Abstract
The genome of Aspergillus fumigatus has four genes that encode mitogen-activated protein kinases (MAPKs), sakA/hogA, mpkA, mpkB, and mpkC. The functions of the MpkB and MpkC MAPKs are unknown for A. fumigatus and the closely related and genetically amenable species Aspergillus nidulans. mpkC deletion mutants of A. fumigatus were made and their phenotypes characterized. The mpkC deletion mutants were viable and had normal conidial germination and hyphal growth on minimal or complete media. This is in contrast to deletion mutants with deletions in the closely related MAPK gene sakA/hogA that we previously reported had a nitrogen source-dependent germination phenotype. Similarly, the growth of the mpkC deletion mutants was wild type on high-osmolarity medium. Consistent with these two MAP kinase genes regulating different cellular responses, we determined that the mpkC deletion mutants were unable to grow on minimal medium with sorbitol or mannitol as the sole carbon source. This result implicates MpkC signaling in carbon source utilization. Changes in mRNA levels for sakA and mpkC were measured in response to hypertonic stress, oxidative stress, and a shift from glucose to sorbitol to determine if there was overlap in the SakA and MpkC signaling pathways. These studies demonstrated that SakA- and MpkC-dependent patterns of change in mRNA levels are distinct and have minimal overlap in response to these environmental stresses.
Aspergillus fumigatus is a filamentous fungus that is ubiquitous in the environment. It, like many other saprophytic fungi, contributes to the turnover of nitrogen and carbon in the environment through degradation of biological polymers such as cellulose and lignin. A. fumigatus is also the cause of significant morbidity and mortality in patients with compromised immune systems (18). This fungus causes the life-threatening disease invasive pulmonary aspergillosis, a major clinical concern for patients with leukemia and lymphoma and bone marrow transplant patients. Despite the clinical importance of this fungus, it has not been the subject of intensive biological investigation. If novel therapies for invasive pulmonary aspergillosis are to be developed, a much better understanding of the basic biological processes of A. fumigatus is required.
Mitogen-activated protein kinases (MAPKs) play a central role in regulating fungal cell physiology in response to nutritional status and environmental stresses, including hypertonic shock, heat shock, and oxidative stress (11, 19, 26, 28). MAPKs also function in sexual and asexual development and in fungal cell morphogenesis (19, 26). Thus, defects in MAPK signaling pathways could decrease the virulence of the fungus in a host. A number of studies of plant and animal fungal pathogens have shown that mutants defective in MAPK signaling have reduced virulence (2, 5, 7, 10, 13, 16, 19, 26, 27).
The genome of A. fumigatus has four genes that code for MAPKs, sakA/hogA, mpkA, mpkB, and mpkC (23). sakA encodes the stress-activated kinase that responds to hypertonic stress, reactive oxygen species, and heat shock (28). This highly conserved MAPK's functions are also well conserved among fungi (1, 11, 12, 14, 19, 26, 28). mpkA is the ortholog of the SLT2 MAPK gene in Saccharomyces cerevisiae that is responsible for monitoring cell wall integrity. Deletion of mpkA in Aspergillus nidulans produces a strain with delayed conidial germination and swelling at the hyphal tips. These phenotypes are largely remedied by growth on high-osmolarity complex media (3). Orthologs of this MAPK in plant-pathogenic fungi have been demonstrated to play an important role in pathogenesis (26). Similarly, the Candida albicans ortholog MKC1 has been shown to play a role in virulence (6, 21, 22). mpkB codes for a MAPK that is most similar to the MAPKs encoded by the S. cerevisiae genes FUS3 and KSS1. The Fus3p and Kss1p MAPKs function in the pheromone mating response pathway and the filamentation/invasive growth pathways, respectively. Homologues of these MAPK systems in other fungi have been shown to be important for pathogenesis in plant and animal models (7, 19, 26). Finally, mpkC has no clearly orthologous gene in S. cerevisiae, but the protein is most similar in sequence to Hog1p and related stress-activated protein kinases.
These four MAPK genes also define the complete set for A. nidulans and Aspergillus oryzae, suggesting that they are all that is necessary to regulate filamentous-fungal-cell physiology (9, 20, 23). Nevertheless, the functions of the mpkA, mpkB, and mpkC MAPK genes in A. fumigatus remain unknown. Because of their central role in regulating fungal physiology and their role in virulence in other fungi, they should be investigated in A. fumigatus. Thus, it is important that we determine how each of the MAPKs regulates fungal cell physiology in the human pathogen A. fumigatus, as it will provide insight into the development of potentially novel antifungal drugs.
In this paper, we report the construction and phenotypic characterization of mpkC deletion mutants. We determined that mpkC is required for growth on media where sorbitol or mannitol is the sole carbon source. These results define a new and novel MAPK signaling pathway that regulates conidial germination in response to the carbon source in the medium.
MATERIALS AND METHODS
Aspergillus fumigatus growth media, strains, and transformation.
A. fumigatus strain Af293 was grown on minimal medium (MM) that was composed of 7 mM KCl, 2 mM MgSO4, 20 mM ammonium tartrate, 12 mM KPO4 (pH 6.8), 1% (wt/vol) glucose, vitamin mix (1 μg per ml each of p-aminobenzoic acid, choline HCl, niacin, pyridoxine HCl, riboflavin, and thiamine HCl and 2 ng/ml d-biotin), and trace elements (added to 650 ml of water: 100 ml 0.25 M EDTA, pH 8.0, 1 g FeSO4 7H2O, 8.8 g ZnSO4 7H2O, 0.4 g CuSO4 5H2O, 0.15 g MnSO4 H2O, 0.1 g Na2B4O7 10H2O, and 0.1 g NaMoO4 2H2O; each chemical was dissolved completely in order and brought to a final volume of 1 liter; final volume of medium, 1 ml per liter) or complete medium (CM) that consisted of MM plus 0.1% (wt/vol) yeast extract, 0.2% (wt/vol) peptone, and 0.1% (wt/vol) tryptone (Difco Laboratories, Inc., Sparks, MD). To test growth on different carbon sources, glucose was omitted and the alternative carbon source was added to a final concentration of 100 mM. The alternative carbon sources that we tested were ethanol, fructose, galactose, glycerol, lactose, maltose, mannitol, raffinose, and sorbitol. To test growth on different nitrogen sources, ammonium tartrate was omitted and the alternative nitrogen source sodium nitrate, sodium nitrite, proline, or phenylalanine was substituted at a 20 mM final concentration. The solid media for the plates incorporated 1.5% (wt/vol) agar (Difco Laboratories, Inc., Sparks, MD). Transformation of the pyrG1 mutant strain Af293.1 was performed as previously described (25). The pyrG1 mutation was determined to be a G-to-T transversion of the 5′ donor splice site in intron 1 of the gene by sequencing the PCR-amplified gene from Af293.1 using oligonucleotides AfpyrG-1 and AfpyrG-2. The names and sequences of the oligonucleotides used in these investigations are given in Table 1. The sakA/hogA deletion strain used in these studies was previously described (28). The Aspergillus nidulans strain R153 (wA3 pyroA4) was used to prepare RNA for reverse transcription-PCR (RT-PCR) as described below.
TABLE 1.
Oligonucleotides used in the preparation of plasmids
| Oligonucleotide | Sequence |
|---|---|
| mpkC-5′-1 | GGGGACAACTTTGTATAGAAAAGTTGCGACCGAAGGATTTGATGGA |
| mpkC-5′-2 | GGGGACTGCTTTTTTGTACAAACTTGGTGGATAGGGCCGAGCGGTC |
| mpkC-3′-1 | GGGGACAGCTTTCTTGTACAAAGTGGGTGTGGCCATCTTCATAGAT |
| mpkC-3′-2 | GGGGACAACTTTGTATAATAAAGTTGCACACCACCTCGAAGCTGCG |
| AfpyrG-1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTGCCTCAAACAATGCTCTTCAC |
| AfpyrG-2 | GGGGACAACTTTGTATAATAAAGTTGATTCTGTCTGAGAGGAGGCAC |
| Actin Fwd | GGTGGTACCACCATGTACCCC |
| Actin Rev | GCACTTGCGGTGAACGATCGAAG |
| sakA Fwd | GGACTTGTCTGCTCTGCAAGAG |
| sakA Rev | ACGACGCCTGCTGAGTGAACG |
| mpkC Fwd | CATTGACCTTCTCAAGAAGATGCTGGTTAT |
| mpkC Rev | AGCGTCTTGAGATGCGCAACCGAGCAGCGT |
| AnmpkC1 | TATGGCCGAGTTCATTCGCTCTGA |
| AnmpkC2 | CTGGCTCTGGCTCTGGCTCTG |
| AnmpkC3 | CTCAACCTTACGAAAAGGAATTGG |
| AnmpkC4 | CACGCTCAAAACAGAATCGAAACTCAT |
Construction of an mpkC deletion mutant.
The plasmid used to delete mpkC was made using the Gateway Multisite plasmid system (Invitrogen, Carlsbad, CA). Briefly, ∼1 kb of the 5′-end-flanking sequence beginning immediately upstream of the predicted start codon of mpkC was PCR amplified from Af293 genomic DNA using oligonucleotides mpkC-5′-1 and mpkC-5′-2 (Table 1) and recombined into pDONRP4-P1R using a BP recombination reaction as described by the manufacturer. Similarly, approximately 1 kb of the 3′-end-flanking sequence beginning immediately after the predicted termination codon of mpkC was PCR amplified from Af293 genomic DNA using oligonucleotides mpkC-3′-1 and mpkC-3′-2 and recombined into pDONRP2R-P3 using a BP recombination reaction. The A. fumigatus pyrG gene was PCR amplified from Af293 genomic DNA with oligonucleotides AfpyrG-1 and AfpyrG-2 and recombined into pDONR221 in a BP recombination reaction. These three plasmids were then recombined into the pDESTR4-R3 vector using an LR recombination reaction as described by the manufacturer to generate a plasmid that consisted of the pyrG gene flanked by the mpkC 5′- and 3′-end-flanking sequences. The mpkC deletion plasmid was transformed into protoplasts of Af293.1, and pyrG+ transformants were selected for as previously described (25). DNA was prepared from transformant strains (28), and deletion mutants were identified by Southern blot analysis.
RNA preparation.
Total RNA was prepared from freeze-dried mycelia using TRIzol reagent with modifications (Invitrogen Corp., Carlsbad, CA). Briefly, dried mycelia were ground to a fine powder using a 1-ml micropipette tip in a microcentrifuge tube and acid-washed glass beads (particle size, 425 to 600 μm) in a volume equal to that of the mycelia. An approximately 50-μl volume of the powder was rapidly vortexed in 1 ml of TRIzol reagent for 1 min. This mixture was allowed to incubate at room temperature for 5 min, 200 μl chloroform was added, the mixture was vortexed, and the phases were separated by centrifugation for 15 min at 14,000 × g at 4°C in a microcentrifuge. The TRIzol reagent phase was recovered, the chloroform extraction was repeated, and the RNA was precipitated from the final TRIzol phase by the addition of 600 μl isopropanol. Poly(A)+ RNA was prepared from total RNA using a polyA-Quick kit (QIAGEN, Valencia, CA).
cDNA library construction and screening.
mpkC cDNA clones were isolated from a library constructed with the UniZAP vector by using poly(A)+ mRNA, using methods described by the manufacturer (Stratagene, La Jolla, CA). A primary library of approximately 1.15 × 106 PFU was plated on 23 plates (150-mm diameter), and positive plaques were identified by hybridization, with mpkC genomic DNA as the probe. Positive plaques were plated again on 150-mm plates, and single positive plaques were purified. Positive lambda clones were excised to plasmid form as described by the manufacturer (Stratagene, La Jolla, CA). The A. nidulans mpkC cDNA was prepared using a SuperScript one-step RT-PCR kit as described by the manufacturer (Invitrogen, Carlsbad, CA), using the oligonucleotide pairs AnmpkC1 and AnmpkC2, AnmpkC1 and AnmpkC3, and AnpmkC1 and Anmpkc4 (Table 1). The final products were TA cloned in pCR2.1 by using the conditions recommended by the supplier (Invitrogen, Carlsbad, CA). The cloned PCR products were sequenced on both strands. This sequence was used to deduce the complete amino acid sequence of A. nidulans MpkC (AnMpkC) for comparison to MpkC from A. fumigatus. The DNA sequence assembly and analysis were done using DNASTAR software (DNASTAR Inc., Madison, WI). The same software package was used to determine MpkC's mass and isoelectric point.
Culture manipulations for gene expression studies.
For exposure to hypertonic conditions or hydrogen peroxide, mycelia were grown in CM at 37°C for 10 to 12 h with shaking at 250 rpm from 5 × 106 conidia per ml in 300 ml in a 1-liter flask. For the hypertonic-stress assay, a sample (50 ml) was removed (0 min), an equal volume of CM plus 2 M KCl at 37°C was added to the culture, and incubation continued at 37°C with shaking at 250 rpm. Samples (100 ml) were removed at 10, 20, 30, and 60 min of hypertonic stress. For the oxidative-stress assay, a sample (50 ml) was removed (0 min), hydrogen peroxide was added to the culture to a final concentration of 17 mM, and incubation continued at 37°C with shaking at 250 rpm. The conditions for hypertonic and oxidative stress were similar to those previously described (12, 14, 28). Samples (50 ml) were removed at 10, 20, 30, and 60 min of exposure to hydrogen peroxide. For the glucose-to-sorbitol shift experiments, mycelia were grown for 16 h at 37°C in 400 ml of MM from 5 × 106 conidia per ml. The 0-min sample (100 ml) was harvested; the remainder of the culture was collected by filtration on Miracloth (Calbiochem, La Jolla, CA), rinsed with MM without a carbon source, and returned to the rinsed flask containing 300 ml of MM with 100 mM sorbitol. Samples were then harvested at 15, 30, and 60 min of growth at 37°C with shaking at 250 rpm. All samples of mycelia were rapidly harvested by filtration on Miracloth, rinsed with water, rapidly frozen in liquid nitrogen, and freeze-dried. Total RNA was prepared as described above.
Northern blot analysis was conducted as previously described (28). First-strand cDNA was primed for synthesis with oligo(dT)12-18 using SuperScript II reverse transcriptase under conditions described by the manufacturer (Invitrogen, Carlsbad, CA). This cDNA was used to prime quantitative RT-PCR (qRT-PCR) using Power SYBR green reagents on an Applied Biosystems 7900HT fast real-time PCR system using conditions specified by the manufacturer (Applied Biosystems, Foster City, CA). Oligonucleotide primer pairs for actin, sakA, and mpkC mRNA analysis are given in Table 1, and the expected product sizes were 230, 300, and 350 base pairs, respectively. The data were then analyzed using the comparative cycle threshold method as described by the manufacturer, and results are presented as change relative to the value at time zero. Each qRT-PCR was done in triplicate, and the mean value was plotted ± the standard deviation.
Conidial germination on different carbon sources.
Freshly prepared wild-type or mpkC deletion mutant conidia (4 × 107) were placed into 100-mm petri dishes containing 20 ml of MM made with 100 mM glucose or 100 mM sorbitol as the sole carbon source. The plates had five sterile 18- by 18-mm cover glasses submerged in the medium, on which the conidia settled during germination. Plates were incubated without agitation at 37°C, and at 6 and 8 h, a cover glass was removed. The cover glass was inverted on a drop of 4% formaldehyde in phosphate-buffered saline (PBS) for 5 min and then rinsed in PBS and mounted on a slide in PBS-50% (vol/vol) glycerol. The absence or presence of a germ tube was then scored for at least 200 cells and the percent germination scored at each time point.
Nucleotide sequence accession numbers.
The A. fumigatus mpkC cDNA sequence was deposited at the NCBI and assigned accession number DQ402475. The A. nidulans mpkC cDNA sequence was deposited at the NCBI and assigned accession number DQ402476.
RESULTS
Analysis of the mpkC gene and the predicted polypeptide.
The structure of the mpkC gene and the properties of the predicted MpkC polypeptide were determined by the isolation and sequencing of mpkC cDNA clones. A total of eight positive cDNA clones were sequenced at their 5′ ends, and all contained sequences from mpkC. In addition, six of the eight clones contained the entire MpkC open reading frame (Fig. 1). Interestingly, the longest cDNA extended 241 base pairs 5′ of the initiation codon and the six longest clones all included an upstream micro-open reading frame 21 amino acids in length (Fig. 1). Two of the cDNA clones were completely sequenced (accession number DQ402475). The cDNA and genomic DNA sequences were used to derive the mpkC gene structure and MpkC protein sequence. The mpkC coding region was interrupted by eight introns ranging in size from 51 to 91 base pairs in length (Fig. 1). The cDNAs that we isolated had variable 5′ ends, suggesting the presence of multiple sites for the initiation of transcription. We also identified two different classes of transcripts that differed at their 3′ polyadenylation sites (Fig. 1). These multiple start and stop sites for transcription may explain why the mpkC mRNA was not readily detectable by Northern analysis (see below).
FIG. 1.
Sequence and structure of the Aspergillus fumigatus MAPK gene mpkC. Protein-encoding DNA sequences are in uppercase and noncoding sequences in lowercase. The deduced amino acid sequence is below the DNA sequence in single-letter code. The DNA sequence coding for the upstream micro-open reading frame in the 5′ leader region of cDNA clones is in bold. The 5′-end nucleotides of cDNAs are in bold and double underlined, and the sites of 3′ polyadenylation are bold and underlined.
The MpkC protein sequence elucidated from the cDNA has a predicted molecular mass of approximately 43,000 Da, with an isoelectric point of 5.5. The MpkC sequence is most closely related to other members of the stress-activated kinase family of MAP kinases. These include the SakA proteins of A. fumigatus and of A. nidulans, to which it is most similar, and Hog1p of S. cerevisiae. During our comparative analysis, we noted that the predicted AnMpkC protein sequence in the sequence database was very divergent at the carboxyl terminus from the one that we had determined for A. fumigatus MpkC. Further analysis of the genomic sequence suggested that the predicted AnMpkC sequence may be incorrect because an alternative model of the gene's structure would increase the amino acid similarity at the carboxyl terminus. We therefore RT-PCR amplified and cloned AnmpkC cDNAs and confirmed the alternative gene structure and predicted protein sequence (NCBI accession number DQ402476).
mpkC deletion mutant germinates poorly on and is unable to utilize polyalcohol sugars as sole carbon sources.
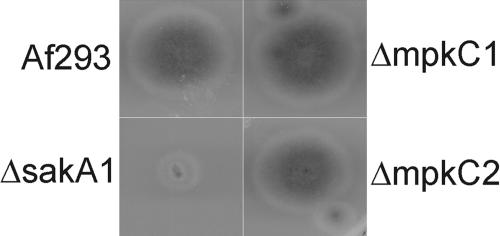
A deletion mutant that replaced the mpkC coding region with the nutritionally selective gene pyrG was made. Two independent mpkC deletion mutants were isolated, and all phenotypic studies were conducted with both strains. In all these studies, the ΔmpkC1 and ΔmpkC2 mutants behaved identically. Our initial phenotypic studies, based on the assumption that SakA and MpkC signaling pathways may overlap, focused on growth conditions that we previously determined for the sakA deletion mutants and produced an identifiable phenotype. We tested for defects in growth at high osmolarity (1.4 M KCl) (Fig. 2), survival at high temperatures, and exposure to hydrogen peroxide. None of the conditions tested resulted in an observable phenotype for the mpkC deletion mutants. These studies demonstrate that SakA and MpkC MAPK signaling pathways are differentially utilized and have little or no overlap in nutritional sensing.
FIG. 2.
An mpkC deletion mutant is viable and is not growth inhibited on high-osmolarity medium. Growth of Af293, a sakA deletion mutant (ΔsakA1), and two mpkC deletion mutants (ΔmpkC1 and ΔmpkC2) on CM containing 1.4 M KCl, showing that the loss of MpkC function does not confer reduced growth on high-osmolarity medium.
Failing to identify growth conditions under which the mpkC deletion mutant displays a phenotype based on our studies of sakA deletion mutants, we investigated the growth of the wild type, a sakA deletion mutant, and mpkC deletion mutants on a range of media in which we replaced glucose with different carbon sources and ammonium tartrate with different nitrogen sources (described in Materials and Methods). These growth studies determined that the mpkC deletion mutants were unable to grow on MM with sorbitol or mannitol as the sole carbon source (Fig. 3).
FIG. 3.
mpkC deletion mutants are unable to grow on MM with either sorbitol or mannitol as the sole carbon source. The plates were inoculated with conidia by using a toothpick and incubated for 48 h at 37°C.
We measured conidial germination on MM-sorbitol compared to that on MM-glucose media for the mpkC deletion mutant and the wild type (Af293). We found that the deletion mutant germinated poorly in MM-sorbitol compared to Af293. The mpkC deletion mutant showed only 2.5% germination on sorbitol compared to 86% germination for Af293. In contrast, both strains germinated well on MM-glucose, with each strain exhibiting about 60% germination on glucose at 8 h (Table 2). After 16 h in MM-sorbitol, the mpkC deletion mutant still had large numbers of conidia that did not germinate. We also demonstrated that mpkC is required for growth on sorbitol, by first germinating conidia overnight in MM-glucose and then transferring an agar plug of mycelia to a plate of MM-sorbitol, where the mycelium was unable to grow. These experiments demonstrate that MpkC function is required for efficient conidial germination on sorbitol as well as for hyphal growth.
TABLE 2.
The ΔmpkC mutant is defective for conidial germination on MM-sorbitol
| Fungus | Growth (h) | Germination on glucose (%) | Germination on sorbitol (%) |
|---|---|---|---|
| Af293 | 6 | 10 | 0 |
| ΔmpkC mutant | 6 | 7 | 0 |
| Af293 | 8 | 66 | 86 |
| ΔmpkC mutant | 8 | 56 | 3 |
mpkC mRNA increases in abundance in response to the carbon source in the medium and oxidative stress but not following osmotic stress.
Since the mpkC deletion mutants were unable to grow on MM with sorbitol as the sole carbon source, we reasoned that a growth shift from glucose to sorbitol would activate the MpkC signaling pathway and might result in increased mpkC mRNA levels. We tested this by probing Northern blots of total RNA prepared at intervals following a shift from MM-glucose to MM-sorbitol from the wild type (Af293) and a mpkC deletion mutant (ΔmpkC1). We found that sakA mRNA levels appeared to transiently increase in response to the glucose-to-sorbitol shift, that actin mRNA levels remained unchanged, and that mpkC mRNA was virtually undetectable, producing a very weak signal in the wild-type 60-min sample and none at all, as would be expected, in the deletion mutant strain (Fig. 4a). We think that the low-level expression of mpkC combined with the multiple transcription start and stop sites identified from our analysis of cDNA sequences make detection by Northern blot analysis difficult. It has also been reported that AnmpkC mRNA was not detectable in A. nidulans (8). We therefore used RT-PCR and qRT-PCR to detect and quantify the mpkC mRNA and compared that amount to the levels of sakA and actin mRNA (Fig. 4b). We readily identified an mpkC RT-PCR product in total RNA from the wild type but not in RNA from the mpkC deletion strain. In addition, we easily detected RT-PCR products for sakA and actin in the same RNA samples. By qRT-PCR, we readily detected an mpkC-specific product, which increased in abundance in RNA prepared after the shift to sorbitol-containing medium. Thus, we used this qRT-PCR assay to assess changes in mpkC mRNA levels in response to changes in culture conditions.
FIG. 4.
Changes in mRNA levels for mpkC, sakA, and actin in the wild type (Af293) and the mpkC deletion mutant (ΔmpkC1) following a shift from glucose to sorbitol. (a) Northern blots were probed for mpkC, sakA, and actin mRNAs. The probe used for hybridization is indicated to the right of each row. (b) qRT-PCR was used to measure changes in the levels of sakA and mpkC relative to that of actin. Plotted on the graph are the average changes (n-fold) determined for triplicate samples, with the standard deviations indicated by error bars.
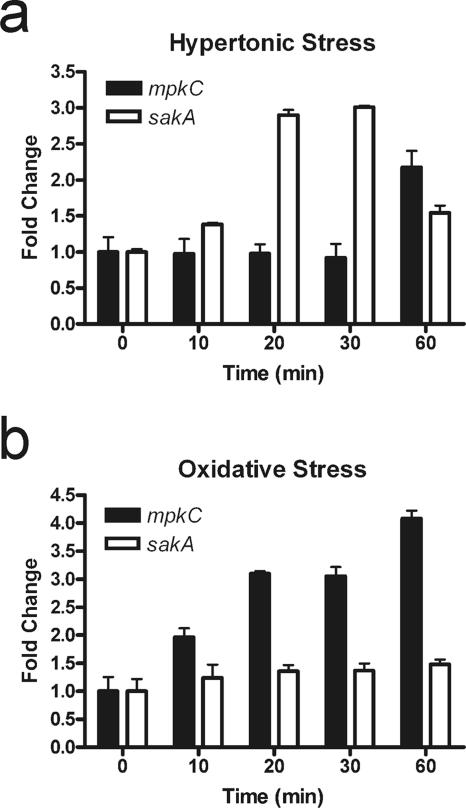
To further investigate whether mpkC may share functions with sakA, we examined changes in sakA and mpkC mRNA levels by qRT-PCR following hypertonic shock (1 M KCl) and oxidative stress (H2O2). We previously showed that sakA mRNA levels are increased following hypertonic stress (28). Since protein sequence analysis suggests that mpkC encodes a MAPK that is related to the SakA MAPK, it was possible that mpkC mRNA levels would increase following hypertonic or oxidative stress. Furthermore, the AnMpkC is reportedly activated by the same MAPK kinase, PbsB, in A. nidulans (8). Using qRT-PCR, we found that mpkC mRNA levels were slightly reduced following a shift to growth medium with 1 M KCl (Fig. 5). Interestingly, mpkC mRNA levels displayed a rapid and sustained increase following exposure to H2O2. In contrast, sakA exhibited only a modest increase in response to oxidative stress. Thus, it appears that while MpkC and SakA are similar in sequence, they do not respond transcriptionally in the same way to various environmental stresses.
FIG. 5.
qRT-PCR was used to measure mpkC and sakA mRNA levels relative to the actin mRNA level in response to (a) hypertonic stress (1 M KCl) and (b) oxidative stress (17 mM H2O2). Plotted on the graphs are the average changes (n-fold) determined for triplicate samples, with the standard deviations indicated by error bars.
DISCUSSION
We have deleted the mpkC gene of A. fumigatus and determined that it is not essential for fungal cell growth in culture. We did determine that MpkC is required for growth on the polyalcohol sugars sorbitol and mannitol, suggesting a role for MpkC in carbon source sensing. We have thus identified a novel MAPK signaling pathway in A. fumigatus that responds to specific carbon sources. It will be interesting to see if the mpkC deletion mutant of A. nidulans also fails to grow on these carbon sources (8). We also found that mpkC produces a number of different classes of transcripts and is expressed at very low levels. While the complexity of transcripts in A. nidulans has not been reported for mpkC in this fungus, it has been shown that it is expressed at very low levels (8). We found that the mpkC mRNA level increases in the wild type in response to a shift from glucose to sorbitol as the sole carbon source, consistent with a role in sensing alternative carbon sources. Interestingly, we found that while the predicted MpkC protein is similar in sequence to SakA, the mRNA levels of mpkC do not increase in response to hypertonic stress but do in response to oxidative stress. Overall, these results suggest that SakA and MpkC signaling minimally overlap, based on our limited study.
Recent studies of the genetically tractable fungus Neurospora crassa and the plant-pathogenic fungus Colletotrichum lagenarium have shown that the stress-activated MAPK signaling pathway is the target of the phenylpyrrole fungicide fludioxonil (17, 29). sakA/hogA MAPK mutants are resistant to fludioxonil, suggesting that the chemical acts through constitutive activation of the sakA/hogA pathway in fungi. We have also shown that the sakA/hogA deletion mutant of A. fumigatus that we made is resistant to fludioxonil (15) but that the mpkC deletion mutants are not (unpublished data). Thus, it appears that this MAPK pathway is universally important to fungi and is a target for antifungal compounds. These results suggest that other MAPK signaling pathways would be useful targets for new antifungal drugs.
MAPK signaling pathways in fungi stimulate the transcription of specific sets of genes that alter fungal cell physiology in response to environmental change (4, 11, 19, 24). MAPK pathways in yeast are the best studied but are poor models for the full range of MAPK functions in filamentous fungi. This is because filamentous fungi engage in lifestyles that are very different from that of yeast. Filamentous fungi include plant pathogens that must respond to plant host defenses in order to be successful pathogens (19, 26). Therefore, it is likely that filamentous fungal MAPK signaling pathways have evolved to respond to these cues. Similarly, other filamentous fungi are saprophytes that survive by acquisition of carbon and nitrogen through the process of decay. These organisms are also able to utilize a wide range of carbon sources that are often very complex, for example, lignin and cellulose, and to penetrate solid substrates. Thus, understanding MAPK signaling pathways in filamentous fungi will illustrate how different species use similar MAPKs to regulate different and novel cellular functions. The results from our study of MpkC in A. fumigatus illustrate this by demonstrating that this MAPK is required for germination and growth on sorbitol or mannitol as a sole carbon source. This extends our previous studies of SakA, in which we identified a new role for this MAPK in regulating conidial germination in response to environmental nitrogen (28). Finally, these experiments demonstrate a role for MAPK function in the differential use of carbon sources in this fungus. To our knowledge, this is the first case of a fungal MAPK having such a function.
Acknowledgments
This research was supported by grants AI051144, AI052236, and N01-AI-30041 (G.S.M.) from the National Institutes of Health and by M. D. Anderson Cancer Center support grant CA16672 for the DNA sequencing core facility.
We also acknowledge that preliminary sequence data were obtained from The Institute for Genomic Research website at http://www.tigr.org.
Footnotes
Published ahead of print on 22 September 2006.
REFERENCES
- 1.Alonso-Monge, R., F. Navarro-García, E. Román, A. I. Negredo, B. Eisman, C. Nombela, and J. Pla. 2003. The Hog1 mitogen-activated protein kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans. Eukaryot. Cell 2:351-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brachmann, A., J. Schirawski, P. Muller, and R. Kahmann. 2003. An unusual MAP kinase is required for efficient penetration of the plant surface by Ustilago maydis. EMBO J. 22:2199-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bussink, H. J., and S. A. Osmani. 1999. A mitogen-activated protein kinase (MPKA) is involved in polarized growth in the filamentous fungus, Aspergillus nidulans. FEMS Microbiol. Lett. 173:117-125. [DOI] [PubMed] [Google Scholar]
- 4.Chen, D., W. M. Toone, J. Mata, R. Lyne, G. Burns, K. Kivinen, A. Brazma, N. Jones, and J. Bahler. 2003. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14:214-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dhillon, N. K., S. Sharma, and G. K. Khuller. 2003. Signaling through protein kinases and transcriptional regulators in Candida albicans. Crit. Rev. Microbiol. 29:259-275. [DOI] [PubMed] [Google Scholar]
- 6.Diez-Orejas, R., G. Molero, F. Navarro-García, J. Pla, C. Nombela, and M. Sánchez-Perez. 1997. Reduced virulence of Candida albicans MKC1 mutants: a role for mitogen-activated protein kinase in pathogenesis. Infect. Immun. 65:833-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Pietro, A., F. I. Garcia-MacEira, E. Meglecz, and M. I. Roncero. 2001. A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39:1140-1152. [PubMed] [Google Scholar]
- 8.Furukawa, K., Y. Hoshi, T. Maeda, T. Nakajima, and K. Abe. 2005. Aspergillus nidulans HOG pathway is activated only by two-component signalling pathway in response to osmotic stress. Mol. Microbiol. 56:1246-1261. [DOI] [PubMed] [Google Scholar]
- 9.Galagan, J. E., S. E. Calvo, C. Cuomo, L. J. Ma, J. R. Wortman, S. Batzoglou, S. I. Lee, M. Basturkmen, C. C. Spevak, J. Clutterbuck, V. Kapitonov, J. Jurka, C. Scazzocchio, M. Farman, J. Butler, S. Purcell, S. Harris, G. H. Braus, O. Draht, S. Busch, C. D'Enfert, C. Bouchier, G. H. Goldman, D. Bell-Pedersen, S. Griffiths-Jones, J. H. Doonan, J. Yu, K. Vienken, A. Pain, M. Freitag, E. U. Selker, D. B. Archer, M. A. Penalva, B. R. Oakley, M. Momany, T. Tanaka, T. Kumagai, K. Asai, M. Machida, W. C. Nierman, D. W. Denning, M. Caddick, M. Hynes, M. Paoletti, R. Fischer, B. Miller, P. Dyer, M. S. Sachs, S. A. Osmani, and B. W. Birren. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105-1115. [DOI] [PubMed] [Google Scholar]
- 10.Guhad, F. A., C. Csank, H. E. Jensen, D. Y. Thomas, M. Whiteway, and J. Hau. 1998. Reduced pathogenicity of a Candida albicans MAP kinase phosphatase (CPP1) mutant in the murine mastitis model. APMIS 106:1049-1055. [DOI] [PubMed] [Google Scholar]
- 11.Gustin, M. C., J. Albertyn, M. Alexander, and K. Davenport. 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1264-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han, K. H., and R. A. Prade. 2002. Osmotic stress-coupled maintenance of polar growth in Aspergillus nidulans. Mol. Microbiol. 43:1065-1078. [DOI] [PubMed] [Google Scholar]
- 13.Hou, Z. M., C. Y. Xue, Y. L. Peng, T. Katan, H. C. Kistler, and J. R. Xu. 2002. A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant-Microbe Interact. 15:1119-1127. [DOI] [PubMed] [Google Scholar]
- 14.Kawasaki, L., O. Sanchez, K. Shiozaki, and J. Aguirre. 2002. SakA MAP kinase is involved in stress signal transduction, sexual development and spore viability in Aspergillus nidulans. Mol. Microbiol. 45:1153-1163. [DOI] [PubMed] [Google Scholar]
- 15.Kim, J. H., B. C. Campbell, N. Mahoney, K. L. Chan, and G. S. May. 2006. Targeting antioxidative signal transduction and stress response system: control of pathogenic Aspergillus with phenolics that inhibit mitochondrial function. J. Appl. Microbiol. 101:181-189. [DOI] [PubMed] [Google Scholar]
- 16.Kohler, J. R., and G. R. Fink. 1996. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc. Natl. Acad. Sci. USA 93:13223-13228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kojima, K., Y. Takano, A. Yoshimi, C. Tanaka, T. Kikuchi, and T. Okuno. 2004. Fungicide activity through activation of a fungal signalling pathway. Mol. Microbiol. 53:1785-1796. [DOI] [PubMed] [Google Scholar]
- 18.Latge, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harashima, W. C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol Rev. 64:746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Machida, M., K. Asai, M. Sano, T. Tanaka, T. Kumagai, G. Terai, K. Kusumoto, T. Arima, O. Akita, Y. Kashiwagi, K. Abe, K. Gomi, H. Horiuchi, K. Kitamoto, T. Kobayashi, M. Takeuchi, D. W. Denning, J. E. Galagan, W. C. Nierman, J. Yu, D. B. Archer, J. W. Bennett, D. Bhatnagar, T. E. Cleveland, N. D. Fedorova, O. Gotoh, H. Horikawa, A. Hosoyama, M. Ichinomiya, R. Igarashi, K. Iwashita, P. R. Juvvadi, M. Kato, Y. Kato, T. Kin, A. Kokubun, H. Maeda, N. Maeyama, J. Maruyama, H. Nagasaki, T. Nakajima, K. Oda, K. Okada, I. Paulsen, K. Sakamoto, T. Sawano, M. Takahashi, K. Takase, Y. Terabayashi, J. R. Wortman, O. Yamada, Y. Yamagata, H. Anazawa, Y. Hata, Y. Koide, T. Komori, Y. Koyama, T. Minetoki, S. Suharnan, A. Tanaka, K. Isono, S. Kuhara, N. Ogasawara, and H. Kikuchi. 2005. Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157-1161. [DOI] [PubMed] [Google Scholar]
- 21.Navarro-Garcia, F., R. Alonso-Monge, H. Rico, J. Pla, R. Sentandreu, and C. Nombela. 1998. A role for the MAP kinase gene MKC1 in cell wall construction and morphological transitions in Candida albicans. Microbiology 144:411-424. [DOI] [PubMed] [Google Scholar]
- 22.Navarro-Garcia, F., M. Sanchez, J. Pla, and C. Nombela. 1995. Functional characterization of the MKC1 gene of Candida albicans, which encodes a mitogen-activated protein kinase homolog related to cell integrity. Mol. Cell. Biol. 15:2197-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nierman, W. C., A. Pain, M. J. Anderson, J. R. Wortman, H. S. Kim, J. Arroyo, M. Berriman, K. Abe, D. B. Archer, C. Bermejo, J. Bennett, P. Bowyer, D. Chen, M. Collins, R. Coulsen, R. Davies, P. S. Dyer, M. Farman, N. Fedorova, T. V. Feldblyum, R. Fischer, N. Fosker, A. Fraser, J. L. Garcia, M. J. Garcia, A. Goble, G. H. Goldman, K. Gomi, S. Griffith-Jones, R. Gwilliam, B. Haas, H. Haas, D. Harris, H. Horiuchi, J. Huang, S. Humphray, J. Jimenez, N. Keller, H. Khouri, K. Kitamoto, T. Kobayashi, S. Konzack, R. Kulkarni, T. Kumagai, A. Lafton, J. P. Latge, W. Li, A. Lord, C. Lu, W. H. Majoros, G. S. May, B. L. Miller, Y. Mohamoud, M. Molina, M. Monod, I. Mouyna, S. Mulligan, L. Murphy, S. O'Neil, I. Paulsen, M. A. Penalva, M. Pertea, C. Price, B. L. Pritchard, M. A. Quail, E. Rabbinowitsch, N. Rawlins, M. A. Rajandream, U. Reichard, H. Renauld, G. D. Robson, S. Rodriguez de Cordoba, J. M. Rodriguez-Pena, C. M. Ronning, S. Rutter, S. L. Salzberg, M. Sanchez, J. C. Sanchez-Ferrero, D. Saunders, K. Seeger, R. Squares, S. Squares, M. Takeuchi, F. Tekaia, G. Turner, C. R. Vazquez de Aldana, J. Weidman, O. White, J. Woodward, J. H. Yu, C. Fraser, J. E. Galagan, K. Asai, M. Machida, N. Hall, B. Barrell, and D. W. Denning. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151-1156. [DOI] [PubMed] [Google Scholar]
- 24.O'Rourke, S. M., and I. Herskowitz. 2004. Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol. Biol. Cell 15:532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osherov, N., D. P. Kontoyiannis, A. Romans, and G. S. May. 2001. Resistance to itraconazole in Aspergillus nidulans and Aspergillus fumigatus is conferred by extra copies of the A. nidulans P-450 14alpha-demethylase gene, pdmA. J. Antimicrob. Chemother. 48:75-81. [DOI] [PubMed] [Google Scholar]
- 26.Xu, J. R. 2000. Map kinases in fungal pathogens. Fungal Genet. Biol. 31:137-152. [DOI] [PubMed] [Google Scholar]
- 27.Xu, J. R., and J. E. Hamer. 1996. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 10:2696-2706. [DOI] [PubMed] [Google Scholar]
- 28.Xue, T., C. K. Nguyen, A. Romans, and G. S. May. 2004. A mitogen-activated protein kinase that senses nitrogen regulates conidial germination and growth in Aspergillus fumigatus. Eukaryot. Cell 3:557-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, Y., R. Lamm, C. Pillonel, S. Lam, and J.-R. Xu. 2002. Osmoregulation and fungicide resistance: the Neurospora crassa os-2 gene encodes a HOG1 mitogen-activated protein kinase homologue. Appl. Environ. Microbiol. 68:532-538. [DOI] [PMC free article] [PubMed] [Google Scholar]