Abstract
Symbiotic associations involving intracellular microorganisms and animals are widespread, especially for species feeding on poor or unbalanced diets. Buchnera aphidicola, the obligate intracellular bacterium associated with most aphid species, provides its hosts with essential amino acids (EAAs), nutrients in short supply in the plant phloem sap. The Buchnera genome has undergone severe reductions during intracellular evolution. Genes for EAA biosynthesis are conserved, but most of the transcriptional regulatory elements are lost. This work addresses two main questions: is transcription in Buchnera (i) regulated and (ii) scaled to aphid EAA demand? Two microarray experiments were designed for profiling the gene expression in Buchnera. The first one was characterized by a specific depletion of tyrosine and phenylalanine in the aphid diet, and the second experiment combined a global diminution of EAAs in the aphid diet with a sucrose concentration increase to manipulate the aphid growth rate. Aphid biological performance and budget analysis (the balance between EAAs provided by the diet and those synthesized by Buchnera) were performed to quantify the nutritional demand from the aphids toward their symbiotic bacteria. Despite the absence of known regulatory elements, a significant transcriptional regulation was observed at different levels of organization in the Buchnera genome: between genes, within putative transcription units, and within specific metabolic pathways. However, unambiguous evidence for transcriptional changes underpinning the scaling of EAA biosynthesis to aphid demand was not obtained. The phenotypic relevance of the transcriptional response from the reduced genome of Buchnera is addressed.
Cooperative relationships are founded on reciprocity: each organism provides resources to its partner(s) that are more valuable to the partner(s) than to the donor. Furthermore, the participating organisms are predicted to scale the provisioning of resources to the demand of their partner(s), i.e., to increase provisioning when demand is high and reduce provisioning when demand is low. One system for which there is good evidence for scaling is the insect-microbial relationship between aphids and their intracellular γ-proteobacteria, Buchnera aphidicola (7). In this symbiosis, Buchnera provides the insect with essential amino acids (EAAs), the nine amino acids that contribute to protein but cannot be synthesized by most animals, enabling aphids to utilize their phloem sap diet, which is deficient in these nutrients (7, 8, 13, 18, 27). In particular, Febvay et al. (14) demonstrated very significant increases in the incorporation of carbon from dietary sucrose by Buchnera in all EAAs but methionine when the pea aphid, Acyrthosiphon pisum, was reared on diets with an amino acid composition that mimicked phloem sap relative to that on a diet optimized to aphid amino acid requirements.
Buchnera has undergone a severe genome reduction, attributable largely to genetic drift as a consequence of its lifestyle as an obligatory, vertically transmitted symbiont with a very small effective population size. As a result, Buchnera sp. APS lacks many metabolic capabilities, including the capacity to synthesize all non-EAAs apart from cysteine, glycine, and arginine. However, a functional set of enzymes for EAA biosynthesis has been selected for the maintenance of the symbiosis (28, 33). Amino acid biosynthesis in Escherichia coli, a free-living relative of Buchnera, is regulated by a complex network that includes transcriptional regulation, attenuation, and allosteric control by feedback inhibition (22). Although global regulators, sometimes described as “histone-like” proteins (6, 15, 17), are conserved in Buchnera, no identifiable specific transcriptional regulators, including attenuation systems, controlling amino acid biosynthesis are apparent in the Buchnera in pea aphids (21).
These genomic and transcriptional analyses raise two issues, both of which are general to bacteria with much-reduced genomes. First, is transcription by Buchnera regulated in the sense that mRNA abundance varies predictably across different genes? Second, is the level of mRNAs for genes in the EAA biosynthetic pathways scaled to aphid demand?
Relevant to these questions, recent studies by Moran and coworkers have revealed a small or no transcriptional response, apart from those predicted from the few gene-specific regulatory mechanisms present in Buchnera (the sigma factor σ32 and metR) in aphids exposed to high temperature (33, 35) and plants under different nutritional regimens (20). To explore these questions more precisely, we adopted an experimental approach specifically to identify very small transcriptional differences between treatments and to link transcriptional responses to precisely calculated differences in aphid demand for Buchnera-derived EAAs. Specifically, we developed an oligonucleotide microarray analysis dedicated to the Buchnera from A. pisum. The 35-mer probe set was designed to guarantee the gene specificity and covers the 617 genes of the bacterium predicted from its genome sequence (28). Our experimental design was shaped by two issues. First, the need to quantify aphid demand for Buchnera-derived EAAs was met by using aphids reared on chemically defined diets of precisely known composition. The second issue was the requirement that the aphid performance be equivalent on those diets imposing high and low aphid demand for Buchnera-derived EAAs; if aphid performances differed among the treatments, then it would be impossible to discriminate whether any observed transcriptional differences reflected a response to the aphid demands or to the differences in growth rates.
This study includes two experimental designs, building on our previous experience with the nutritional physiology of the symbiosis between Buchnera and pea aphids (8, 9, 13, 14). In experiment 1, two aromatic amino acids, the essential phenylalanine and the nonessential tyrosine, were omitted from an otherwise complete diet, imposing increased demand for Buchnera-derived phenylalanine (tyrosine is synthesized by the aphid from phenylalanine, and Buchnera lacks the genes for tyrosine synthesis). This dietary challenge is not representative of natural amino acid profiles in plant phloem sap, but it has the experimental value of explicit predictions: if Buchnera scaling to aphid demand is underlaid by transcriptional responses, it is predicted to include the upregulation of expression of genes carried in the phenylalanine (including chorismate) biosynthetic pathway. In experiment 2, the dietary amino acids had an EAA/non-EAA ratio of either 1:1 or 1:3, giving low and high aphid demand for Buchnera-derived EAAs, respectively. This comparison was set in a two-by-two factorial design with sucrose concentrations at 0.5 M and 1.0 M, supporting high and low aphid growth rates, so that the generality of the impact of dietary amino acid composition on Buchnera transcription patterns could be explored in two distinct nutritional regimens. The nutritional challenges posed by diets with an amino acid composition of 1:3 EAA/non-EAA and both sucrose concentrations were representative of the natural diet of plant phloem sap (34).
MATERIALS AND METHODS
Aphid performances and budget analysis.
A long-established parthenogenetic clone, LL01, of A. pisum Harris was maintained at 21°C with a 16-h light photoperiod on Vicia faba. The clone was shown to be free of any of the five taxa of secondary endosymbionts identified to date. In experiment 1, the aphids were reared from birth to day 7 on the AP3 (A. pisum diet 3) diet described by Febvay et al. (14), either with the full complement of amino acids or lacking phenylalanine and tyrosine (the YF0 diet). In experiment 2, aphids were reared from day 2 to day 7 on the diet described by Prosser and Douglas (24), with sucrose at 0.5 M or 1.0 M and amino acids at 0.15 M and with the EAA/non-EAA ratio at 1:1 or 1:3 in a two-by-two factorial design. Aphids were weighed individually on the day they reached adulthood (experiment 1) or day 7 (experiment 2). The quantities of EAAs provided by Buchnera to the aphids were estimated from the protein growth rates of the aphids on all diets by a procedure modified from Douglas et al. (8). In each experiment, the initial and final weights of each of 40 or 20 replicate aphids were scored and converted to the protein content, using conversion factors of 60 and 53 μg protein mg−1 fresh weight for 0- to 2-day-old and 7-day-old aphids, respectively. Protein growth was determined by subtraction, and the increase in each protein EAA was determined from its percentage of contribution to aphid protein, as determined experimentally (8). The Buchnera-derived EAAs were calculated as the difference between the increase in protein amino acid and the amount ingested, as determined from the volume of diet ingested. Ingestion was estimated from the method described by Febvay et al. (14) for experiment 1 and quantified by the radiolabeled inulin method described by Karley et al. (16), with 14 replicate aphids for each of the four diets for experiment 2.
Microarray experiments.
The details for the microarray protocols are described by Calevro et al. (2). The Buchnera microarray is composed of 6,144 spots, including two to three probes per Buchnera gene, designed using ROSO software (25). The 35-mer oligonucleotide probes were printed in duplicate and by two different pins onto Quantifoil MicroTool aldehyde slides (Interchim, France). Positive and negative controls were distributed throughout the slide.
For experiment 1, two aliquots of three independent RNA preparations for each condition were labeled with Cy3- or Cy5-dUTP, and a dye swap design including six slides was made (see Table S1 in the supplemental material). For experiment 2, a loop design was built including two independent repeats of eight slides (see Table S2 in the supplemental material).
An equal amount of Cy3- and Cy5-dUTP-labeled target, based on the incorporated dye concentration, was applied to each slide. Manual hybridization for the measurement of tRNA expression (3) took place under a glass coverslip in a humidified slide chamber (Proteigene, Saint Marcel, France) at 50°C for 16 h. We also ran automatic hybridizations in an automated Ventana Discovery station (Ventana Medical Systems, Illkirch, France) at 45°C for 8 h. Following hybridization, the slides were washed at room temperature with solutions of various stringencies (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 5 min; 0.2× SSC, 2 min; 0.05× SSC, dip in and out 10 times), dried by centrifugation, and then scanned for fluorescence by using a GeneTAC LSIV scanner (Genomic Solutions, Huntingdon, United Kingdom). Spot pixel medians were recorded with GenePix 4.0 image analysis software (Axon Instruments, Foster City, CA). For each slide, a quality analysis was performed, and a flag was determined for each spot by visual inspection and dedicated Genepix filters.
Data analysis.
Normalization and statistical analysis of microarray data were performed with Bioconductor libraries of the R software (http://www.r-project.org). Normalization was accomplished with an invariant set of genes that was determined a posteriori following the nonparametric method of Tseng et al. (31). A global Loess normalization was applied to the two data sets. After normalization, red and green fluorescence intensities of repeated spots were averaged separately within the technical repeat groups and within the probe groups for each gene. Individual spots (or probe groups) with bad quality flags were not included in subsequent calculations. Gene expression levels are given using the M and A values as defined in reference 11, where the absolute gene expression is defined as A = log2 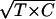 (T is the treatment fluorescence signal and C is the control fluorescence signal of an individual slide) and the relative gene expression is defined as M = log2 (T/C).
(T is the treatment fluorescence signal and C is the control fluorescence signal of an individual slide) and the relative gene expression is defined as M = log2 (T/C).
Two statistical tests were performed, the modified t test (32) and analysis of variance (ANOVA) (36). For experiment 2, P values were calculated by permutation and corrected with the James-Stein shrinkage estimates of the error variance (4). For both experiments, genes were selected as differentially expressed (see Tables S3 to S5 in the supplemental material) if they fulfilled the following three criteria: (i) a P value of <0.05 in one test; (ii) an increase of >1.20-fold for upregulated genes or <0.83-fold for downregulated genes; and (iii) good quality analysis (no flag over the slide set). To complete the analysis, we also tested the significance of differential expression within groups of genes involved in the same pathway with the nonparametric rank test of Wilcoxon (corresponding P values are denoted Pw in the text).
The numbers of false positives (false discovery rate [FDR]) were estimated for the two experiments based on a mixture of controls and simulated data sets. The whole statistical procedure and plot of FDR-versus-differential expression ratio and P value thresholds are presented in the supplemental material (Fig. S1). The FDR was 25% for experiment 1 and 5% for experiment 2, with a P value of <0.05 and a threshold ratio beyond 0.8 to 1.2. We did not seek to reduce the FDR in experiment 1 by increasing the change in threshold (n-fold) or by reducing the critical probability as this would have drastically increased the false-negative rate.
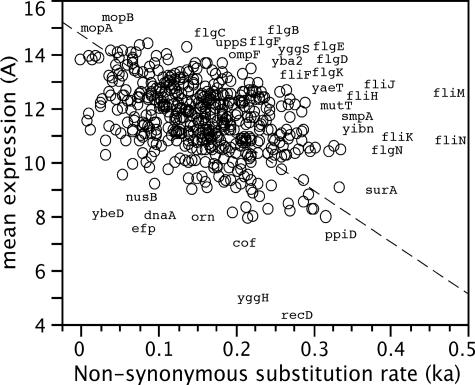
FIG. 1.
Plot of nonsynonymous substitution rates (Ka) versus expression levels (A) for Buchnera genes. Ka values are frequencies of nonsynonymous substitutions per site accumulated by Buchnera in the lineages of the aphids Acyrthosiphon pisum and Schizaphis graminum since their divergence 50 to 70 million years ago (29). Mean expression is the constant term of the ANOVA model of experiment 1. The dotted line is the orthogonal regression adjustment. Outlier gene names (outside of the 95% density ellipse) are recorded on the figure.
Quantitative PCR. (i) Buchnera density estimation.
Real-time PCR (TaqMan) reactions were set up in 96-well reaction plates. Cycling and data collection were performed using an ABI Prism 7900HT sequence detection system (Applied Biosystems, Warrington, United Kingdom). Primers and probe data, designed for a 120-bp region of the dnaK gene of Buchnera sp. APS, are provided in Table S6 in the supplemental material.
(ii) Microarray validation.
Primers were designed to amplify short regions of selected Buchnera genes (see Table S7 in the supplemental material). In addition to the target genes, we included three control genes (rpsF, rplO, and atpA). Quantitative real-time reverse transcription-PCR (RT-PCR) was carried out with a Roche LightCycler and reagents of the LightCycler FastStart DNA Master SYBR green I kit by Roche. Amplification conditions were as follows: 95°C for 10 min and then 45 cycles of 95°C for 10 s, 47 or 51°C for 4 s, and 72°C for 8 s. An internal standard curve was generated for each of the reactions by using serial dilutions of purified PCR products amplified starting from Buchnera genomic DNA. The same RNA extraction was used for control and test genes, and the template copy number was estimated from the standard curve for each gene.
Molecular evolution and functional analysis.
Putative transcription units (pTUs) in Buchnera have been identified manually by comparison with the E. coli genome. Each orthologue of an E. coli gene in Buchnera was searched in the EcoCyc database (http://ecocyc.org/), and groups of Buchnera genes corresponding to transcription units in E. coli were collected. After the removal of singletons, 81 pTUs in Buchnera were identified, including 291 different genes, with each pTU consisting of 2 to 13 genes.
Functional analysis was performed using fatiGO (http://fatigo.bioinfo.cnio.es/), a web tool for gene list projection of gene ontology (GO), and Blastsets (http://cbi.labri.fr/outils/BlastSets), a web tool for comparing groups of genes within different classifications.
Microarray data accession numbers.
Microarray data were deposited in the Array Express database under accession numbers E-TABM-83 and E-TABM-85.
RESULTS
Significant gene transcription regulation in Buchnera.
Our first approach was to explore the global pattern of gene expression across the Buchnera genome. As with other bacteria and microorganisms (10, 26), Buchnera displayed a negative correlation between the absolute level of expression of protein-coding genes and the nonsynonymous substitution rate (Ka) (29) (Pearson's correlation r = −0.3, P = 10−5): highly conserved genes are more highly expressed than rapidly evolving genes (Fig. 1). Thirty-three of the 540 protein-coding genes scored were outliers (outside of the 95% density ellipse on Fig. 1), and of the 24 genes with expression levels higher than those predicted from Ka (putative positively selected genes), 13 were fli or flg genes, with a role in the flagellar apparatus. The Buchnera genome lacks the fliC gene coding for the flagellar filament required for motility (28). We hypothesize that the Buchnera incomplete flagellum may function as a transporter, for example, of polypeptides. This hypothesis is supported by the work of Majander et al. (19) showing that ΔfliC mutants of E. coli (conserving only untranslated regulatory domains of the fliC gene) were able to export several reporter polypeptides through the flagellum channel. Other transport and surface genes (ompF, yaeT, and smpA) were also outliers in Fig. 1.
We used pTUs of Buchnera, identified by comparison with those of E. coli, as a framework to explore the variations in absolute and relative gene expression levels of Buchnera genes (A and M values, respectively; see Materials and Methods for definitions). For both A and M values, the variations within pTUs were significantly smaller than those between pTUs, indicating that genes belonging to the same pTU tend to be expressed at similar absolute rates and to be coregulated following nutritional changes in the aphid diet. Representative results from experiment 2 are presented in Table 1, and similar results were obtained for all the other diet comparisons (data not shown). Overall, the partitioning of genes into pTUs accounts for 40 to 63% of the total variation in absolute expression values and 23 to 36% of the variation in relative expression values, providing evidence that the expression of Buchnera genes is regulated at an operon-like level, even though most known regulatory proteins other than “global regulators” are lacking in Buchnera. Small RNAs yet to be discovered may also be responsible for this observed regulation.
TABLE 1.
Variability analysis of absolute and relative gene expression levels within and between Buchnera pTUs for experiment 2 with 0.5 M sucrose
| Expression level | Source | df | MSEa | R2 | P value |
|---|---|---|---|---|---|
| Absolute | Between pTUs | 80 | 5.95 × 109 | 0.54 | 10−11 |
| Within pTUs | 210 | 1.04 × 109 | |||
| Relative | Between pTUs | 80 | 0.356 | 0.35 | 0.03 |
| Within pTUs | 210 | 0.253 |
MSE, mean square error.
Manipulation of host demand for Buchnera-derived nutrients.
The comparison in experiment 1 was between aphids on the complete diet and those on the diet lacking amino acids phenylalanine and tyrosine (YF0 diet). The aphids displayed high growth rates, showing full symbiotic compensation for aromatic shortage on these diets (see Table S6 in the supplemental material), with the phenylalanine and tyrosine in the protein growth YF0 diet of necessity derived from Buchnera. The mean protein growth per aphid on the YF0 diet was 68.2 μg, including an estimated 23.9 nmol phenylalanine and 5.7 nmol tyrosine that must have been derived from Buchnera as 29.6 nmol phenylalanine.
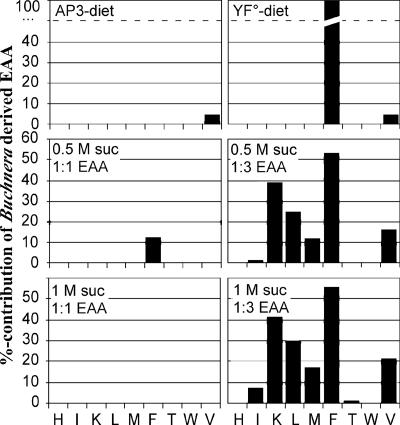
The growth rates of the aphids in experiment 2 did not differ significantly with dietary amino acid composition but were reduced significantly (due to decreasing feeding rate) on diets with 1 M sucrose relative to those on diets with 0.5 M sucrose (see Table S6 in the supplemental material). Buchnera is not implicated in aphid osmoregulation, which occurs mainly in the digestive tract (9). The contribution of Buchnera-derived EAAs to the host's growth requirement for each amino acid was obtained from the difference between protein growth and the amount of each amino acid obtained from the diet, as determined from the dietary concentration and aphid feeding rate. These calculations revealed that the aphids on the diets with 1:1 EAA/non-EAA derived a sufficiency of all EAAs except for phenylalanine on the 0.5 M sucrose diet, for which there was a dietary shortfall of 13% made up by Buchnera function. On the diets with 1:3 EAA/non-EAA, the aphids required a contribution from Buchnera of all amino acids except histidine and tryptophan, and threonine for aphids on the 0.5 M sucrose diet. The contribution of Buchnera-derived amino acids to the total aphid requirement varied from just 1% for threonine on the 1.0 M sucrose diet to more than 50% for phenylalanine on both dietary sucrose concentrations (Fig. 2).
FIG. 2.
Budget analysis for the six diets of the two experiments. The y axis represents the percentage of the contribution of Buchnera-derived EAAs compared to the total aphid amino acid requirement for protein growth (amino acids are presented on the x axes according to their one-letter codes).
Functional analysis of differentially expressed genes. (i) Experiment 1.
The expression of 52 (8.5%) of a total of 617 Buchnera genes was significantly altered when aphids were fed on the YF0 diet relative to the complete diet. Thirty-one genes were upregulated and 21 genes downregulated, with eight and nine genes showing changes of ≥2-fold and ≤0.5-fold, respectively (see Table S3 in the supplemental material). Quantitative RT-PCR was performed to evaluate the accuracy of the microarray differential gene list because a high rate of false positives was suspected in this experiment (see Fig. S1 in supplemental material). The changes (n-fold) did not differ significantly between the microarray results and those of the quantitative RT-PCR for 10 of the 12 genes tested (see Table S7 in the supplemental material), giving broad confidence in the differentially expressed list.
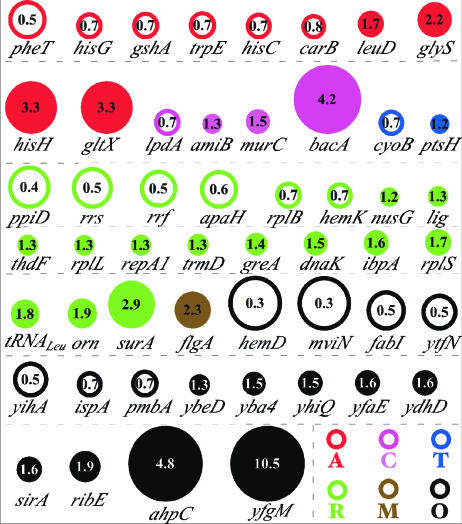
Figure 3 summarizes the differentially expressed genes grouped by functional category. Few genes were found within the amino acid metabolism class, and a general trend in the transcriptional response is difficult to draw based on the individual gene analysis. These results are consistent with published analysis of the transcriptional response of Buchnera to variations in plant nutrition (20).
FIG. 3.
Differentially expressed genes of experiment 1. Open circles are downregulated genes, and plain circles are upregulated genes. Circle diameters are proportional to the change (n-fold) of the gene (except yfgM). Changes (n-fold) to one decimal place are also reported within circles. Gene colors correspond to their GO classes. A, amino acids and derivative metabolism, GO0006519; C, carbohydrate metabolism, GO0005975; T, transport, GO0006810; R, RNA, nucleic acid, and protein metabolism, GO0006139, GO0016070, GO0019219, GO0019538; M, cellular morphogenesis, cell motility, GO0000902, GO0006928; O, others.
(ii) Experiment 2.
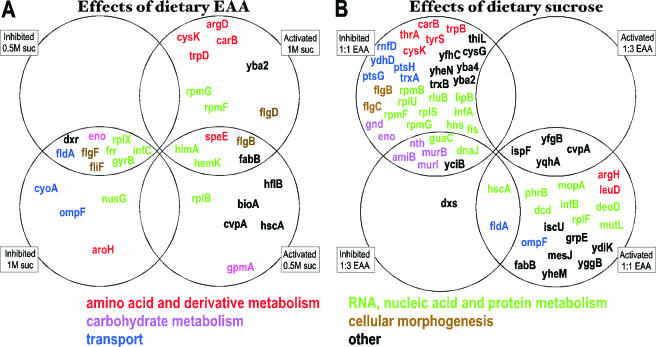
An FDR of 5% was estimated for this experiment (see Fig. S1 in supplemental material). From the Buchnera gene expression levels corresponding to the four diet compositions of experiment 2, it was possible to calculate four differential expression ratios: amino acid compositions at 0.5 M (high growth rate) and 1 M (low growth rate) sucrose and sucrose concentrations at 1:1 and 1:3 non-EAA/EAA. Changes in the differential gene expression ratios varied between 0.4-fold and 1.8-fold (see Table S4 and Table S5 in the supplemental material). Results are summarized in Fig. 4. Buchnera in aphids on diets with low EAA content displayed a significant reduction in the expression of 13 genes and an increased expression of 19 genes relative to those on the 1:1 diet. All the individual significant genes of the amino acid and derivative metabolism class were found to be downregulated at 1 M sucrose with the 1:3 diet. Despite the fact that the aphid demands for EAAs are globally similar at low and high growth rates (Fig. 2), only 14 of 32 genes are common for both conditions, including none associated with the metabolism of amino acids and their derivatives. Furthermore, aphids in both experiments 1 and 2 share a high phenylalanine demand, but only four genes are differentially expressed in both experiments, and none of them either contribute to the EAA metabolism or vary concordantly between the two experiments. Elevated dietary sucrose (which reduced aphid growth rate) was characterized by a stronger response of Buchnera transcriptome, with 18 genes upregulated and 38 genes downregulated (Fig. 4B). Here again the response of Buchnera is not strongly directed toward EAA metabolism, and a general trend is difficult to draw from this response, which includes genes from transport, general metabolism, stress, and hypothetical functional classes.
FIG. 4.
Differentially expressed genes of experiment 2. (A) Effects of dietary amino acid concentrations in the aphid diets. (B) Effects of dietary sucrose concentrations in the aphid diets. Inhibited 0.5 M suc, Buchnera genes downregulated in aphids on 1:3 diet related to 1:1 (EAA/non-EAA) diet on the 0.5 M sucrose diet; Activated 1:1EAA, Buchnera genes upregulated in aphids on 1 M sucrose related to the 0.5 M sucrose diet at 1:1 EAA/non-EAA. Complete names of GO classes are given in Fig. 3.
No specific transcriptional regulators controlling the amino acid metabolism pathways are apparent in the Buchnera genome sequence. However, three transcriptional regulators encoding major “histone-like” proteins were upregulated in our experiments (Fig. 4): IHF (himA upregulated by low EAA concentration in experiment 2; P = 0.02) and FIS and H-NS (both downregulated by high sucrose concentration in experiment 2; P = 0.06 and 0.05, respectively). himD, which encodes the partner of HimA in the IHF factor, was not significantly induced.
(iii) Global transcriptional regulation within metabolic pathways.
Analysis of tRNA threshold changes (n-fold) in experiment 1 revealed upregulation for 26 of the 32 tRNAs of Buchnera, although only one was individually detected as differentially expressed (tRNALeucine). The global Wilcoxon rank test is highly significant (Pw = 10−5), supporting the observation that such a ratio distribution is not random. The same observation was not possible for experiment 2, for which the hybridization was automated (see Materials and Methods). A similar response has also been observed in Bacillus subtilis and E. coli subjected to amino acid-depleted culture medium (5, 12). Five other genes related to the tRNA metabolism were upregulated, glnS, gltX, glyS, tdhF, and trmD, but pheT was significantly downregulated under the YF0 diet conditions.
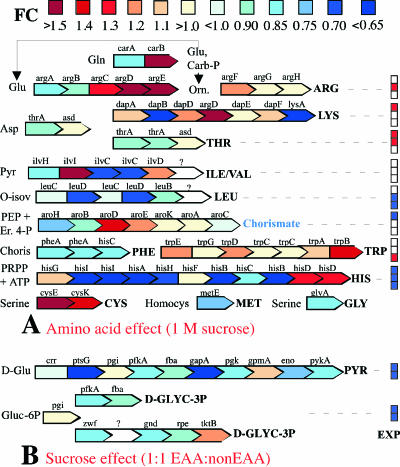
EAA depletion in the aphid diet induced significant global changes in the expression of genes of all amino acid biosynthesis pathways (Fig. 5A), other than isoleucine and valine biosynthesis. The terminal branch of tryptophan biosynthesis was significantly upregulated in experiment 2 with 1 M sucrose (Pw = 0.008), as well as the arginine pathway with 0.5 M sucrose (Pw = 0.012), the lysine pathway in experiment 1 (Pw = 0.01) and the threonine pathway in experiment 1 (Pw = 0.03), and in experiment 2 with 0.5 M sucrose (Pw = 0.03). The histidine biosynthesis pathway was downregulated under the three conditions tested (experiment 1, Pw = 0.02; experiment 2 with 0.5 M sucrose, Pw = 0.01; and experiment 2 with 1 M sucrose, Pw = 0.03). The seven genes present on the leucine plasmid were downregulated in experiment 1 (Pw = 0.01) and with 1 M sucrose in experiment 2 (Pw = 0.01), although no gene was individually significant. The latter downregulation was present but not significant with 0.5 M sucrose.
FIG. 5.
Average effects of treatments on transcript levels of amino acid (A) and sucrose (B) metabolism genes of Buchnera, based on the results of experiment 2. Colored squares (experiments [EXP]) correspond to significant tests (Wilcoxon signed-rank test) for global pathway regulation (red, upregulation; blue, downregulation) under (A) the three experimental conditions of EAA composition (experiment 1, experiment 2 at 0.5 M and 1 M sucrose from the top downward) and (B) the two EAA conditions (1:1 EAA and 1:3 EAA from the top downward). FC, change (n-fold). (Adapted from reference 20 with permission.)
The reduced growth rate of the aphids on diets with 1 M sucrose was correlated with significant downregulation of the expression of genes in the glycolysis and pentose phosphate pathways for both amino acid conditions (Fig. 5B). Genes encoding sugar transport systems in Buchnera sp. PTS were downregulated accordingly. Genes amiB, murB, and murI, involved in the peptidoglycan biosynthesis, were also significantly downregulated, and four additional genes of this pathway (murC, murG, murF, and murE from BU215 to BU223) followed the same trend, albeit not significantly.
DISCUSSION
The first issue addressed in this study was the global pattern of Buchnera transcription. We demonstrated here that gene transcription is regulated in Buchnera and that this regulation is correlated to the genome organization in pTUs. Functionally important flagellar genes were found to be highly expressed, and mutations of these genes are suspected to be positively selected, perhaps as an evolving transport system in Buchnera. Low differential expression values were observed for Buchnera, but these changes (n-fold) are comparable to those observed for most genes in E. coli or in B. subtilis subjected to moderate nutritional stresses (1, 30). Moreover, a direct comparison between free-living bacteria and endosymbionts is questionable for various reasons: (i) dietary experimental conditions could be buffered by aphid homeostasis; (ii) measures do not result from a homogeneous time process because they are the outcome from the whole aphid developmental period; and (iii) the response is averaged from different Buchnera populations (embryo and maternal bacteriocytes) with putative differential metabolisms. Moreover, the transcription system of Buchnera has evolved in the stable intracellular environment and has probably been attenuated, producing less adaptive fluctuations in gene expression. The striking result of this work is the regulation of entire pathways indicative of the coregulation of pTUs in Buchnera. Topoisomerase (gyrB), acting on DNA supercoiling, as well as “histone-like” global regulators (6, 15, 17) was significantly regulated and that might explain global regulations in the absence of specific regulators. However, their role in the control of transcription in Buchnera requires further work, including research of the binding sites and spatial patterns of transcriptional activity in the chromosome of Buchnera (work presently in progress).
The second question explored in this study related to the scaling, at the transcription level, of EAA production by Buchnera in response to aphid demand for Buchnera-derived EAAs. Analysis of single significant differential genes in our two experiments does not support a functional and adaptive transcriptional response of Buchnera to the EAA demand of the aphids. Global analysis of the expression data set, however, revealed elevated expression of tRNAs in experiment 1, as well as small successive coordinate changes (n-fold) along specific pathways in EAA and sugar metabolism (Fig. 5). Small changes (n-fold) along an entire metabolic pathway might have a cumulative effect, resulting in significant changes in the corresponding metabolic flux. This is illustrated by the correlation between the low aphid growth rates on the high sucrose diets and the significant down-regulation of genes in central carbohydrate metabolism, including glycolysis and pentose phosphate pathways, as well as the PTS strain's sugar transport system in Buchnera (Fig. 4B and 5B). In other words, some genes in carbohydrate metabolism may be regulated according to Buchnera growth rate. This may be mediated by csrA, which regulates expression of glycolysis genes in E. coli and is present in Buchnera (23), although the expression of csrA did not vary significantly in our experiments.
A similar link between transcriptional responses and aphid demand for EAAs was not consistently evident. Some operons have genes that are both up- and downregulated under the same experimental condition. Even so, the activation of the lysine biosynthesis is consistent with the demand for this amino acid, as well as for cysteine (for methionine biosynthesis) and arginine (one of the major protein amino acids). The activation of cysteine biosynthesis might be explained by the lack of the MetR regulator in Buchnera sp. APS, which is responsible for the activation of metE in E. coli, or Buchnera sp. SG (20). However, despite the fact that the demand is high for phenylalanine and branched amino acids (isoleucine, valine, and leucine), expression of pheA was not induced, and repression of the terminal leucine pathway was unexpected. The very high expression level of the ilvHI operon (two of the most highly expressed genes in Buchnera) might explain the constant and high production of branched-chain amino acids by Buchnera.
These considerations raise the possibilities that the transcriptional response of Buchnera to variations in aphid demand for EAAs may not have a strong influence of EAA biosynthetic rates and that the scaling of EAA biosynthesis to aphid demand is underpinned by translational or posttranslational regulation or potentially by the flux of precursors for EAA synthesis from the host cell. Further research and modeling of the coupling of host and bacterial metabolisms will be needed to explore this complex system of symbiotic interactions.
Acknowledgments
This work was supported by the BQR INSA-UCBL-ECL (2001), the action bioinformatique/NSA 2003-2006, the Emergence program (2003) from the Région Rhône-Alpes, the programme fédérateur INRA de Biologie Intégrative AgroBI 2006, and the short-term EMBO fellowship ASTF 278-2003.
We thank A. Groppi and A. Barre for the development of specific Buchnera tools for the Blastsets software, G. Duport and A. Vallier for the material preparation and qPCR experiments, J. Bernillon for the microarray manufacturing at the DTAMB Rhône-Alpes Genopole platform, and C. M. Allen and C. L. M. J. François for technical support.
Footnotes
Published ahead of print on 13 October 2006.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Berka, R. M., X. Cui, and C. Yanofsky. 2003. Genomewide transcriptional changes associated with genetic alterations and nutritional supplementation affecting tryptophan metabolism in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 100:5682-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calevro, F., H. Charles, N. Reymond, V. Dugas, J. P. Cloarec, J. Bernillon, Y. Rahbe, G. Febvay, and J. M. Fayard. 2004. Assessment of 35mer amino-modified oligonucleotide based microarray with bacterial samples. J. Microbiol. Methods 57:207-218. [DOI] [PubMed] [Google Scholar]
- 3.Charles, H., F. Calevro, J. Vinuelas, J. M. Fayard, and Y. Rahbe. 2006. Codon usage bias and tRNA over-expression in Buchnera aphidicola after aromatic amino acid nutritional stress on its host Acyrthosiphon pisum. Nucleic Acids Res. 34:4583-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui, X., J. T. Hwang, J. Qiu, N. J. Blades, and G. A. Churchill. 2005. Improved statistical tests for differential gene expression by shrinking variance components estimates. Biostatistics 6:59-75. [DOI] [PubMed] [Google Scholar]
- 5.Dittmar, K., E. Mobley, A. Radek, and T. Pan. 2004. Exploring the regulation of tRNA distribution on the genomic slide. J. Mol. Biol. 337:31-47. [DOI] [PubMed] [Google Scholar]
- 6.Dorman, C. J., and P. Deighan. 2003. Regulation of gene expression by histone-like proteins in bacteria. Curr. Opin. Genet. Dev. 13:179-184. [DOI] [PubMed] [Google Scholar]
- 7.Douglas, A. E. 2003. Buchnera bacteria and other symbionts of aphids, p. 23-38. In K. Bourtzis and T. A. Miller (ed.), Insect symbiosis. CRC Press Inc., Boca Raton, Fla.
- 8.Douglas, A. E., L. B. Minto, and T. L. Wilkinson. 2001. Quantifying nutrient production by the microbial symbionts in an aphid. J. Exp. Biol. 204:349-358. [DOI] [PubMed] [Google Scholar]
- 9.Douglas, A. E., D. R. Price, L. B. Minto, E. Jones, K. V. Pescod, C. L. Francois, J. Pritchard, and N. Boonham. 2006. Sweet problems: insect traits defining the limits to dietary sugar utilisation by the pea aphid, Acyrthosiphon pisum. J. Exp. Biol. 209:1395-1403. [DOI] [PubMed] [Google Scholar]
- 10.Drummond, D. A., J. D. Bloom, C. Adami, C. O. Wilke, and F. H. Arnold. 2005. Why highly expressed proteins evolve slowly. Proc. Natl. Acad. Sci. USA 102:14338-14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dudoit, S., Y. H. Yang, M. J. Callow, and T. Speed. 2002. Statistical methods for identifying genes with differential expression in replicated cDNA microarray experiments. Statistica Sinica 12:111-139. [Google Scholar]
- 12.Emilsson, V., A. K. Naslund, and C. G. Kurland. 1993. Growth-rate-dependent accumulation of twelve tRNA species in Escherichia coli. J. Mol. Biol. 230:483-491. [DOI] [PubMed] [Google Scholar]
- 13.Febvay, G., I. Liadouze, J. Guillaud, and G. Bonnot. 1995. Analysis of energetic amino acid metabolism in Acyrthosiphon pisum: a multidimensional approach to amino acid metabolism in aphids. Arch. Insect Biochem. Physiol. 29:45-69. [Google Scholar]
- 14.Febvay, G., Y. Rahbe, M. Rynkiewicz, J. Guillaud, and G. Bonnot. 1999. Fate of dietary sucrose and neosynthesis of amino acids in the pea aphid, Acyrthosiphon pisum, reared on different diets. J. Exp. Biol. 202:2639-2652. [DOI] [PubMed] [Google Scholar]
- 15.Hommais, F., E. Krin, C. Laurent-Winter, O. Soutourina, A. Malpertuy, J. P. Le Caer, A. Danchin, and P. Bertin. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 40:20-36. [DOI] [PubMed] [Google Scholar]
- 16.Karley, A. J., A. E. Douglas, and W. E. Parker. 2002. Amino acid composition and nutritional quality of potato leaf phloem sap for aphids. J. Exp. Biol. 205:3009-3018. [DOI] [PubMed] [Google Scholar]
- 17.Kelly, A., M. D. Goldberg, R. K. Carroll, V. Danino, J. C. Hinton, and C. J. Dorman. 2004. A global role for Fis in the transcriptional control of metabolism and type III secretion in Salmonella enterica serovar Typhimurium. Microbiology 150:2037-2053. [DOI] [PubMed] [Google Scholar]
- 18.Liadouze, I., G. Febvay, J. Guillaud, and G. Bonnot. 1996. Metabolic fate of energetic amino acids in the aposymbiotic pea aphid Acyrthosiphon pisum (Harris) (Homoptera: Aphididae). Symbiosis 21:115-127. [Google Scholar]
- 19.Majander, K., L. Anton, J. Antikainen, H. Lang, M. Brummer, T. K. Korhonen, and B. Westerlund-Wikstrom. 2005. Extracellular secretion of polypeptides using a modified Escherichia coli flagellar secretion apparatus. Nat. Biotechnol. 23:475-481. [DOI] [PubMed] [Google Scholar]
- 20.Moran, N. A., H. E. Dunbar, and J. L. Wilcox. 2005. Regulation of transcription in a reduced bacterial genome: nutrient-provisioning genes of the obligate symbiont Buchnera aphidicola. J. Bacteriol. 187:4229-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panina, E. M., A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand. 2001. Regulation of aromatic amino acid biosynthesis in gamma-proteobacteria. J. Mol. Microbiol. Biotechnol. 3:529-543. [PubMed] [Google Scholar]
- 22.Panina, E. M., A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand. 2003. Regulation of biosynthesis and transport of aromatic amino acids in low-GC Gram-positive bacteria. FEMS Microbiol. Lett. 222:211-220. [DOI] [PubMed] [Google Scholar]
- 23.Prickett, M. D., M. Page, A. E. Douglas, and G. H. Thomas. 2006. BuchneraBASE: a post-genomic resource for Buchnera sp. APS. Bioinformatics 22:641-642. [DOI] [PubMed] [Google Scholar]
- 24.Prosser, W. A., and A. E. Douglas. 1992. A test of the hypotheses that nitrogen is upgraded and recycled in an aphid (Acyrthosiphon pisum) symbiosis. J. Insect Physiol. 38:93-99. [Google Scholar]
- 25.Reymond, N., H. Charles, L. Duret, F. Calevro, G. Beslon, and J. M. Fayard. 2004. ROSO: optimizing oligonucleotide probes for microarrays. Bioinformatics 20:271-273. [DOI] [PubMed] [Google Scholar]
- 26.Rocha, E., and A. Danchin. 2003. Essentiality, not expressiveness, drives gene-strand bias in bacteria. Nat. Genet. 34:377-378. [DOI] [PubMed] [Google Scholar]
- 27.Sandstrom, J. P., and N. A. Moran. 2001. Amino acid budgets in three aphid species using the same host plant. Physiol. Entomol. 26:202-211. [Google Scholar]
- 28.Shigenobu, S., H. Watanabe, M. Hattori, Y. Sasaki, and H. Ishikawa. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81-86. [DOI] [PubMed] [Google Scholar]
- 29.Tamas, I., L. Klasson, B. Canback, A. K. Naslund, A. S. Eriksson, J. J. Wernegreen, J. P. Sandstrom, N. A. Moran, and S. G. E. Andersson. 2002. 50 million years of genomic stasis in endosymbiotic bacteria. Science 296:2376-2379. [DOI] [PubMed] [Google Scholar]
- 30.Tao, H., C. Bausch, C. Richmond, F. R. Blattner, and T. Conway. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181:6425-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tseng, G. C., M. K. Oh, L. Rohlin, J. C. Liao, and W. H. Wong. 2001. Issues in cDNA microarray analysis: quality filtering, channel normalization, models of variations and assessment of gene effects. Nucleic Acids Res. 29:2549-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilcox, J. L., H. E. Dunbar, R. D. Wolfinger, and N. A. Moran. 2003. Consequences of reductive evolution for gene expression in an obligate endosymbiont. Mol. Microbiol. 48:1491-1500. [DOI] [PubMed] [Google Scholar]
- 34.Wilkinson, T. L., and A. E. Douglas. 2003. Phloem amino acids and the host plant range of the polyphagous aphid, Aphis fabae. Entomol. Exp. Appl. 106:103-113. [Google Scholar]
- 35.Wilson, A. C., H. E. Dunbar, G. K. Davis, W. B. Hunter, D. L. Stern, and N. A. Moran. 2006. A dual-genome microarray for the pea aphid, Acyrthosiphon pisum, and its obligate bacterial symbiont, Buchnera aphidicola. BMC Genomics 7:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu, S., M. K. Kerr, X. Cui, and G. A. Churchill. 2003. MAANOVA: a software package for the analysis of spotted cDNA microarray experiments, p. 455. In G. Parmigiani, E. Garrett, R. Irizarry, and S. Zeger (ed.), The analysis of gene expression data: methods and software. Springer, New York, N.Y.