 is the only form specialized for sulfur response, other EIL family members from Arabidopsis, EIN3, EIL2, EIL4, and EIL5, were overexpressed in slim1-1 and slim1-2 mutants. The results clearly indicated that SLIM1 is the only form able to restore the –S-responsive expression of GFP from the PSULTR1;2-GFP indicator construct in slim1 backgrounds (Figure 8B). None of the other Arabidopsis EIL proteins showed the same function (Figure 8B).
Figure 8.
SLIM1 Has a Unique Function in the EIL Family.
(A) Phylogenic relationships of SLIM1 and EIL family proteins. Arabidopsis and rice SLIM1 proteins are highlighted in green. Arabidopsis EIN3, EIL1, and EIL2 that mediate ethylene response are highlighted in yellow. Arabidopsis SLIM1 and EIL family proteins are indicated with their Arabidopsis Genome Initiative codes. Locus numbers of rice EILs are indicated according to the Rice Annotation Project (RAP) and The Institute for Genomic Research (TIGR) databases, respectively. The rest of the members are indicated by accession numbers of protein sequences. Bootstrap values are shown at the tree nodes by red characters.
(B) The sulfur limitation response of PSULTR1;2-GFP indicator was recovered by CaMV 35S promoter-driven overexpression of Arabidopsis SLIM1 and its rice homologs, Os SLIM1;1 and Os SLIM1;2. Plants were grown on S1500 or S0 agar medium, and fluorescence of GFP was monitored as in Figure 1. The fluorescence intensity of parental line on S0 is described as 100. Each column indicates independent transgenic or mutant lines. Means ± se (n = 3) for each lines are presented.
The alignment and phylogenetic analysis of EIL family proteins indicated that SLIM1 proteins are ubiquitously found in the plant kingdom (Figure 8A). Arabidopsis SLIM1, two rice (Oryza sativa) homologs, Os SLIM1;1 and Os SLIM1;2, and proteins from petunia (Petunia hybrida) and Fagus sylvatica formed a unique branch that can be distinguished from other group members (Figure 8A). Os SLIM1;1 and Os SLIM1;2 were overexpressed in slim1 mutants and were demonstrated to have capabilities to restore the expression of GFP from the PSULTR1;2-GFP indicator construct in Arabidopsis in response to sulfur limitation (Figure 8B). These results strongly suggested that proteins in the SLIM1/EIL3 subgroup are functionally distinct from other EIL family members mediating ethylene responses and may have unique functions in regulating sulfur responses in various plant species.
DISCUSSION
Significance of SLIM1 in Plant Sulfur Response
In this study, we identified a transcription factor, SLIM1, that regulates the main pathways of sulfate uptake and metabolism under the –S environment in Arabidopsis roots. To our knowledge, this is the first report for the identification of a transcription factor regulating the assimilatory sulfur metabolism in higher plants. The significance of SLIM1's function was evidenced by its ability to control the expression of the SULTR1;2 sulfate transporter, the major sulfate uptake facilitator in Arabidopsis roots. SLIM1 was required for the –S-responsive induction of SULTR1;2 transcripts, leading to a remarkable increase of high-affinity sulfate uptake activities that eventually contribute to the maintenance of plant growth on –S.
The high-affinity sulfate transport system predominates under the –S environment for efficient acquisition of sulfate from the soil. This phenomenon has been well characterized by classical physiological experiments (Clarkson et al., 1983; Deane-Drummond, 1987). More recent molecular biological studies suggested that this –S-inducible transport system can be attributed to the function of two sulfate transporter genes, SULTR1;1 and SULTR1;2, in Arabidopsis (Takahashi et al., 2000; Vidmar et al., 2000; Shibagaki et al., 2002; Yoshimoto et al., 2002). Similar inducible sulfate transporters exist in other plant species as well (Smith et al., 1995, 1997; Vidmar et al., 1999; Howarth et al., 2003; Buchner et al., 2004a, 2004b; Hopkins et al., 2005). Studies on the selenate-resistant mutant and T-DNA knockout of the SULTR1;2 gene further confirmed that the SULTR1;2 high-affinity sulfate transporter predominantly mediates this –S-inducible transport system in Arabidopsis roots (Shibagaki et al., 2002; Maruyama-Nakashita et al., 2003). Furthermore, gene expression studies with promoter-reporter constructs indicated that both SULTR1;1 and SULTR1;2 are regulated under their –S-responsive promoters in response to sulfur nutrition (Maruyama-Nakashita et al., 2004a, 2004b). At least for SULTR1;1, identification of a sulfur-responsive cis-acting element, SURE, in its 5′-region supports the importance of transcriptional regulation in sulfur response (Maruyama-Nakashita et al., 2005). These findings were indicative of the existence of trans-acting regulatory proteins controlling gene expression of high-affinity sulfate transporters in Arabidopsis.
The strategy we took here for the screening of regulatory proteins uses the promoter-GFP fusion gene construct of SULTR1;2 (Maruyama-Nakashita et al., 2004b). Under this method, the reporter activity (i.e., fluorescence of GFP) phenocopies the transcript abundance of SULTR1;2, which in turn reflects the activity of sulfate uptake in Arabidopsis roots. The slim1 mutant described in this work demonstrates the usefulness of this strategy in identifying metabolic mutants that tend to have subtle differences of morphological phenotypes and viabilities. In fact, we found that growth of slim1 mutants is affected under severe –S conditions, showing 30% reduction of their root lengths and 60% decrease of sulfate uptake rates (Figure 4). As suggested from these characteristics of slim1 mutants, the SLIM1 transcription factor is important for regulation of sulfate uptake and assimilation (Figures 4 and 5). From the methodological point of view, such metabolic or biochemical phenotypes related to alterations of metabolism and nutrient transport processes are only measurable by careful evaluation of established mutant lines. On the other hand, initial setups of mutant screening must be simple and traceable enough to report the metabolic changes of mutant candidates from wild-type plants. In this work, we used a fluorescence imaging system to solve this problem and eventually identified the function of SLIM1, which is a transcription factor demonstrated to play important roles in regulating sulfate uptake and metabolism in higher plants.
SLIM1, a Unique EIL Family Protein, Controls Sulfur Metabolism
The Arabidopsis EIL family consists of six distinct members (EIN3 and EIL1 to EIL5) (Figure 8A; Guo and Ecker, 2004). The functions of EIN3 and EIL1 have been extensively studied for the control of ethylene-responsive genes in Arabidopsis (Chao et al., 1997; Solano et al., 1998; Guo and Ecker, 2004). EIN3 is the member whose function was first characterized by the ethlylene-insensitive phenotypes of ein3 mutants (Chao et al., 1997). EIL1 and EIL2 are the closest homologs of EIN3 (Figure 8B) and were able to complement the phenotype of the ein3 mutant (Chao et al., 1997). More recently, the functional redundancy of EIL1 with EIN3 has been proved genetically (Alonso et al., 2003). Molecular studies of ethylene-responsive promoter elements suggest that EIN3, EIL1, and EIL2, but not EIL3, can bind to the upstream EIN3 binding sequence in the ethylene-responsive ERF1 gene (Solano et al., 1998). From these observations, the functions of EIL3/SLIM1, EIL4, and EIL5 are still unclear as to whether they participate in ethylene response or in totally unrelated pathways (Guo and Ecker, 2004).
This work confirmed that EIL3, renamed SLIM1 according to its functionality, has specific function in regulating sulfate uptake and metabolism in Arabidopsis. SLIM1, but no other EIL proteins from Arabidopsis, was able to restore the sulfur limitation responseless phenotypes of slim1 mutants (Figure 8B). Accordingly, SLIM1 is suggested to be specialized for sulfur response. In fact, the absence of EIL3 from ethylene response (Solano et al., 1998; Guo and Ecker, 2004) was indicative of its distinct function. In addition, the –S-responsive SLIM1-dependent genes (Figure 5A) were not regulated by 1-aminocyclopropane 1-carboxylic acid treatment (see Supplemental Figure 3 online), suggesting that SLIM1-mediated regulation can be separated from the ethylene signaling pathways. We further demonstrated that SLIM1 homologs from rice can complement the Arabidopsis slim1 mutants, suggesting their functional identities with the Arabidopsis SLIM1 gene product in regulation (Figure 8). This further emphasizes the significance of this new functional category of EIL family proteins and suggests generality of their functions in plant sulfur response both in dicotyledonous and monocotyledonous plant species. Contrary to the genetically confirmed evidence showing its significance in sulfur response, SLIM1 mRNA itself was not modulated by the changes of sulfur conditions (see Supplemental Figure 1 online). Presumably, SLIM1 may require a posttranscriptional mechanism for its regulation, like in the case of EIN3, whose protein level is strictly regulated by ethylene and carbon status (Guo and Ecker, 2003; Potuschak et al., 2003; Yanagisawa et al., 2003). However, unlike the case in EIN3, nuclear localization of GFP-SLIM1 protein was not affected by sulfur conditions (data not shown). Further studies are required for investigation of posttranscriptional modification of its functionalities under the –S environment.
A recent study reported that synthetic oligonucletides designed for a conserved EIL binding sequence, AYGWAYCT (Solano et al., 1998; Kosugi and Ohashi, 2000), were able to interact with an in vitro–synthesized Ser-162 to Gln-288 region, including the putative DNA binding domains III and IV of EIL3 (Yamasaki et al., 2005). This contrasts with the previous findings showing inability of EIL3 protein to bind to the corresponding nucleotide sequences in the ERF1 gene promoter (Solano et al., 1998). Binding of EIL3 to this conserved sequence can be unstable, as it is only detectable with surface plasmon resonance but not by electro-mobility shift assay (Yamasaki et al., 2005). Differences in binding kinetics may imply distinct function of SLIM1/EIL3 in transcriptional regulation, separated from the ethylene signaling pathways. We searched for the existence of this conserved EIL binding sequence in the upstream regions of –S-responsive genes categorized in transcriptome analysis (Figure 5A). The AYGWAYCT sequences were most frequently found in the SLIM1-dependent genes whose transcript levels in the parental line were greater in S15 than in S1500 (see Supplemental Table 2 online). Judging at least from the frequencies of putative SLIM1 binding sequences within the upstream regions, the role of SLIM1 in transcriptional regulation is likely delimited to –S-responsive upregulation. The putative binding sequences were present in SULTR4;2 but absent from SULTR1;1 and SULTR1;2 (see Supplemental Table 3 online). Genes with no conservation of binding sequences but showing clear SLIM1-dependent expression, such as SULTR1;1 and SULTR1;2, presumably require additional regulatory elements downstream of SLIM1 for immediate activation of their gene expression under –S conditions. In fact, SLIM1 was predominantly expressed in the vasculatures (see Supplemental Figure 2 online), suggesting that horizontal signal transfer would become necessary to induce the expression of high-affinity sulfate transporters that localize at surface cell layers of roots. The spatial localization of SLIM1 suggested that at least for the SLIM1-mediated regulation, sulfur status is primarily sensed in the central vascular region of plants, rather than in the root surface where SULTR1;2 sulfate transporter is present and is in direct contact with the soil environment (Yoshimoto et al., 2002). Identification of immediate targets of SLIM1 and downstream molecules transferring signals from vasculature to epidermis awaits further investigation.
SLIM1 Controls Sulfur Metabolism
The transcriptome data clearly suggested that SLIM1 functions as a hub regulatory protein, participating universally in the regulation of –S-responsive genes that play essential roles in optimizing transport and internal utilization of sulfate in Arabidopsis (Figure 5). As highlighted in the metabolic pathways in Figure 9, not exclusively all but the majority of –S-responsive genes playing pivotal roles in sulfur assimilation and metabolism were controlled under SLIM1. All these pathways need to be properly regulated for acquisition of sulfate and efficient utilization of internal sulfur pools when Arabidopsis plants are starved for sulfur.
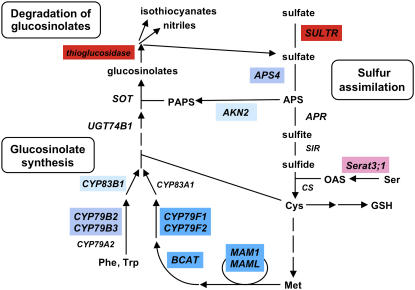
Figure 9.
SLIM1-Mediated Regulation of Sulfur Assimilation and Glucosinolate Metabolism.
SLIM1-dependent genes are highlighted by color indexes as in Figure 5A. Red and blue indicate the SLIM1-dependent pathways regulated positively and negatively, respectively, by sulfur limitation. AKN2, 5′adenylylsulfate kinase; APR, 5′adenylylsulfate reductase; APS4, ATP sulfurylase; BCAT, branched-chain amino acid aminotransferase; CS, Cys synthase; CYP, cytochrome P450; MAM, methyl(thio)alkylmalate synthase; Serat, Ser acetyltransferase; SIR, sulfite reductase; SOT, desulfoglucosinolate sulfotransferase; SULTR, sulfate transporter; UGT74B1, UDP-glucose:thiohydroximic acid S-glucosyltransferase; APS, adenosine 5′-phosphosulfate; PAPS, 3′-phosphoadenosine-5′-phosphosulfate.
Under the –S environment, the wild-type Arabidopsis plants maximize their sulfur use efficiencies through the induction of SULTR1;1 and SULTR1;2 for sulfate uptake (Shibagaki et al., 2002; Yoshimoto et al., 2002) and SULTR4;1 and SULTR4;2 for release of vacuolar sulfate in root tissues (Kataoka et al., 2004b). These pathways were almost completely turned down in slim1 mutants (Figure 5B). In addition, modulation of sulfate contents suggests disturbance of vacuolar sulfate recycling in the roots of slim1 mutants (Figure 6). Although transcripts of these sulfate transporters were modulated by SLIM1, the effects of this transcription factor was not exclusive for the control of SULTR1;1 that also follows –S-responsive regulation under a previously identified cis-acting element, SURE (Maruyama-Nakashita et al., 2005). In fact, SULTR1;1 mRNA was partially induced by –S even in the slim1 mutants (Figure 5B). By contrast, absence of SURE from the promoter region of SULTR1;2 suggests that this major sulfate uptake facilitator is controlled predominantly by SLIM1 under –S conditions. Corollary of the SLIM1-mediated regulation of SULTR1;2, sulfate uptake activity was significantly decreased by slim1 mutations (Figure 4A), which was comparable to the results of deletion of SULTR1;2 (Shibagaki et al., 2002). As –S-responsive gene expression is critical for plant survival in the –S condition, it is reasonable that plants have evolved multiplex regulatory mechanisms to control gene expression. Further identification of signaling components should unravel the full-set mechanisms underlying this hypothesis.
The metabolite analysis further suggested that slim1 mutants may suffer from severer shortage of sulfur, as indicated by significant decrease of GSH content and overaccumulation of OAS in their shoots (Figure 6). These metabolite profiles are typical of –S-cultured plants and are suggested to be caused by insufficient supply of sulfur to sulfate reduction pathway and Cys synthesis in shoots. The –S-inducible isoform of Ser acetyltransferase, Serat3;1 (Kawashima et al., 2005), which may have specific function in Cys synthesis under –S environment, showed SLIM1-dependent expression (Figure 5A). In contrast with these metabolic genes, the –S response of adenylylsulfate reductase (APR2 and APR3) was unaffected by slim1 mutations (Figure 5A). This enzyme is suggested to be important for the flux control of sulfate reduction, and its transcripts are abundantly accumulated under –S conditions (Gutierrez-Marcos et al., 1996; Setya et al., 1996; Vauclare et al., 2002). The mechanism of its regulation is likely different from the SLIM1-mediated pathway identified in this study.
Feeding experiments suggest that excess loading of GSH or OAS to plants can regulate sulfur metabolic genes, mimicking the transcript levels observed under the conditions where plants may receive adequate or limiting amount of sulfate, respectively (Hirai et al., 2004; Maruyama-Nakashita et al., 2004b). Contrary to what one might have expected for their postulated actions in regulation, neither the increase of GSH nor decrease of OAS was observed by slim1 mutation on –S (Figure 6), although slim1 mutants resulted in showing typical –S responseless expression profiles of transcripts (Figure 5). Although possibilities of local metabolite accumulation remain unverified, SLIM1 is not suggested to be located upstream of the previously hypothesized regulatory functions of OAS and GSH in plant sulfur signaling. Furthermore, comparison of macroarray data of OAS response (Hirai et al., 2003) and our GeneChip analysis identified no significant overlaps between OAS-responsive upregulation and SLIM1-dependent –S upregulation; however, aconitase family protein (At4g13430) and iron superoxide dismutase (At4g25100) were found in both OAS-responsive downregulation and SLIM1-dependent –S downregulation. At present, our data are not supportive to prove clear relation between OAS and SLIM1 in –S signaling.
Degradation of glucosinolate is another important aspect of sulfur limitation response. The degradation process is catalyzed by thioglucosidase (or myrosinase) that releases the aglycon of glucosinolate. The aglycon is subsequently broken down to isothiocyanate, oxazolidine-2-thione, nitrile, epithionitrile, or thiocyanate (Grubb and Abel, 2006; Halkier and Gershenzon, 2006). Sulfate can be released upon this chemical rearrangement of the aglycon and is possibly reused in primary metabolism. Induction of putative thioglucosidase on –S is suggested to be a significant metabolic response that may serve for reuse of the sulfur pool in glucosinolates (Figure 9), though its physiological effects on total sulfur metabolic flux await further investigation. In slim1 mutants, expression of thioglucosidase gene was eliminated (Figure 5B), and glucosinolates were abundantly accumulated even under –S conditions (Figure 7). Our results strongly suggested that SLIM1 coregulates this sulfur recycling process in parallel with sulfate transport systems under the –S environment (Figures 5 and 9).
The slim1 mutations additionally affected the expression of metabolic and regulatory genes of glucosinolate biosynthetic pathways, such as Met chain-elongation enzymes (Field et al., 2004; Textor et al., 2004), CYP79F1/F2 (Hansen et al., 2001b; Reintanz et al., 2001; Chen et al., 2003), CYP79B2/B3 (Hull et al., 2000; Mikkelsen et al., 2000), CYP83B1 (Bak et al., 2001; Hansen et al., 2001a), and the Myb34 (ATR1) transcription factor (Celenza et al., 2005) (Figure 5). In addition, the –S-downregulated forms of adenyllylsulfate kinase (AKN2) and ATP sulfurylase (APS4) were coclassified in this group, suggesting possible contribution of these particular isoenzymes to generating a sulfate donor, 3′-phosphoadenosine-5′-phosphosulfate, of sulfation reaction, part of which may be used for glucosinolate biosynthesis (Piotrowski et al., 2004; Hirai et al., 2005). The slim1 mutations generally acted suppressive to APS4 and glucosinolate biosynthetic genes, branched-chain amino acid aminotransferase, CYP79B2, and CYP79F2 under sulfur-sufficient conditions (Figure 5B, S1500). On the other hand, their transcripts were slightly enhanced by slim1 mutations under –S conditions (Figure 5B, S15). As a result, these contrasting profiles gave relatively lower S15:S1500 ratios for the parental line (Figure 5A). The effects of SLIM1 on these metabolic genes were rather moderate compared with its major contribution to –S-responsive upregulation of sulfate acquisition and catabolic sulfur recycling from glucosinolates (Figure 5B). Frequent occurrence of putative SLIM1 binding sites in the –S-upregulated SLIM1-dependent genes (see Supplemental Table 2 online) supports predominant roles of SLIM1-mediated regulation in these metabolic processes under the –S environment.
Our findings demonstrate that the SLIM1 transcription factor participates in the main pathway of –S-responsive regulation of sulfate acquisition and metabolism in Arabidopsis. This work provides perspectives in investigating the regulatory networks of plant sulfur response and metabolism. Identification of SLIM1 will lead in-depth analysis of downstream regulatory elements of –S-responsive gene regulation. Findings on SLIM1 functions may further facilitate general improvement of sulfur-use efficiencies and engineering of glucosinolate production in cruciferous plants.
METHODS
Plant Growth
Arabidopsis thaliana plants were grown at 22°C under 16-h-light/8-h-dark cycles. Plants were grown on mineral nutrient media (Hirai et al., 1995) containing 1% sucrose. For preparation of agar medium, agar was washed twice with 1 liter of deionized water and vacuum filtrated. S0 agar medium was prepared by complete replacement of MgSO4 to MgCl2. S1500, S300, S100, S30, and S15 agar media were prepared by adding MgSO4 to the S0 medium. Mg concentration was adjusted to 1500 μM by adding MgCl2.
Isolation of slim1
The homozygous progeny of PSULTR1;2-GFP transgenic plants (Columbia-0 background) (Maruyama-Nakashita et al., 2004b) was used as a parental line. Approximately 15,000 seeds were mutagenized with 0.3% ethyl methanesulfonate for 16 h and directly sowed on soil. M2 seeds were harvested separately as 36 pools. Approximately 2000 M2 seedlings from each pool (72,000 in total) were screened for alteration of GFP fluorescence on S0 agar medium. GFP was detected in 11-d-old seedlings using the FluorImager 595 image analyzer (Molecular Dynamics). The M2 seedlings showing reduced GFP fluorescence were selected as mutant candidates. M3 seeds were collected from 209 independent mutant candidates. Eighty lines with reduced GFP fluorescence were reselected by comparing fluorescence on S0 and S1500 agar media and backcrossed with the parental PSULTR1;2-GFP plant. F2 progenies of the backcrossed lines were assayed following the same condition. Finally, 60 lines were selected as sulfur limitation responseless mutants. A family of allelic mutant lines was named slim1 and analyzed in this study. The slim1-1 and slim1-2 plants were backcrossed three times with the parental line and used for phenotypic analyses.
Positional Identification of slim1
F2 plants that derive from the crosses between slim1 mutants and Landsberg erecta were used for map-based cloning. F2 seedlings showing kanamycin resistance and significant reduction in GFP fluorescence were selected on S0 agar medium containing 10 μM kanamycin sulfate. Chromosome-containing slim1 was determined using the genetic markers nga68, nga111, nga361, nga162, nga6, nga8, and nga139. slim1 was mapped between nga111 and ATPASE markers and narrowed down to the region between the BAC clones T9L24 and F1M20 using single nucleotide polymorphism markers generated by Inplanta Innovations (http://www.inplanta.jp/eng/service/mapping.html). We generated additional molecular markers summarized in Supplemental Table 4 online. Using these markers, slim1 was mapped to the position between At1g73660 and At1g73850. Mutations in slim1-1 and slim1-2 were identified on At1g73730 by sequencing 21 genes between At1g73660 and At1g73850.
Transgenic Plants
The SLIM1 overexpression construct (35S-SLIM1) was created by cloning the SLIM1 coding region under CaMV 35S promoter in pSMAH621 vector (Kubo et al., 2005). Constructs for overexpression of other EIL family genes and Oryza sativa SLIM1 genes were prepared in pH35GS (Kubo et al., 2005), a binary vector containing a Gateway cassette (Invitrogen) in the sense orientation under CaMV 35S promoter in pSMAH621. GFP-SLIM1 fusion construct (35S-GFP-SLIM1) was prepared by cloning a translational fusion gene of sGFP (Chiu et al., 1996) and SLIM1 under CaMV 35S promoter in pBI121 (Clontech). SLIM1 promoter-GFP (PSLIM1-GFP) construct was prepared by cloning a 2087-bp 5′-promoter region of SLIM1 in pBI101-GFP vector (Yoshimoto et al., 2003) that has replacement of β-glucuronidase gene in pBI101 (Clontech) with sGFP. All DNA fragments cloned in transformation vectors were amplified by PCR using high-fidelity KOD-Plus DNA polymerase (Toyobo). Primer sequences used for the PCR are summarized in Supplemental Table 5 online. Binary plasmids were transferred to Agrobacterium tumefaciens GV3101 (pMP90) (Koncz and Schell, 1986) and transformed to Arabidopsis plants according to the floral dip method (Clough and Bent, 1998). Transgenic plants were selected with 25 mg L−1 hygromycin B (for pSMAH621 and pH35GS constructs) or 50 mg L−1 kanamycin sulfate (for pBI constructs), and T2 or T3 progenies were used for the analysis.
Phylogenetic Analysis
BLASTP programs at The Arabidopsis Information Resources (http://www.arabidopsis.org/Blast/), RAP (http://rapdb.lab.nig.ac.jp/blast/index.html), TIGR (http://tigrblast.tigr.org/euk-blast/index.cgi?project=osa1), and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/) were used for database searches of SLIM1 homologs. Protein sequences were aligned using ClustalW (Thompson et al., 1994), and the phylogenetic tree was created by the neighbor-joining method (Saitou and Nei, 1987) using the programs at the DDBJ (http://www.ddbj.nig.ac.jp/search/clustalw-j.html). Bootstrap analysis was conducted with 1000 replicates. The sequence alignment (see Supplemental Figure 4 online) was edited using Jalview (http://www.jalview.org/) (Clamp et al., 2004). The unrooted phylogenetic tree (Figure 8A) was drawn using TreeView (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) (Page, 1996).
Imaging of GFP Expression
Expression of GFP in intact plants was visualized using the FluorImager 595 image analyzer under 488-nm excitation (Molecular Dynamics). Relative intensities of GFP signals were quantified using ImageQuant (Molecular Dynamics). The laser scanning confocal microscopy system FluoView 500 (Olympus) was used for microscopy analysis of SLIM1 promoter-GFP plants and nuclear localization of GFP-SLIM1 fusion protein.
Sulfate Uptake
Plants were vertically grown for 10 d on S15 media (15 μM sulfate). The roots were submerged in nutrient solution containing 15 μM [35S] sodium sulfate (Amersham Biosciences) and incubated for 30 min. Washing and measurement were performed as described previously (Kataoka et al., 2004b; Maruyama-Nakashita et al., 2004b).
GeneChip Hybridization and Data Analysis
Parental line, slim1-1, and slim1-2 mutants were grown for 10 d under S15 (15 μM sulfate) or S1500 (1500 μM sulfate) conditions. Duplicate RNA samples were prepared from roots, and hybridization of ATH-1 array (Affymetrix) was performed according to the manufacturer's protocol. Microarray Suite 5.0 (Affymetrix) and GeneSpring 7.2 (Silicon Genetics) programs were used for the data analysis. All data of GeneChip experiments have been deposited in the Gene Expression Omnibus database under accession number GSE4455 (www.ncbi.nlm.nih.gov/geo).
Each chip was normalized to the 50th percentile of the measurements taken from that chip (per chip normalization). Per gene normalization was performed using the measurements of the S1500 samples of each plant line as controls (e.g., signals of each gene on the arrays of the parental line [Parental-S1500-a, Parental-S1500-b, Parental-S15-a, and Parental-S15-b] were divided by the median of that gene's measurements in the S1500 samples [Parental-S1500-a and Parental-S1500-b]). The array data of slim1 mutants were normalized similarly.
Array elements showing present or marginal calls at least in one of the four parental line chip experiments (16,731 genes) were selected for further gene classification. The –S-responsive genes were first selected from the parental line chip data. Student's t test (P value cutoff: 0.01) was performed between the S1500 and S15 samples of the parental line, and 469 genes were obtained as –S-responsive genes. SLIM1-dependent genes were further extracted by a multiple comparison of parental line, slim1-1, and slim1-2 mutants. The normalized values of S15 samples (S15/S1500 ratios) of –S-responsive genes were subjected to a one-way analysis of variance test applying Benjamini and Hochberg multiple testing correction (Benjamini and Hochberg, 1995) with a false discovery rate of 0.05 and Tukey post hoc test. As a result, 79 genes were listed as –S-responsive SLIM1-dependent genes that show statistically significant differences between the parental line and two mutant alleles (Figure 5; see Supplemental Table 1 online). Values in Supplemental Table 1 online are the normalized data and corrected P values calculated according to this procedure.
Quantitative Real-Time RT-PCR
RNA preparation and reverse transcription were performed as reported previously (Maruyama-Nakashita et al., 2004b). Real-time PCR was performed using the SYBR Green Perfect Real Time kit (Takara) and GeneAmp 5700 sequence detection system (Applied Biosystems). The mRNA contents were calculated using ubiquitin as an internal standard. Gene-specific primers are described in Supplemental Table 6 online.
Metabolite Analysis
Plant tissues were harvested and frozen in liquid nitrogen before extraction. Frozen tissues (100 mg FW) were homogenized at 4°C in 5× volume (500 μL) of extraction solvent (80% methanol: 20% milliQ water [Millipore]) using a mixer mill MM300 (Retsch). After 3 min of homogenization, cell debris was removed by centrifugation to have cleared extract. Three hundred microliters of extract was vacuum-evaporated to dryness and resolved in 100 μL of milliQ water, followed by ultrafiltration through NANOSEP MF GHP 0.45 μm (PALL Life Sciences). This filtrate was immediately used for the analysis.
Glucosinolates were analyzed by liquid chromatography–mass spectrometry modifying the methods described by Mellon et al. (2002). Ten microliters of sample was applied to the Acquity UPLC system (Waters) and separated on SunFire C18 column (150 × 2.1-mm diameter; Waters) under a linear gradient elution program with solvent A (0.1% trifluoroacetic acid in water) and solvent B (0.1% trifluoroacetic acid in methanol): 0 to 2% solvent B (8 min), 2 to 30% solvent B (12 min), 30 to 100% solvent B (18 min), 100% solvent B (7 min), and 100% solvent A (5 min). Elution was operated at 0.2 mL/min and 30°C. Metabolites were detected by photodiode array scanning of UV absorption (210 to 400 nm), and [M-H]− ions for specific glucosinolates were detected in a Q-Tof Premier time-of-flight mass analyzer (Micromass). The electrospray probe was operated at 2.8 kV. The source and desolvation temperatures were 100 and 120°C, respectively. The identities of 4-methylsulfinylbutyl and 4-methylthiobutyl glucosinolates with the standard compounds were confirmed by elution time, UV absorption, m/z value, and mass fragmentation patterns. Other methylsulfinylalkyl and methylthioalkyl gluosinolates, and indole glucosinolates were identified by their UV absorption, m/z values, and mass fragmentation patterns. The glucosinolate contents were calculated by comparing the levels of [M-H]− ions of each compound with that of sinigrin as a standard.
Cys and GSH contents were determined by monobromobimane (Molecular Probes) labeling of thiols after reduction of the extracts by DTT. The labeled products were separated by HPLC using a Symmetry C18 column (150 × 4.6-mm diameter; Waters) and detected with a Waters 474 scanning fluorescence detector, monitoring fluorescence of thiol-bimane adducts at 482 nm under excitation at 390 nm. Met and OAS were analyzed by capillary electrophoresis–mass spectrometry (Agilent Technologies) (Sato et al., 2004). Sulfate content was determined by a capillary electrophoresis-photodiode array detection system according to the manufacturer's protocol (Agilent Technologies).
Accession Numbers
Microarray data from this article can be found in the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo) under accession number GSE4455.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. List of Genes Regulated by Sulfur and SLIM1.
Supplemental Table 2. Existence of the Putative SLIM1/EIL3 Binding Site AYGWAYCT in the Upstream Regions.
Supplemental Table 3. AYGWAYCT in the Upstream Regions of –S-Upregulated SLIM1-Dependent Genes.
Supplemental Table 4. Molecular Markers Generated for Identification of slim1.
Supplemental Table 5. Primers for Construction of Plant Transformation Vectors.
Supplemental Table 6. Gene-Specific Primers for Quantitative Real-Time RT-PCR.
Supplemental Figure 1. SLIM1 mRNA Content Is Not Modulated by Sulfur.
Supplemental Figure 2. Tissue and Cellular Distribution of SLIM1.
Supplemental Figure 3. ACC Response of –S-Regulated SLIM1-Dependent Genes.
Supplemental Figure 4. Multiple Sequence Alignment of EIL Family Proteins.
Supplementary Material
Acknowledgments
We thank Akiko Watanabe-Takahashi, Eri Inoue, and Rie Niida for excellent technical support; Takeshi Nakano, Tetsuya Ishida, and Eiji Nambara for advices on mapping strategy; and Taku Demura and Minoru Kubo for providing pH35GS and pSMAH621 vectors. We thank Jonathan Gershenzon for providing standards of glucosinolates and Yasuo Niwa for providing sGFP(S65T) vector. This work was supported by the RIKEN President's Discretionary Fund and by Grants-in-aid for Scientific Research in Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hideki Takahashi (hideki@riken.jp).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso, J.M., Stapanova, A.N., Solano, R., Wisman, E., Ferrari, S., Ausubel, F.M., and Ecker, J.R. (2003). Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA 100 2992–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak, S., Tax, F.E., Feldmann, K.A., Galbraith, D.W., and Feyereisen, R. (2001). CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell 13 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false positive discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57 289–300. [Google Scholar]
- Buchner, P., Stuiver, C.E., Westerman, S., Wirtz, M., Hell, R., Hawkesford, M.J., and De Kok, L.J. (2004. a). Regulation of sulfate uptake and expression of sulfate transporter genes in Brassica oleracea as affected by atmospheric H2S and pedospheric sulfate nutrition. Plant Physiol. 136 3396–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner, P., Takahashi, H., and Hawkesford, M.J. (2004. b). Plant sulphate transporters: Co-ordination of uptake, intracellular and long-distance transport. J. Exp. Bot. 55 1765–1773. [DOI] [PubMed] [Google Scholar]
- Celenza, J.L., Quiel, J.A., Smolen, G.A., Merrikh, H., Silvestro, A.R., Normanly, J., and Bender, J. (2005). The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 137 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, Q., Rothenberg, M., Solano, R., Roman, G., Terzaghi, W., and Ecker, J.R. (1997). Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89 1133–1144. [DOI] [PubMed] [Google Scholar]
- Chen, S., Glawischnig, E., Jorgensen, K., Naur, P., Jorgensen, B., Olsen, C.E., Hansen, C.H., Rasmussen, H., Pickett, J.A., and Halkier, B.A. (2003). CYP79F1 and CYP79F2 have distinct functions in the biosynthesis of aliphatic glucosinolates in Arabidopsis. Plant J. 33 923–937. [DOI] [PubMed] [Google Scholar]
- Chiu, W.L., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6 325–330. [DOI] [PubMed] [Google Scholar]
- Clamp, M., Cuff, J., Searle, S.M., and Barton, G.J. (2004). The Jalview Java alignment editor. Bioinformatics 20 426–427. [DOI] [PubMed] [Google Scholar]
- Clarkson, D.T., Smith, F.W., and Vanden Berg, P.J. (1983). Regulation of sulfate transport in a tropical legume, Macroptilium atropurpureum cv. Siratro. J. Exp. Bot. 34 1463–1483. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Crawford, N.M., Kahn, M.L., Leustek, T., and Long, S.R. (2000). Nitrogen and sulfur. In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: American Society of Plant Biologists), pp. 824–849.
- Deane-Drummond, C.E. (1987). The regulation of sulfate uptake following growth of Pisum sativum L. seedlings in S nutrient limiting conditions. Interaction between nitrate and sulphate transport. Plant Sci. 50 27–35. [Google Scholar]
- Field, B., Cardon, G., Traka, M., Botterman, J., Vancanneyt, G., and Mithen, R. (2004). Glucosinolate and amino acid biosynthesis in Arabidopsis. Plant Physiol. 135 828–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb, C., and Abel, S. (2006). Glucosinolate metabolism and its control. Trends Plant Sci. 11 89–100. [DOI] [PubMed] [Google Scholar]
- Guo, H., and Ecker, J.R. (2003). Plant responses to ethylene gas are mediated by SCFEBF1/EBF2-dependent proteolysis of EIN3 transcription factor. Cell 115 667–677. [DOI] [PubMed] [Google Scholar]
- Guo, H., and Ecker, J.R. (2004). The ethylene signaling pathway: New insights. Curr. Opin. Plant Biol. 7 40–49. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Marcos, J.F., Roberts, M.A., Campbell, E.J., and Wray, J.L. (1996). Three members of a novel small gene-familiy from Arabidopsis thaliana able to complement functionally an Escherichia coli mutant defective in PAPS reductase activity encode proteins with a thioredoxin-like domain and “APS reductase” activity. Proc. Natl. Acad. Sci. USA 93 13377–13382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkier, B.A., and Gershenzon, J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57 303–333. [DOI] [PubMed] [Google Scholar]
- Hansen, C.H., Du, L., Naur, P., Olsen, C.E., Axelsen, K.B., Hick, A.J., Pickett, J.A., and Halkier, B.A. (2001. a). CYP83B1 is the oxime-metabolizing enzyme in the glucosinolate pathway in Arabidopsis. J. Biol. Chem. 276 24790–24796. [DOI] [PubMed] [Google Scholar]
- Hansen, C.H., Wittstock, U., Olsen, C.E., Hick, A.J., Pickett, J.A., and Halkier, B.A. (2001. b). Cytochrome P450 CYP79F1 from Arabidopsis catalyzes the conversion of dihomomethionine and trihomomethionine to the corresponding aldoximes in the biosynthesis of aliphatic glucosinolates. J. Biol. Chem. 276 11078–11085. [DOI] [PubMed] [Google Scholar]
- Hirai, M.Y., Fujiwara, T., Awazuhara, M., Kimura, T., Noji, M., and Saito, K. (2003). Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-L-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J. 33 651–663. [DOI] [PubMed] [Google Scholar]
- Hirai, M.Y., Fujiwara, T., Chino, M., and Naito, S. (1995). Effects of sulfate concentrations on the expression of a soybean seed storage protein gene and its reversibility in transgenic Arabidopsis thaliana. Plant Cell Physiol. 36 1331–1339. [PubMed] [Google Scholar]
- Hirai, M.Y., et al. (2005). Elucidation of gene-to-gene and metabolite-to-gene networks in Arabidopsis by integration of metabolomics and transcriptomics. J. Biol. Chem. 280 25590–25595. [DOI] [PubMed] [Google Scholar]
- Hirai, M.Y., Yano, M., Goodenowe, D.B., Kanaya, S., Kimura, T., Awazuhara, M., Arita, M., Fujiwara, T., and Saito, K. (2004). Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 101 10205–10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins, L., Parmar, S., Blaszczyk, A., Hesse, H., Hoefgen, R., and Hawkesford, M.J. (2005). O-acetylserine and the regulation of expression of genes encoding components for sulfate uptake and assimilation in potato. Plant Physiol. 138 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth, J.R., Fourcroy, P., Davidian, J.-C., Smith, F.W., and Hawkesford, M.J. (2003). Cloning of two contrasting high-affinity sulphate transporters from tomato induced by low sulphate and infection by the vascular pathogen Verticillium dahlia. Planta 218 58–64. [DOI] [PubMed] [Google Scholar]
- Hull, A.K., Vij, R., and Celenza, J.L. (2000). Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. USA 97 2379–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka, T., Hayashi, N., Yamaya, T., and Takahashi, H. (2004. a). Root-to-shoot transport of sulfate in Arabidopsis: Evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol. 136 4198–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka, T., Watanabe-Takahashi, A., Hayashi, N., Ohnishi, M., Mimura, T., Buchner, P., Hawkesford, M.J., Yamaya, T., and Takahashi, H. (2004. b). Vacuolar sulfate transporters are essential determinants controlling internal distribution of sulfate in Arabidopsis. Plant Cell 16 2693–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima, C.G., Berkowitz, O., Hell, R., Noji, M., and Saito, K. (2005). Characterization and expression analysis of a serine acetyltransferase gene family involved in a key step of the sulfur assimilation pathway in Arabidopsis. Plant Physiol. 137 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of T-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel types of Agrobacterium binary vector. Mol. Gen. Genet. 204 383–396. [Google Scholar]
- Kosugi, S., and Ohashi, Y. (2000). Cloning and DNA binding properties of a tobacco Ethylene-Insensitive3 (EIN3) homolog. Nucleic Acids Res. 28 960–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, M., Udagawa, M., Nishikubo, N., Horiguchi, G., Yamaguchi, M., Ito, J., Mimura, T., Fukuda, H., and Demura, T. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutz, A., Müller, A., Hennig, P., Kaiser, W.M., Piotrowski, M., and Weiler, E.W. (2002). A role for nitrilase 3 in the regulation of root morphology in sulphur-starving Arabidopsis thaliana. Plant J. 30 95–106. [DOI] [PubMed] [Google Scholar]
- Leustek, T., Martin, M.N., Bick, J.A., and Davies, J.P. (2000). Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51 141–165. [DOI] [PubMed] [Google Scholar]
- Levy, M., Wang, Q., Kaspi, R., Parrella, M.P., and Abel, S. (2005). Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J. 43 79–96. [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita, A., Inoue, E., Watanabe-Takahashi, A., Yamaya, T., and Takahashi, H. (2003). Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effect on sulfur nutrition on multiple metabolic pathways. Plant Physiol. 132 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita, A., Nakamura, Y., Watanabe-Takahashi, A., Inoue, E., Yamaya, T., and Takahashi, H. (2005). Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J. 42 305–314. [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita, A., Nakamura, Y., Watanabe-Takahashi, A., Yamaya, T., and Takahashi, H. (2004. a). Induction of SULTR1;1 sulfate transporter in Arabidopsis roots involves protein phosphorylation/dephosphorylation circuit for transcriptional regulation. Plant Cell Physiol. 45 340–345. [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita, A., Nakamura, Y., Yamaya, T., and Takahashi, H. (2004. b). A novel regulatory pathway of sulfate uptake in Arabidopsis roots: Implication of CRE1/WOL/AHK4-mediated cytokinin-dependent regulation. Plant J. 38 779–789. [DOI] [PubMed] [Google Scholar]
- Mellon, F.A., Bennett, R.N., Holst, B., and Williamson, G. (2002). Intact glucosinolate analysis in plant extracts by programmed cone voltage electrospray LC/MS: Performance and comparison with LC/MS/MS methods. Anal. Biochem. 306 83–91. [DOI] [PubMed] [Google Scholar]
- Mikkelsen, M.D., Hansen, C.H., Wittstock, U., and Halkier, B.A. (2000). Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J. Biol. Chem. 275 33712–33717. [DOI] [PubMed] [Google Scholar]
- Nikiforova, V., Freitag, J., Kempa, S., Adamik, M., Hesse, H., and Hoefgen, R. (2003). Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: Interlacing of biosynthetic pathways provides response specificity. Plant J. 33 633–650. [DOI] [PubMed] [Google Scholar]
- Page, R.D.M. (1996). TREEVIEW: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12 357–358. [DOI] [PubMed] [Google Scholar]
- Piotrowski, M., Schemenewitz, A., Lopukhina, A., Müller, A., Janowitz, T., Weiler, E.W., and Oecking, C. (2004). Desulfoglucosinolate sulfotransferases from Arabidopsis thaliana catalyze the final step in the biosynthesis of the glucosinolate core structure. J. Biol. Chem. 279 50717–50725. [DOI] [PubMed] [Google Scholar]
- Potuschak, T., Lechner, E., Parmentier, Y., Yanagisawa, S., Grava, S., Koncz, C., and Pascal, G. (2003). EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115 679–689. [DOI] [PubMed] [Google Scholar]
- Reintanz, B., Lehnen, M., Reichelt, M., Gershenzon, J., Kowalczyk, M., Sandberg, G., Godde, M., Uhl, R., and Palme, K. (2001). bus, a bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. Plant Cell 13 351–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, K. (2004). Sulfur assimilatory metabolism. The long and smelling road. Plant Physiol. 136 2443–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Sato, S., Soga, T., Nishioka, T., and Tomita, M. (2004). Simultaneous determination of the main metabolites in rice leaves using capillary electrophoresis mass spectrometry and capillary electrophoresis diode array detection. Plant J. 40 151–163. [DOI] [PubMed] [Google Scholar]
- Setya, A., Murillo, M., and Leustek, T. (1996). Sulfate reduction in higher plants: Molecular evidence for a novel 5′-adenylylsulfate reductase. Proc. Natl. Acad. Sci. USA 93 13383–13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibagaki, N., Rose, A., Mcdermott, J.P., Fujiwara, T., Hayashi, H., Yoneyama, T., and Davies, J.P. (2002). Selenate-resistant mutants of Arabidopsis thaliana identify SULTR1;2, a sulfate transporter required for efficient transport of sulfate into roots. Plant J. 29 475–486. [DOI] [PubMed] [Google Scholar]
- Skirycz, A., Reichelt, M., Burow, M., Birkemeyer, C., Rolcik, J., Kopka, J., Zanor, M.I., Gershenzon, J., Strnad, M., Szopa, J., Mueller-Roeber, B., and Witt, I. (2006). DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. 47 10–24. [DOI] [PubMed] [Google Scholar]
- Smith, F.W., Ealing, P.M., Hawkesford, M.J., and Clarkson, D.T. (1995). Plant members of a family of sulfate transporters reveal functional subtypes. Proc. Natl. Acad. Sci. USA 92 9373–9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, F.W., Hawkesford, M.J., Ealing, P.M., Clarkson, D.T., Vanden Berg, P.J., Belcher, A.R., and Warrilow, A.G.S. (1997). Regulation of expression of a cDNA from barley roots encoding a high affinity sulfate transporter. Plant J. 12 875–884. [DOI] [PubMed] [Google Scholar]
- Solano, R., Stepanova, A., Chao, Q., and Ecker, J.R. (1998). Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, H., Watanabe-Takahashi, A., Smith, F.W., Blake-Kalff, M., Hawkesford, M.J., and Saito, K. (2000). The role of three functional sulfate transporters involved in uptake and translocation of sulfate in Arabidopsis thaliana. Plant J. 23 171–182. [DOI] [PubMed] [Google Scholar]
- Takahashi, H., Yamazaki, M., Sasakura, N., Watanabe, A., Leustek, T., de Almeida Engler, J., Engler, G., Van Montagu, M., and Saito, K. (1997). Regulation of sulfur assimilation in higher plants: A sulfate transporter induced in sulfate starved roots plays a central role in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94 11102–11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talalay, P., and Fahey, J.W. (2001). Phytochemicals from cruciferous plants protect against cancer by modulating carcinogen metabolism. J. Nutr. 131 3027S–3033S. [DOI] [PubMed] [Google Scholar]
- Textor, S., Bartram, S., Kroymann, J., Falk, K.L., Hick, A., Pickett, J.A., and Gershenzon, J. (2004). Biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana: Recombinant expression and characterization of methylthioalkylmalate synthase, the condensing enzyme of the chain-elongation cycle. Planta 218 1026–1035. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vauclare, P., Kopriva, S., Fell, D., Suter, M., Sticher, L., von Ballmoos, P., Krähenbühl, U., den Camp, R.O., and Brunold, C. (2002). Flux control of sulphate assimilation in Arabidopsis thaliana: Adenosine 5′-phosphosulphate reductase is more susceptible than ATP sulphurylase to negative control by thiols. Plant J. 31 729–740. [DOI] [PubMed] [Google Scholar]
- Vidmar, J.J., Schjoerring, J.K., Touraine, B., and Glass, A.D.M. (1999). Regulation of the hvst1 gene encoding a high-affinity sulfate transporter from Hordeum vulgare. Plant Mol. Biol. 40 883–892. [DOI] [PubMed] [Google Scholar]
- Vidmar, J.J., Tagmount, A., Cathala, N., Touraine, B., and Davidian, J.-C.E. (2000). Cloning and characterization of a root specific high-affinity sulfate transporter from Arabidopsis thaliana. FEBS Lett. 475 65–69. [DOI] [PubMed] [Google Scholar]
- Yamasaki, K., et al. (2005). Solution structure of the major DNA-binding domain of Arabidopsis thaliana Ethylene-insensitive3-like3. J. Mol. Biol. 348 253–264. [DOI] [PubMed] [Google Scholar]
- Yanagisawa, S., Yoo, S.-D., and Sheen, J. (2003). Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425 521–525. [DOI] [PubMed] [Google Scholar]
- Yoshimoto, N., Inoue, E., Saito, K., Yamaya, T., and Takahashi, H. (2003). Phloem-localizing sulfate transporter, Sultr1;3, mediates re-distribution of sulfur from source to sink organs in Arabidopsis. Plant Physiol. 131 1511–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto, N., Takahashi, H., Smith, F.W., Yamaya, T., and Saito, K. (2002). Two distinct high-affinity sulfate transporters with different inducibilities mediate uptake of sulfate in Arabidopsis roots. Plant J. 29 465–473. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.