Abstract
Ace2 transcription factor family genes are found in many fungal genomes and are required for regulation of expression of genes involved in cell separation. We used transcriptional profiling to identify the targets of Ace2 in Candida albicans, and we show that these include several cell wall components, such as glucanases and glycosylphosphatidylinositol-anchored proteins. Expression is downregulated in ace2 deletion mutants in both yeast and hyphal cells. In addition, deleting ace2 results in dramatic changes in expression of metabolic pathways. Expression of glycolytic enzymes is reduced, while expression of respiratory genes (including those involved in the tricarboxylic acid cycle, oxidative phosphorylation, and ATP synthesis) is increased. Similar changes occur in both yeast and hyphal cells. In contrast, genes required for acetyl-coenzyme A and lipid metabolism are upregulated in an ace2 deletion mutant grown predominantly as yeast cells but are downregulated in hyphae. These results suggest that in wild-type strains, Ace2 acts to increase glycolysis and reduce respiration. This is supported by the observation that deleting ace2 results in increased resistance to antimycin A, a drug that inhibits respiration. We also show that Ace2 is required for filamentation in response to low oxygen concentrations (hypoxia). We suggest that filamentation is induced in wild-type cells by reducing respiration (using low oxygen or respiratory drugs) and that mutants with increased respiratory activity fail to undergo filamentation under these conditions.
In Saccharomyces cerevisiae, ACE2 and SWI5 encode two of a set of nine transcription factors that control the mitotic cell cycle (40). The nine regulators are SBF (Swi4 and Swi6) and MBF (Swi6 and Mbp1), which control the expression of genes in G1 and S phase (18); Ndd1, Fkh1, Fkh2, and Mcm1, which regulate the expression of genes at the G2/M border; and Swi5 and Ace2 (and Mcm1), which control the expression of genes in late M and early G1. The transcription factors act in a cascade: SBF and MBF regulate expression of NDD1, the G2 activators control expression of ACE2 and SWI5, and these in turn are required for exit from mitosis and subsequent activation of SBF and MBF.
Ace2 and Swi5 share many functional similarities in S. cerevisiae. The proteins are 37% identical and have the same DNA-binding sites in vitro (29). They also regulate the expression of many of the same genes (9). However, there are substantial differences. There is only ∼22% overlap between the groups of genes regulated by the two factors (40). Swi5 remains cytoplasmic until the end of M phase, when it enters the nucleus (33). Ace2 also enters the nucleus at the end of mitosis (35) but is rapidly exported from (or degraded in) the nucleus of the mother cell and remains only in the nucleus of the daughter cell (7, 46). Localization is regulated by components of the RAM pathway (34, 38). Expression of many Ace2 targets is therefore restricted to the daughter cell, and one of their main functions is to enable the separation of mother and daughter cells following cell division (7, 46).
Ace2 and Swi5 arose from a genome duplication event in the evolution of Saccharomyces, and there are orthologs of both in species that evolved following this event (47, 48). In species that split from the Saccharomyces lineage before polyploidization, there is likely to be a single ortholog. The Candida glabrata ortholog of ACE2 was originally identified as a virulence factor (20). Deleting CgACE2 causes a dramatic increase in virulence in immunosuppressed mice (20). In contrast, deleting the single ACE2 ortholog in Candida albicans results in attenuation of virulence (21). There is also a single ACE2 ortholog, which regulates cell separation, in Schizosaccharomyces pombe, although its effect on virulence has not been tested (1, 28). It is therefore likely that the functions of the proteins differ significantly in yeast species, although all are involved in regulating genes involved in cell separation (1, 20, 21).
We used transcriptional profiling to determine the role of ACE2 in C. albicans. We confirmed that Ace2 regulates the expression of genes involved in cell separation. However, we also show that deleting ace2 results in downregulation of expression of glycolytic genes and in upregulation of expression of many genes required for mitochondrial function and that Ace2 is required for filamentation in response to hypoxic conditions.
MATERIALS AND METHODS
Strains and media.
The yeast strains used in this study are shown in Table 1. Strains were grown in liquid YPD (1% yeast extract, 2% Bacto peptone, 2% glucose) at 30°C to induce yeast cells and in YPD plus 10% fetal calf serum at 37°C to induce hyphae. Antimycin A was added to YPD plates at a concentration of 20 μg ml−1 where indicated. Growth under hypoxic conditions was monitored on a number of additional media, including YNB (0.67% yeast nitrogen base without amino acids, 0.75% amino acid dropout mix) supplemented at 2% with either glucose, acetate, or glycerol. For growth under hypoxic conditions, overnight cultures were diluted, spotted onto agar plates, and incubated in one of four hypoxic chambers (In Vivo2 400 workstation) at 37°C with 5% CO2. The oxygen concentration was maintained at 10%, 5%, 3%, and 1% by varying the nitrogen concentration. Control plates were incubated in a tissue culture incubator with 20% oxygen. For embedded conditions, overnight cultures were subcultured and grown in YPD for 3 to 4 h. Approximately 100 CFU were spread on the surfaces of YPD or YPS (1% yeast extract, 2% Bacto peptone, 2% sucrose) plates, and a thin layer of YPD or YPS agar (10 to 12 ml) was poured onto the dry plates. The plates were incubated at 25°C, 30°C, and 37°C in standard incubators.
TABLE 1.
Candida albicans strains used in this study
RNA extraction and real-time PCR.
For microarray analysis, RNAs were extracted using a RiboPure yeast kit (Ambion) according to the manufacturer's instructions. Strains were cultured overnight in YPD medium, diluted to an A600 of 0.2 in 50 ml of YPD at 30°C or in YPD supplemented with 10% fetal calf serum at 37°C, and grown for 4 h with shaking. All 50 ml of culture was used for RNA extraction. Approximately 100 ng of each RNA sample was tested using an Agilent 2100 bioanalyzer to determine accurate concentrations and RNA quality. Only samples with a 28S/18S rRNA ratio of between 1.6 and 2.2 were used for subsequent analysis. RNAs were maintained at a concentration of 4 μg μl−1.
For real-time PCR, RNAs were extracted from cultures grown in synthetic complete medium (supplemented with 10% fetal calf serum where indicated), and cDNAs were generated as described previously (21). Primers were designed using Primer Express software (Table 2), and actin was used as an endogenous control. For analysis of cell wall genes, 0.5 μg of cDNA was used as a template for amplification, using a 2× SYBR green PCR master mix from Applied Biosystems and a 0.3 μM concentration of each of two specific primers in a total volume of 10 μl. Samples were run in duplicate. The PCR program was carried out on an ABI 7900HT sequence detection system and consisted of denaturing and activation steps at 50°C for 2 min and 95°C for 10 min, respectively, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. For glycolytic and mitochondrial genes, duplicate reactions were carried out using 2 μg cDNA and 10 μl of Platinum SYBR green qPCR SuperMix (Invitrogen) in a final volume of 20 μl. Specific primers were added at a concentration of 0.2 μM. PCR was carried out on a Stratagene MX3000P system, using a denaturation step at 95°C for 10 min followed by 40 cycles of 95°C for 15 s, 60°C for 1 min, and 72°C for 30 s. Amplification of specific transcripts was confirmed by melting-curve analysis at the end of each run, and the efficiency of the assay was verified with a standard curve (data not shown). For analysis, the cycle threshold (CT) value for the ACT1 gene was subtracted from that for the gene of interest to obtain a ΔCT value. The ΔCT value for an arbitrary wild-type sample was selected as a calibrator, and the gene expression level relative to the calibrator is expressed as 2−ΔΔCT.
TABLE 2.
Oligonucleotide primers used for real-time PCR
| Primer name | Primer sequence |
|---|---|
| ACT1F | TTGGTGATGAAGCCCAATCC |
| ACT1R | CCATATCGTCCCAGTTGGAAA |
| GSY1F | TGCCTTGCCATTATGTAGAAAAAG |
| GSY1R | TCGGTACTACCAGCACACAAGTATC |
| PFK1F | TGATGCTCCTGGTATGAATCCA |
| PFK1R | CGGCATAAACATCACAACCATAA |
| TPS3F | AGCACAATCCCCGTTGGTATAG |
| TPS3R | TCCGCCATTCTGTGACTTGTT |
| ACO1F | TCCACCAGAAGACCGTGCTT |
| ACO1R | TTACCATCCCATGGTTTGAATG |
| IDH2F | CTGGGAACCAGTCGATGTCA |
| IDH2R | ACGGAGTCAACAGCTGGTTGT |
| FUM12.3F | TGATGTTGGTCACTGCATTGAA |
| FUM12.3R | TTTTGTGAGCGTTCTTTGCAA |
| ORF19.5267F | CAACTACCACTGTTCTTGCCAGTAC |
| ORF19.5267R | TGTTATCATCACTACTTGAGGGATCAG |
| CHT3F | AGAGCCGCTGGATCAGGTTA |
| CHT3R | TGAAACATCCCACATTGAAATACC |
| DSE1F | CCCAGAAAAAAACCATAGCAGTACTA |
| DSE1R | CTTGTCATGGATTTGTGCAGTATTATAA |
| PGA38F | TCAACTACCATTTCCTCACTATCATCA |
| PGA38R | GATGACAAGTTCGGTGCAACA |
| SCW11F | CTACACTTCCAGCACATCCTCAA |
| SCW11R | GGCCGGAATATCTCCACTAGAA |
Transcriptional profiling.
DNA microarrays representing 6,039 open reading frames (ORFs) from C. albicans SC5314 were purchased from Eurogentec (Seraing, Belgium). Twenty-five micrograms of total RNA was labeled in a total volume of 40 μl including 5 ng LuxA control RNA (Eurogentec), 1× first-strand buffer (Invitrogen), 0.2 pmol C. albicans-specific primer (Eurogentec), 1.5 mM (each) dATP, dTTP, and dGTP, 25 μM dCTP, 10 mM dithiothreitol, 1 μl RNasin, and 37.5 μM Cy3-dCTP or 37.5 μM Cy5-dCTP (Amersham). The mixture was incubated at 65°C for 5 min and then at 42°C for 5 min. One microliter of RNasin (Promega) and 1 μl Superscript II reverse transcriptase (Invitrogen) were added and incubated at 42°C for 1 h. Following the addition of another 1 μl of Superscript II, the incubation was continued for 1 h at 42°C. The reaction was stopped by the addition of 5 mM EDTA, pH 8.0, and 0.5 M NaOH and incubation at 65°C for 20 min and was neutralized by adding 0.5 M acetic acid. The labeled cDNAs were purified using a QIAquick PCR purification kit (QIAGEN) and concentrated by drying under vacuum to a volume of approximately 2 μl.
Before hybridization, one of the labeled cDNA samples was resuspended in 45 μl of hybridization buffer (1× Dig Ease buffer [Roche] containing 0.5 mg ml−1 baker's tRNA and 0.5 mg ml−1 salmon sperm DNA) and then added to the second labeled cDNA. The mixture was heat denatured at 95°C for 2 min, quickly cooled on ice, applied to the DNA microarray, and covered with a 24-mm by 60-mm coverslip. Slides were placed in a hybridization chamber (Corning, Palo Alto, CA) with 9 μl of 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) in each of the wells and incubated at 37°C for 16 h by immersion in a water bath. Following hybridization, the slides were immersed in 1× SSC at 50°C until the coverslip fell off, washed twice for 15 min each in 1× SSC-0.1% sodium dodecyl sulfate (SDS) at 50°C and once in 0.1× SSC-0.1% SDS for 20 min at 50°C, and rinsed three times in 1× SSC at 50°C and, finally, once in 0.2× SSC at room temperature. The slides were dried by centrifugation at 900 rpm for 5 min. Seven independent comparisons (incorporating three dye swaps) were performed for yeast growth conditions, and five independent comparisons (with two dye swaps) were performed for growth under hypha-inducing conditions.
The microarrays were scanned using an Axon 4000B scanner at a 10-μm resolution. Low-quality spots were automatically flagged by Genepix Pro 5.0. In addition, spots that were saturated, spots that did not have 50% of pixels with a value of at least 1 standard deviation above the background, and spots that did not have a background-corrected intensity of >20 in the Cy3 or Cy5 channel were flagged for quality control. Lowess normalization was carried out in GeneSpring, and spots which were not flagged in six of seven yeast arrays and in four of five hyphal arrays were included in the subsequent analysis. The high-quality data were analyzed statistically using SAM software (significance analysis of microarrays) (44), using the one-class response, K-nearest neighbor settings (10 neighbors), and a random number seed (58,874,688 for yeast arrays and 41,250,246 for hyphal arrays). Four hundred permutations were carried out for the yeast arrays, and 120 permutations were carried out for the hyphal arrays. Delta values were selected to calculate low false discovery rates (FDRs) for the median d scores. The selected FDRs were 0.23% for the yeast arrays (363 genes) and 0.99% for the hyphal arrays (347 genes). For some analyses, an FDR of 0.9% was also used for the yeast arrays (645 genes). Data from duplicate spots on the arrays were averaged. Hierarchical clustering was carried out using Eisen's cluster program (similarity metric, correlation [uncentered]; clustering method, average linkage) and was viewed with Treeview (11).
Gene ontology analysis.
Coding sequences of the C. albicans genes present on the microarrays were obtained from CandidaDB (http://genolist.pasteur.fr/CandidaDB/). S. cerevisiae orthologs of 3,685 C. albicans genes were identified using BLASTX analysis, with a cutoff of 1e−10, and were used to define the reference data set. Two hundred eighty-three orthologs of the 363 genes with altered expression in yeast cells and 248 orthologs of 347 genes with altered expression in hyphal cells were identified and defined the test data sets. Genes with upregulated expression were analyzed separately from genes with downregulated expression. The data were analyzed using the Fisher exact test in GoToolBox (http://139.124.62.227/GOToolBox/).
Promoter analysis.
Promoter motifs were identified using MEME software (2; http://meme.sdsc.edu/meme/intro.html). The intergenic region (or 1,000 bp [whichever was the shortest]) was extracted from upstream of the ATG codon for orf19.3066 (DSE4), orf19.3629 (DSE1), orf19.6586 (CHT3), orf19.5343 (ASH1), orf19.7218 (PRY2), orf19.4438 (RME1), and orf19.220 (PIR1). Up to four motifs with lengths of 6 to 8 bp that occurred in any number of repetitions were identified. The selected motif had an E value of 4.6e−001. For glycolytic and tricarboxylic acid (TCA) cycle genes, 1,000 bp of sequence upstream of the ATG codon was obtained using tools available at the BRI website of the National Research Council Canada (http://candida.bri.nrc.ca/).
In vivo microscopy.
For infection studies, murine RAW264.7 macrophage cells were incubated with yeast at a 1:1 ratio in Dulbecco's modified Eagle's medium in a Perfusion-Open-Closed (P.O.C.) minikit (Zeiss) and were maintained in a small incubation chamber at 37°C and 5% CO2. The progress of the infection was monitored on a Zeiss Axiovert 200 (inverted) microscope, and images were taken every 15 min.
RESULTS
Identification of targets of Ace2.
To identify the role of Ace2 in C. albicans, we compared the transcriptional profiles of wild-type cells and cells deleted for ace2 grown under conditions that are preferential for either yeast growth (YPD at 30°C) or hyphal growth (YPD plus 10% serum at 37°C). The array experiments were carried out using MK106, an ace2 deletion mutant generated by replacing both alleles in C. albicans SC5314 with an FLP recombinase recognition target recombination site (21). For most other experiments, both this strain and an isolate (MK62) generated by replacing ACE2 with URA3 and HIS1 were used (21). The results for MK106 only are shown, as MK62 had identical phenotypes in all assays. Full lists of all genes with reproducible changes in expression are provided in Tables S1, S2, and S3 in the supplemental material. We used Gene Ontology (GO) analysis to identify processes that are altered in the mutant. Since few GO terms are assigned to C. albicans genes at present, we identified the S. cerevisiae orthologs where possible and used the associated GO categories. Terms that were statistically overrepresented or underrepresented in the ace2 deletion mutant were identified using GOToolBox (27). Full lists are available in Tables S4 to S7 in the supplemental material, and some processes with altered expression are shown in Fig. 1A.
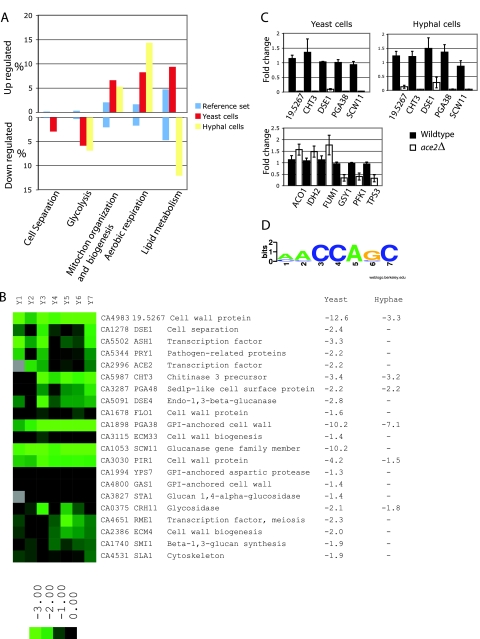
FIG. 1.
Transcriptional profiling of ace2 deletion strains. (A) The S. cerevisiae orthologs of C. albicans genes with altered expression in an ace2 deletion strain (C. albicans MK106) were identified, and GO categories that are overrepresented were determined. The percentages of genes in some categories overrepresented in upregulated genes are shown on the upper graph, and categories overrepresented in downregulated genes are shown in the lower graph. The percentage of genes in each category in the reference set (i.e., S. cerevisiae orthologs of C. albicans genes present on the arrays) is indicated in blue in both the upper and lower graphs. The associated P values are as follows: cell separation during cytokinesis, 0.001 (yeast cells) and not significant in hyphal cells; glycolysis, 9.13e−06 (yeast cells) and 1.4e−07 (hyphal cells); mitochondrial (mitochon) membrane organization and biogenesis, 6.99e−05 (yeast cells) and 0.027 (hyphal cells); aerobic respiration, 3.77e−06 (yeast cells) and 9.36e−11 (hyphal cells); and lipid metabolism, 0.01 (yeast cells) and 0.005 (hyphal cells). (B) Cluster of cell wall genes with reduced expression in an ace2 deletion strain (C. albicans MK106). The expression levels relative to those of the wild type in seven independent experiments with yeast cells are shown. Gene identifications and descriptions were obtained from CandidaDB, and the names of the S. cerevisiae orthologs are given. CA4983 (orf19.5267) shares some similarity with DAN4 but is unlikely to be an ortholog. The average x-fold changes in expression in yeast and hyphal cells are shown. —, changes in gene expression of these genes are not statistically reproducible with the methods used. GPI, glycosylphosphatidylinositol. (C) Expression of several potential cell wall genes was determined, using real-time PCR for yeast and hyphal cells (top panels) and for glycolytic and TCA cycle genes (bottom panel). SC5314 (WT) and C. albicans MK106 (ace2) cells were grown to exponential phase in SC medium at 30°C and supplemented with 10% fetal calf serum at 37°C for hyphal cells, and the results of three biological replicates (each performed in duplicate) are shown. The changes in expression (relative to an arbitrarily chosen wild-type sample) are shown with the associated standard deviations. (D) A motif present in the upstream sequences of most members of the cluster shown in panel B was identified using MEME. This represents a candidate binding site for Ace2.
Expression of cell wall genes.
Our previous analysis (21) showed that Ace2 regulates the expression of genes involved in cell separation, such as CHT3 (encoding chitinase), DSE1, and SCW11 (encoding a potential glucan 1,3-beta-glucosidase). The microarray results and GO term analysis supported these results, as genes involved in cell separation and cytokinesis were overrepresented in the ace2 deletion mutant grown under conditions favoring yeast growth (Fig. 1A). We identified 20 genes (as well as ACE2) involved in cell wall metabolism or in regulation of expression of cell wall genes whose expression was reduced in the ace2 knockout (Fig. 1B). These included orthologs of additional genes known to be regulated by Ace2 in S. cerevisiae (such as ASH1, DSE1, DSE4, PIR1, PRY2, and RME1) and others that are not known to be regulated by Ace2 in S. cerevisiae, such as SED1, FLO1, and GAS1. orf19.5267 (CA4983) had the most dramatic reduction in expression in C. albicans yeast cells (Fig. 1B). This is annotated as a possibly spurious short ORF (encoding 161 amino acids) in the Can dida Genome Database (http://www.candidagenome.org), whereas CandidaDB (http://genolist.pasteur.fr/CandidaDB/) suggests that it encodes an ortholog of S. cerevisiae DAN4/YJR151C, a putative cell wall mannoprotein (see Table S1 in the supplemental material). There is an ortholog of orf19.5267 in the Candida dubliniensis genome (http://www.sanger.ac.uk/sequencing/Candida/dubliniensis/), suggesting that it is a valid open reading frame. orf19.5267 encodes a threonine-rich protein which, in our opinion, shows borderline similarity to several cell wall proteins. We used real-time PCR to confirm that orf19.5267 is indeed transcribed, and we showed that it is also regulated by Ace2 (Fig. 1C). It is therefore likely to encode a cell wall protein, like most of the other targets in Fig. 1B. We also used real-time PCR to confirm that expression of PGA48, encoding a putative glycosylphosphatidylinositol-anchored protein, is regulated by Ace2 (Fig. 1C).
Some genes (including CHT3 and PIR1) had reduced expression in the ace2 deletion mutant in both yeast and hyphal cells (Fig. 1B). However, most of the cell wall genes identified as downregulated in predominantly yeast-like cells were not identified under hypha-inducing conditions in the array experiments. These included DSE1 and SCW11, which we have previously shown to be regulated by Ace2 under both yeast- and hypha-inducing conditions (21). We assumed that our failure to detect reproducible changes in expression of these genes was due to the application of stringent statistical analyses, resulting in the loss of some positive results. In fact, if we increase the false discovery rate, SCW11 is present in the data set with reduced expression under hyphal conditions (data not shown). We therefore used real-time PCR to confirm that expression of the cell wall genes CHT3, DSE1, and SCW11 is reduced in both yeast and hyphal cells when ace2 is deleted (Fig. 1C). The change in expression is less in hyphal cells (not shown), perhaps because the expression of many genes (such as DSE1 and FLO1) is downregulated in hyphae, even in wild-type cells (21, 32). Our results are therefore in agreement with the reverse transcription-PCR experiments we reported previously (21).
In S. cerevisiae, the consensus binding site for Ace2/Swi5 is RRCCAGC (9, 42). We used the MEME algorithm (2) to search for motifs of 6 to 8 bp that are overrepresented in the upstream regions of some of the target genes we identified in C. albicans. We identified a motif (MMCCASC) that is very similar to the binding site in S. cerevisiae (Fig. 1C). Although we have no biochemical evidence showing that this is indeed a binding site in C. albicans, the results suggest that Ace2 may bind directly to these promoters.
Expression of metabolic genes.
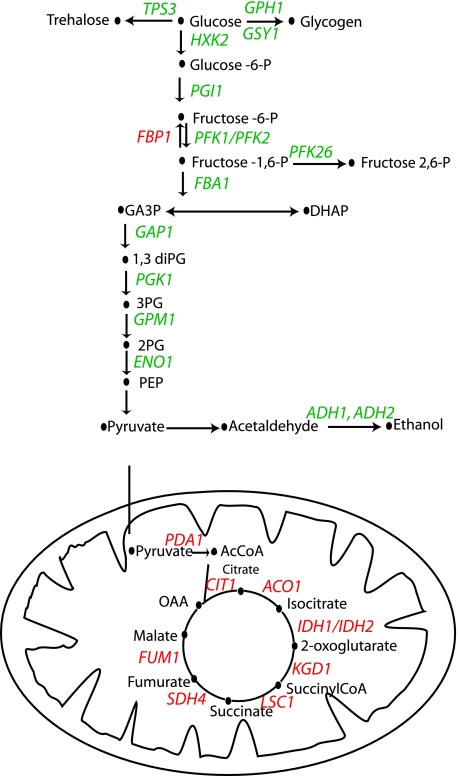
Many genes involved in glycolysis are overrepresented in the categories that have downregulated expression in the ace2 deletion mutant, in both yeast and hyphal cells (Fig. 1 and 2; Table 3). Several related categories (carbohydrate metabolism, glucose metabolism, and monosaccharide catabolism) are also overrepresented (see Tables S3 and S4 in the supplemental material). Although these genes encode enzymes that are required for both glycolysis and gluconeogenesis, expression of the glycolysis-specific genes PFK1 and PFK2 was reduced, and expression of the gluconeogenesis-specific genes FBP1 and PCK1 was increased, in hyphal cells (Fig. 2 and Table 3). We used real-time PCR to confirm the reduction in expression of three genes, namely, GSY1, PFK1, and TPS3 (Fig. 1C). It is therefore likely that glycolysis, not gluconeogenesis, is downregulated in the ace2 deletion mutant.
FIG. 2.
Ace2-dependent regulation of genes involved in carbon metabolism. The cartoon represents the steps in glycolysis occurring in the cytosol (upper section) and the TCA cycle within the mitochondrion. Genes shown in green are downregulated, and genes shown in red are upregulated in an ace2 deletion strain.
TABLE 3.
Changes in expression of metabolic genes in an ace2 deletion strain under yeast- and hypha-inducing conditions
| Systematic name and metabolic categorya | Standard nameb | Description | Change in expression (fold)c
|
|
|---|---|---|---|---|
| Yeastd | Hyphae | |||
| Glycolysis and reserve carbohydrates | ||||
| orf19.5348 | TPS3 | Regulatory subunit of trehalose-phosphate synthase | −3.0 | −1.9 |
| orf19.7021 | GPH1 | Glycogen phosphorylase | −2.4 | — |
| orf19.3278 | GSY1 | Glycogen synthase | −4.8‡ | −1.8 |
| orf19.3325 | GLG2 | Self-glucosylating initiator of glycogen synthesis | −3.2 | −1.5 |
| orf19.2020 | HXT6 | Glucose transporter | −2.4 | −2.0 |
| orf19.542 | HXK2 | Hexokinase II | −2.3 | −1.5 |
| orf19.3888 | PGI1 | Glucose-6-phosphate isomerase | −1.8 | −1.6 |
| orf19.3967 | PFK1 | Phosphofructokinase (alpha subunit) | — | −1.7 |
| orf19.6540 | PFK2 | Phosphofructokinase (beta subunit) | −2.1 | — |
| orf19.4753 | PFK26 | 6-Phosphofructose-2-kinase | −1.5 | — |
| orf19.6814 | TDH3 | Glyceraldehyde-3-phosphate dehydrogenase | −1.7‡ | −1.7 |
| orf19.3651 | PGK1 | 3-Phosphoglycerate kinase | −1.9‡ | −1.7 |
| orf19.4618 | FBA1 | Fructose-bisphosphate aldolase | −2.0 | −1.5 |
| orf19.903 | GPM1 | Phosphoglycerate mutase | −1.6 | −1.6 |
| orf19.395 | ENO1 | Enolase I | −1.8‡ | −1.6 |
| orf19.5113 | ADH2 | Alcohol dehydrogenase | −2.0 | −1.9 |
| orf19.3997 | ADH1 | Alcohol dehydrogenase | −1.8 | — |
| Gluconeogenesis | ||||
| orf19.6178 | FBP1 | Fructose-1,6-bisphosphatase | — | +1.8 |
| orf19.7514 | PCK1 | Phosphoenolpyruvate carboxykinase | — | +3.7 |
| TCA cycle | ||||
| orf19.3097 | PDA1 | Pyruvate dehydrogenase (alpha subunit) | — | +1.9 |
| orf19.4393 | CIT1 | Citrate synthase | +2.5‡ | +2.3 |
| orf19.6385 | ACO1 | Aconitase | +1.5‡ | +4.3 |
| orf19.4826 | IDH1 | Isocitrate dehydrogenase subunit | — | +1.9 |
| orf19.5791 | IDH2 | Isocitrate dehydrogenase subunit | — | +2.75 |
| orf19.6165 | KGD1 | Alpha-ketoglutarate dehydrogenase | +1.5‡ | +1.8 |
| orf19.3358 | LSC1 | Succinyl-CoA ligase (alpha subunit) | — | +1.5 |
| orf19.4022 | SDH4 | Succinate dehydrogenase | +1.6‡ | — |
| orf19.543 | FUM1† | Fumarate hydratase | +1.6 | +1.6 |
| Oxidative phosphorylation and ATP synthesis | ||||
| orf19.3577 | COQ5 | Methyltransferase of ubiquinone (coenzyme Q) biosynthesis | +1.5 | — |
| orf19.3008 | COQ4 | Ubiquinone biosynthesis | +1.6 | — |
| COQ6 | Ubiquinone biosynthesis | — | −1.5 | |
| orf19.7049 | CYB5 | Cytochrome b5 | +1.7 | — |
| orf19.2201 | CBP6† | Cytochrome b protein synthesis | +1.9 | — |
| orf19.1770 | CYC1 | Cytochrome c | +2.0 | — |
| orf19.3527 | CYT1 | Cytochrome c1 | — | +1.6 |
| orf19.4578 | CYT2 | Cytochrome c1 heme lyase | +1.8 | — |
| orf19.1416 | COX11 | Cytochrome c oxidase assembly | +1.7 | — |
| orf19.2006.1 | COX17† | Cytochrome c oxidase assembly | +2.3 | — |
| orf19.3946 | COX18† | Cytochrome c oxidase assembly | +1.8 | — |
| orf19.4967 | COX19 | Required for cytochrome c oxidase activity | +1.6‡ | — |
| orf19.2644 | QCR2 | Ubiquinol-cytochrome c reductase 40-kDa chain II | — | +2.1 |
| orf19.5893 | RIP1 | Ubiquinol cytochrome c reductase | — | +1.9 |
| orf19.4929 | PET309† | Translational activator of COX1 | +1.6 | — |
| orf19.7577 | MSS51 | Required for translation of COX1 | +2.3 | — |
| orf19.6565 | OXA1† | Assembly of cytochrome c oxidase, ATP synthase, and ubiquinol-cytochrome c oxidoreductase | +2.2 | +1.4 |
| orf19.6854 | ATP1 | F1Fo ATPase complex | — | +1.9 |
| orf19.5653 | ATP2 | F1Fo ATPase complex | — | +2.3 |
| orf19.3579 | ATP4 | F1FoATPase complex | — | +1.7 |
| orf19.5419 | ATP5 | F1Fo ATPase complex | — | +1.8 |
| orf19.2785 | ATP7 | F1Fo ATPase complex | +1.7‡ | +2.0 |
| CA4830* | ATP8 | F1Fo ATPase complex | +1.4 | — |
| orf19.6916 | ATP11† | F1Fo ATPase complex | +2.0 | — |
| orf19.3686 | ATP12 | F1Fo ATPase complex | +1.6‡ | — |
| Lipid and fatty acid metabolism | ||||
| orf19.5577 | YDR531W† | Pantothenate kinase | +1.7 | −1.7 |
| orf19.3171 | ACH1 | Acetyl-CoA hydrolase | +2.0 | — |
| orf19.7466 | ACC1 | Acetyl-CoA carboxylase | +1.9 | — |
| orf19.7043.1 | ACB1 | Acyl-CoA binding protein | +1.4 | −1.6 |
| ARE2 | Acyl-CoA:sterol acyltransferase | — | −2.8 | |
| orf19.979 | FAS1 | Fatty acid synthase (beta subunit) | +1.7 | — |
| orf19.5949 | FAS2 | Fatty acid synthase (alpha subunit) | +1.5 | −1.6 |
| orf19.7592 | FAA4 | Long-chain-fatty acyl-CoA synthetase | +1.4 | — |
| orf19.5818 | SUR2 | Hydroxylation of C-4 of sphingoid moiety of ceramide | — | −1.9 |
| orf19.6860 | PIS1 | Phosphatidylinositol synthase | +1.5 | — |
| Ergosterol metabolism | ||||
| orf19.406 | ERG1 | Squalene epoxidase | — | −1.9 |
| orf19.6026 | ERG2 | C-8 sterol isomerase | +1.6 | — |
| orf19.5178 | ERG5 | C-22 sterol desaturase | — | −1.5 |
| orf19.3616 | ERG9 | Farnesyl-diphosphate farnesyltransferase | +1.3‡ | — |
| orf19.1598 | ERG24 | C-14 sterol reductase | +1.3 | — |
| orf19.1591 | ERG10 | Acetyl-CoA acetyltransferase | +1.7 | — |
| orf19.922 | ERG11 | Lanosterol 14-alpha-demethylase | — | −2.4 |
| orf19.4631 | ERG251 | C-4 methylsterol oxidase activity | — | −1.7 |
The systematic (orf19) name from http://www.candidagenome.org is used where possible; one ORF was not represented in the Candida Genome Database, and the CandidaDB (http://genolist.pasteur.fr/CandidaDB/) designation was used (asterisk).
The standard name from the Candida Genome Database is used where possible; where the standard name is the same as the systematic name, the name of the S. cerevisiae ortholog was substituted (†).
—, changes in gene expression were not statistically reproducible with the methods used. This does not suggest that gene expression is unchanged.
The genes with altered expression in yeast cells were mostly obtained using an FDR of 0.23%. The indicated genes (‡) were present only at an FDR of 0.99%.
Two hundred twenty-one genes were identified as having increased expression in an ace2 deletion strain grown under conditions favoring yeast growth (see Table S1 in the supplemental material). Surprisingly, of these genes, 130 (59%) are predicted to be involved in mitochondrial function. Similarly, under hypha-inducing conditions, 75 of 172 genes with increased expression have predicted roles in the mitochondria. These include a large number of proteins with structural roles in mitochondrial organization and biogenesis (Fig. 1A), such as mitochondrial ribosomal proteins, and proteins involved in mitochondrial protein import (see Table S3 in the supplemental material). However, many genes required for aerobic respiration are also overrepresented (Fig. 1A). These include enzymes required for the tricarboxylic acid cycle (Fig. 2) and several components required for electron transfer, such as coenzyme Q biosynthesis, cytochrome b, cytochrome c oxidase, and the ATPase complex (Table 3). We confirmed (using real-time PCR) that expression of three TCA cycle genes (ACO1, IDH2, and FUM1) is increased when ace2 is deleted (Fig. 1C). Overall, these results suggest that fermentation is suppressed and respiration is increased in ace2 deletion strains.
Most of the gene ontology processes that are overrepresented in upregulated or downregulated genes were the same whether the cells were grown under hypha-inducing or noninducing conditions (Fig. 1A; Table 3). One significant difference was in genes involved in lipid metabolism (Fig. 1A; Table 3). In yeast cells, deleting ace2 resulted in increased expression of genes involved in acetyl-coenzyme A (CoA) metabolism (such as orf19.5577 [encoding pantothenate kinase], ACH1, ACC1, ACB1, and ERG10) and in ergosterol synthesis (ERG2, ERG9, ERG10, and ERG24). In hyphal cells, however, expression of orf19.5577 and ACB1 was decreased. Components of the ergosterol biosynthesis pathway (ERG1, ERG5, ERG11, and ERG251) also had reduced expression. In addition, expression of the fatty acid synthase gene (FAS2) was increased in the ace2 deletion mutant grown as yeast cells and decreased in hyphal cells.
Sensitivity to antimycin A.
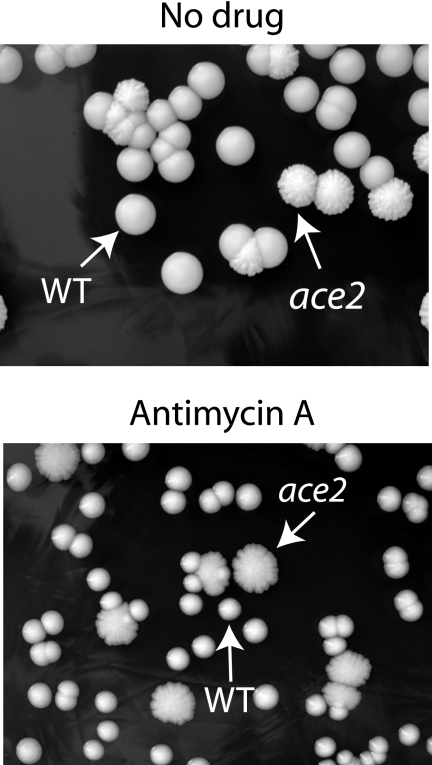
To determine if the increased expression of respiratory and other mitochondrial genes is associated with an increase in respiratory activity, we compared the sensitivities of wild-type and ace2 deletion strains to antimycin A, a drug that inhibits the transport of electrons between cytochrome b and cytochrome c. Because deleting ace2 causes a defect in cell separation (21), it is difficult to use the cell number or serial dilutions to determine sensitivity. However, colonies from ace2 deletion strains can easily be distinguished from lustrous wild-type colonies as a result of their flat, dull appearance. We therefore mixed a population of wild-type and ace2 deletion cells together and plated them on YPD medium in the absence or presence of antimycin A (Fig. 3). In the absence of the drug, the colonies from both strains were of approximately equal sizes. However, in the presence of antimycin A, the ace2 deletion colonies were substantially larger than wild-type colonies (Fig. 3). We interpret this to mean that the ace2 deletion strain is less sensitive to the presence of antimycin A, presumably because it has increased respiration rates.
FIG. 3.
ace2 deletion strains are less sensitive than the wild type to antimycin A. Wild-type (SC5314) and ace2 deletion (MK106) strains were grown to mid-exponential phase, and the cultures were mixed and then plated on YPD in the absence (no drug) or presence of antimycin A at 20 μg ml−1. The ace2 strains can be distinguished from the wild type by their flat, dry appearance.
Ace2 is necessary for filamentation in response to hypoxia.
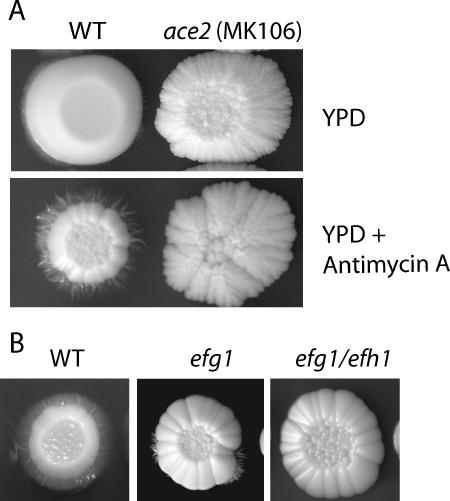
While we were testing the response of C. albicans to the presence of antimycin A, we noticed that the drug induced filamentation in wild-type strains (Fig. 4). These filaments were generated on YPD at 30°C in the absence of any known inducer. This suggests that low levels of respiration induce filamentation. Deleting ace2, however, abolished the ability to form hyphae in response to antimycin A. Deleting efg1 and efh1, which are regulators of filamentation (8), also reduced the response to antimycin A (Fig. 4B). We therefore tested the phenotype of the cells during growth under hypoxic conditions.
FIG. 4.
Antimycin A induces filamentation in wild-type C. albicans isolates but not in ace2 deletion strains. Cultures were grown overnight in YPD and diluted in fresh medium, and 5 μl of the indicated strains was spotted on the indicated media. The plates were incubated at 30°C for 7 days. (A) C. albicans SC5314 (WT) and MK106 (ace2) on YPD and YPD plus antimycin A (20 μg ml−1). (B) C. albicans CAF2-1 (WT), HLC52 (efg1), and H/1.22 (efg1 efh1) on YPD plus antimycin A (20 μg ml−1).
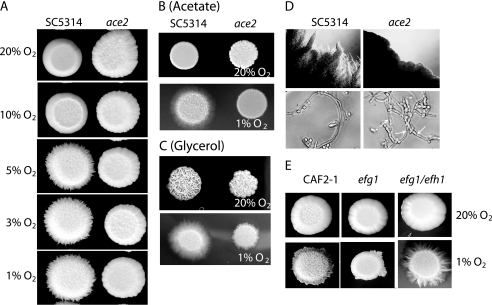
There have been several recent reports that growth in embedded (and therefore hypoxic) media induces filamentation in C. albicans (4, 5, 8, 39). We tested the effect of lowering the oxygen concentration by using hypoxia chambers and also by using cells embedded in agar. The hypoxia chambers were maintained at 37°C with 5% CO2. The O2 concentration was varied by controlling the concentration of nitrogen. As shown in Fig. 5, as the O2 concentration was reduced to 5% and below, wild-type cells responded by undergoing filamentation. Hyphae and pseudohyphae formed all over the colony, forming a fuzzy ball. In contrast, although the ace2 deletion strains grew at the same rate, they failed to generate hyphae, even at 1% O2 (Fig. 5A). The number of pseudohyphae, however, increased on the surface of the colony for both wild-type and ace2 deletion strains as the O2 level declined. Filamentation is not dependent on the medium used. Figure 5B shows growth on minimal medium with acetate, and Fig. 5C shows growth on minimal medium with glycerol. YNB with galactose, Spider medium, and YPD with 10% fetal calf serum gave similar results (not shown). The ace2 deletion mutant did show a small amount of filamentation on glycerol at an O2 concentration of 1%, but it was greatly reduced compared to that of the wild type (Fig. 5C). The presence of true hyphae in the wild type was confirmed by examining the edges of the colonies (Fig. 5D). Nonadherent cells were washed from the plate, and embedded cells were transferred to a microscope slide. The wild-type strain generated long hyphae, whereas in the ace2 deletion strain only pseudohyphae with obvious constrictions were present.
FIG. 5.
Ace2 is required for induction of filamentation in response to hypoxia. (A) Wild-type (SC5314) and ace2 deletion (MK106) strains were grown overnight and spotted onto YPD plates. These were incubated for 5 days in hypoxic chambers at 37°C with 5% CO2 at the indicated oxygen concentrations. (B) Wild-type (SC5314) and ace2 deletion (MK106) strains were spotted onto YNB plates containing acetate as the major carbon source and incubated in hypoxic chambers at 37°C with 5% CO2 at 20% oxygen (top panel) or 1% oxygen (bottom panel). (C) Wild-type (SC5314) and ace2 deletion (MK106) strains were spotted onto YNB plates containing glycerol as the major carbon source and incubated in hypoxic chambers at 37°C with 5% CO2 at 20% oxygen (top panel) or 1% oxygen (bottom panel). (D) The top panels show the edges of colonies from wild-type (SC5314) and ace2 deletion (MK106) strains photographed using a Singer MSM microscope. For the bottom panels, cells were removed from the agar at the edges of the colonies and examined microscopically. The cells from the wild-type strain consist predominantly of very long hyphae. Elongated cells are present in the ace2 deletion strain, but they all have obvious constrictions along their length (indicated with arrows). (E) Wild-type (CAF2-1), efg1 deletion (HLC52), and efg1 efh1 double deletion (H/1.22) strains were spotted onto YPD plates and incubated in hypoxic chambers at 37°C with 5% CO2 at 20% oxygen (top panel) or 1% oxygen (bottom panel).
It has been reported previously that deleting the APSES (Asm1, Phd1, Sok2, Efg1, and StuA family) protein genes EFG1 and EFH1 leads to an increase in filamentation under microaerophilic and embedded hypoxic conditions (8). We determined the phenotypes of these strains under controlled O2 concentrations. Both the wild type and the efg1 efh1 double deletion strain formed filaments at low O2 concentrations at 37°C (Fig. 5E). The double knockout was hyperfilamentous compared to the wild type. In contrast, the efg1 single knockout generated very few filaments. These results are very similar to those reported by Doedt et al. (8) for growth under microaerophilic conditions (approximately 6% O2).
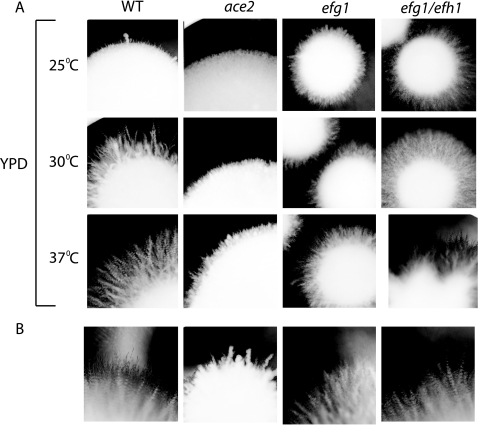
Doedt et al. (8) also reported that Efg1 regulates filamentation in embedded media at 25°C. We compared the phenotypes of ace2, efg1, and efg1 efh1 deletion strains grown in embedded YPD medium at 25°C, 30°C, and 37°C (Fig. 6). Filamentation in the wild-type strain increased as the temperature was elevated. The efg1 deletion strain was hyperfilamentous relative to the wild type at 25°C but generated fewer filaments than the wild type at higher temperatures. This is in agreement with previous reports (8, 15, 39). The efg1 efh1 deletion strain was also hyperfilamentous at low temperatures. In contrast, the ace2 deletion strains produced no filaments at 25°C and few filaments at 30°C, and whereas hyphae were always present at 37°C, they were smaller and fewer than those in the wild type. Most experiments with embedded media are performed using sucrose rather than glucose as a carbon source. We therefore repeated the experiment using YPS medium. All strains formed filaments at all temperatures tested (filamentation at 25°C is shown in Fig. 6B). However, the ace2 deletion strains generated fewer hyphae than did the wild type. The colonies formed by the ace2 strains were small at low temperatures, but they more closely resembled the wild-type colonies as the temperature increased (not shown).
FIG. 6.
Morphology of ace2 deletion strains in embedded medium. The results for one wild-type strain (SC5314) are shown. CAF2-1 has a similar phenotype. (A) Wild-type (SC5314), ace2 deletion (MK106), efg1 deletion (HLC52), and efg1 efh1 deletion (H/1.22) strains were plated on YPD medium, covered with a thin layer of the same medium, and incubated at the indicated temperatures for 5 days. Representative colonies were photographed using a Singer MSM microscope. (B) The strains described above were plated on YPS medium, covered with a thin layer of the same medium, incubated at 25°C for 5 days, and photographed at a magnification of ×20. Similar results were obtained at different incubation temperatures (not shown).
Deleting ace2 does not affect filamentation under all conditions.
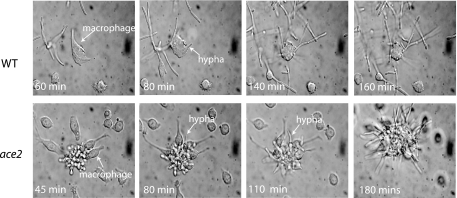
Efg1 is required for hyphal growth under most conditions, particularly during the classical response to induction with serum (reviewed in reference 12). In contrast, we have previously shown that ace2 deletion strains generate hyphae in response to serum induction and also when adhered to plastic surfaces (21). Here we examined the phenotype of ace2 deletion strains following engulfment with mouse macrophages at atmospheric O2 concentrations (Fig. 7). Wild-type and ace2 deletion strains were incubated with murine macrophages (RAW264.7 cells), and the interaction was followed microscopically. As shown in Fig. 7, both strains produced hyphae and escaped from the macrophages.
FIG. 7.
Deleting ace2 does not inhibit filamentation when cells are engulfed by macrophages. Murine macrophage RAW264.7 cells were infected with C. albicans wild-type (SC5314) and ace2 deletion (MK106) cells. Macrophages that engulfed the yeast were followed using in vivo microscopy, and images were taken at the times indicated. Both strains generated hyphae and eventually escaped from the macrophages.
DISCUSSION
Previous analysis has shown that members of the Ace2 transcription factor family are required for regulating the expression of a subset of genes involved in cell wall biosynthesis (1, 9, 20, 21, 40). Our transcriptional profiling indicates that in C. albicans, Ace2 regulates the expression of at least 20 cell wall genes (Fig. 1). Several of these are orthologs of S. cerevisiae genes that are expressed predominantly in daughter, not mother, cells (CHT3, DSE1, DSE4, ASH1, PIR1, and RME1) (Fig. 1B) (7). Since the CaAce2 protein is localized to the nucleus of the daughter cell (similar to Ace2 in S. cerevisiae) (6, 21), it is likely that these genes are also expressed in a daughter-specific manner in C. albicans. Localization of ScAce2 is regulated by the Mob2-Cbk1 kinase complex and other components of the RAM pathway (34, 46). The roles of the RAM orthologs in C. albicans have not yet been determined. However, deleting CaCBK1 leads to a defect in cell separation and decreases expression of some targets of CaAce2 (30), suggesting that the kinase has similar functions in the two yeasts. In addition, Ace2 does not enter the nucleus in cells deficient in the CDC14-encoded phosphatase in either S. cerevisiae (46) or C. albicans (6).
In general, regulation of cell cycle-dependent expression does not appear to be well conserved between S. cerevisiae and C. albicans (17). In S. cerevisiae, ACE2 is expressed at the G2/M boundary, and its expression is coregulated with that of other members of the CLB2 cluster, including CDC5 (49). In C. albicans, CLB2 is also coexpressed with CDC5 (17). However, many of the remaining genes in the clusters differ between S. cerevisiae and C. albicans. The C. albicans CLB2 cluster does not contain ACE2, but it does include several genes associated with control of mitotic exit, such as CDC14 and CBK1 (17). The fact that CaAce2 is localized to the daughter cell nucleus and that localization is dependent on CBK1 and CDC14 suggests that the targets of Ace2 are likely to be expressed at the end of the mitotic cell cycle in C. albicans as well as S. cerevisiae and S. pombe, and possibly also in C. glabrata (20).
Deleting efg1 also leads to a reduction in expression of cell wall genes. In general, the genes are different—Efg1 regulates expression of the ALS family (encoding cell surface glycoproteins involved in adhesion), whereas Ace2 regulates expression of a set of glycosylphosphatidylinositol-anchored protein genes involved in cell wall restructuring (such as CHT and SCW11). There is, however, some overlap, as expression of FLO1 (flocculin) is reduced in both efg1 and ace2 knockouts. The cell wall is dramatically reorganized between yeast and hyphal growth (10), and it is not surprising that interfering with cell wall metabolism affects filamentation under at least some conditions.
We were surprised to observe that deleting ace2 leads to a reduction in expression of glycolytic genes and an increase in expression of genes encoding enzymes in the TCA cycle and components required for oxidative phosphorylation and ATP synthesis. These processes were altered in ace2 deletion strains grown as either yeast or hyphal cells. The same genes were not always identified under both conditions (Table 3), but in the majority of cases the pathways are the same. For example, the expression of components of the ATPase complex was increased in both sets of experiments, but whereas ATP1, ATP2, ATP4, and ATP5 were identified under hypha-inducing conditions, ATP7, ATP8, ATP11, and ATP12 had increased expression in yeast cells. Barrelle et al. (3) have recently shown that the progression of systemic disease caused by C. albicans is dependent on glycolysis. Strains that are deleted for the glycolysis-specific gene PYK1 (encoding pyruvate kinase) have dramatically reduced virulence in animal models (3). In addition, both PYK1 and PFK2 (also specific for glycolysis) are highly expressed during systemic infection. It is therefore possible that the reduction in virulence seen in an ace2 deletion mutant (21) is caused, at least partially, by a downregulation of glycolysis.
A large number of mitochondrial ribosomal protein genes showed altered expression in the ace2 deletion strains, particularly in yeast cells. These tend to dominate the data, possibly swamping other significant results. For this reason, the data from the yeast arrays were evaluated using false discovery rates of both 0.2% (363 genes) and 1% (645 genes). Some additional components required for aerobic respiration were identified in the larger gene set (Table 3). These results indicate that in wild-type cells, Ace2 induces fermentative growth and represses respiration, irrespective of whether the cells are growing as yeast or hyphae. This is supported by the observation that the ace2 deletion strains (with increased respiration) were less susceptible to antimycin A. No similar phenotypes were observed in equivalent deletion mutants of S. cerevisiae or C. glabrata (A. M. Jennings and G. Butler, unpublished observations).
A recent study by Setiadi et al. (39) has shown that in C. albicans, exposure to hypoxic conditions results in upregulation of expression of glycolytic genes and downregulation of expression of genes involved in oxidative metabolism. This is similar to the role suggested here for Ace2 and previously for Efg1 and Efh1 (8). In addition, opaque cells favor oxidative metabolism, while white cells favor fermentation (25). Setiadi et al. (39) also showed that whereas Efg1 regulates the expression of glycolysis and respiration genes under normal oxygen conditions, it has no role in controlling the expression of these genes in hypoxia. It is therefore possible that the effect of Ace2 on metabolism is restricted to normal oxygen conditions, and this remains to be tested. It is unlikely that Ace2 directly regulates the expression of metabolic genes. Whereas a potential Ace2 binding site was identified in the promoters of cell wall biosynthetic genes, no such binding site is present in the promoters of the glycolytic or TCA enzyme genes.
One major difference between the profiling experiments with yeast and hyphal cells is in the effect on sterol, lipid, and fatty acid synthesis. Whereas expression of many components of the ergosterol biosynthetic pathway was increased in an ace2 deletion mutant grown as yeast cells, expression of the oxygen-requiring enzymes (ERG1, ERG5, ERG11, and ERG251) was reduced in the ace2 deletion mutant grown as hyphal cells. Expression of a fatty acid synthase gene (FAS2), a gene required for sphingolipid synthesis (SUR2), and two heme biosynthetic enzyme genes (HEM1 and HEM14) was also reduced in the ace2 deletion strain grown as hyphal cells (Table 3; see Table S3 in the supplemental material). Fatty acid metabolism is increased during hyphal growth (32), and disrupting ergosterol or sphingolipid synthesis leads to defects in filamentation (31, 36, 37). Expression of these oxygen-dependent pathways is generally induced in response to hypoxia in S. cerevisiae (24), S. pombe (43), and C. albicans (19, 22, 39). In S. pombe, decreasing oxygen leads to a decrease in sterol synthesis, which is sensed by the sterol regulatory element binding protein Sre1 (16, 43). Sre1 is the major regulator of the hypoxic response in S. pombe and regulates the expression of >100 genes (43). The biological significance of the reduction in expression of components of the ergosterol synthesis pathway in an ace2 deletion strain grown as hyphal C. albicans cells is not clear, but it does suggest that the membrane composition of the hyphae formed is different from that of hyphae generated by wild-type cells. This may affect the ability of the cells to sense the oxygen concentration in the environment.
The changes in metabolism noted in the ace2 deletion strains prompted us to examine the effects of growth under low-oxygen conditions. As shown in Fig. 5, reducing the oxygen concentration to 5% or below (at a temperature of 37°C) induced filamentation in wild-type isolates. However, very few (or no) hyphae were generated by the ace2 deletion strains, irrespective of the medium used. Deleting efg1 also inhibited filamentation in response to hypoxia under these conditions. There have been several reports that embedding cells in solid media, presumably generating hypoxic conditions, also induces filamentation (8, 15). In our experiments, wild-type strains produce most filaments in embedded media at 37°C. This is likely due to the activation of contact-dependent pathways (5, 23). The regulator CZF1, which is required for filamentation in semisolid media, is most highly expressed at 37°C and is regulated by Efg1p (45). Strains deleted for ace2 did not produce filaments in embedded YPD medium at 25°C and made few filaments at 30°C and 37°C. In contrast, efg1 deletion strains generated most filaments at 25°C and fewer filaments than the wild type at 37°C (Fig. 6) (39), presumably because CZF1 was not expressed. The efg1 efh1 double deletion mutant was hyperfilamentous under most conditions, as previously reported (8), although it did not generate hyphae in the presence of antimycin A (Fig. 4). Surprisingly, the carbon source used has a dramatic effect on filamentation. Replacing glucose (YPD) with sucrose (YPS) led to increased filamentation in all strains (Fig. 6). The ace2 deletion strains did not generate as many hyphae as the wild type, and the colonies were smaller at low temperatures. It therefore appears that several signals are involved in the induction of filamentation in embedded media, including responses to the carbon source, to elevated temperature, to hypoxia, and to contact. Efg1 (not Ace2) is required for the response to temperature, whereas Ace2 is required for a maximal response to the presence of sucrose. It is therefore likely that Ace2 acts through different pathways from those for Efg1 and is not likely to be involved in regulation of expression of CZF1.
Deleting efg1 reduced filamentation under most conditions, including the presence of classical inducers of hyphae, such as serum (12). In contrast, ace2 deletion strains generated true hyphae in response to serum induction, upon adherence to plastic, in embedded sucrose medium, and when engulfed by macrophages (Fig. 6 and 7) (21). These phenotypes are consistent with the transcriptional profiling experiments—Efg1 is required for expression of hypha-induced genes, such as HWP1 and ECE1 (41), and Ace2 is not. In the hypoxic experiments shown in Fig. 5, the temperature and other conditions were constantly maintained, and the only variable was the concentration of oxygen. Under these conditions, the ace2 strains failed to filament, suggesting that this defect is unique to hypoxia. In S. cerevisiae, deleting the hypoxic regulator ROX1 has the opposite effect from that of deleting ACE2 in C. albicans--in the rox1 deletion strain, the expression of glycolytic enzymes is increased and the expression of TCA cycle genes is reduced (24). The ortholog of ROX1 in C. albicans (RFG1) plays no role in regulation of expression in response to hypoxia and, instead, is a regulator of filamentation (19, 22). Whereas in S. cerevisiae ROX1 acts as a repressor of genes induced by hypoxia, in C. albicans ACE2 is more likely to be an activator and is required for expression. This may contribute to the lack of virulence of ace2 deletion strains (21), as the yeast cells are likely to encounter hypoxic conditions when invading tissue.
Our results suggest that filamentation is strongly associated with metabolism and that any conditions that reduce respiration, such as lowering the oxygen concentration or using antirespiratory drugs such as antimycin A, will induce hyphal growth. In contrast, mutants with enhanced respiration and increased resistance to antimycin A (such as ace2 deletion strains) fail to filament in response to hypoxia, possibly because respiration continues at high rates even at low oxygen concentrations. However, the signaling pathway that controls the hypoxic response in C. albicans and the role of Ace2 in regulating gene expression in hyoxia remain to be elucidated.
Supplementary Material
Acknowledgments
We thank Joachim Ernst for providing the efg1 and efg1 efh1 deletion strains, Claire Kenny for carrying out the macrophage infection studies, and Cormac Taylor for assistance with the hypoxia experiments.
This work was supported by the Irish Research Council for Science, Engineering and Technology and by the Wellcome Trust (grant 072420).
Footnotes
Published ahead of print on 22 September 2006.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Alonso-Nunez, M. L., H. An, A. B. Martin-Cuadrado, S. Mehta, C. Petit, M. Sipiczki, F. Del Rey, K. L. Gould, and C. R. Vazquez de Aldana. 2005. Ace2p controls the expression of genes required for cell separation in Schizosaccharomyces pombe. Mol. Biol. Cell 16:2003-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28-36. [PubMed] [Google Scholar]
- 3.Barelle, C. J., C. L. Priest, D. M. Maccallum, N. A. Gow, F. C. Odds, and A. J. Brown. 2006. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell. Microbiol. 8:961-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassilana, M., and R. A. Arkowitz. 2006. Rac1 and Cdc42 have different roles in Candida albicans development. Eukaryot. Cell 5:321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, D. H., Jr., A. D. Giusani, X. Chen, and C. A. Kumamoto. 1999. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 34:651-662. [DOI] [PubMed] [Google Scholar]
- 6.Clemente-Blanco, A., A. Gonzalez-Novo, F. Machin, D. Caballero-Lima, L. Aragon, M. Sanchez, C. R. de Aldana, J. Jimenez, and J. Correa-Bordes. 2006. The Cdc14p phosphatase affects late cell-cycle events and morphogenesis in Candida albicans. J. Cell Sci. 119:1130-1143. [DOI] [PubMed] [Google Scholar]
- 7.Colman-Lerner, A., T. E. Chin, and R. Brent. 2001. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 107:739-750. [DOI] [PubMed] [Google Scholar]
- 8.Doedt, T., S. Krishnamurthy, D. P. Bockmuhl, B. Tebarth, C. Stempel, C. L. Russell, A. J. Brown, and J. F. Ernst. 2004. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol. Biol. Cell 15:3167-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doolin, M. T., A. L. Johnson, L. H. Johnston, and G. Butler. 2001. Overlapping and distinct roles of the duplicated yeast transcription factors Ace2p and Swi5p. Mol. Microbiol. 40:422-432. [DOI] [PubMed] [Google Scholar]
- 10.Ebanks, R. O., K. Chisholm, S. McKinnon, M. Whiteway, and D. M. Pinto. 2006. Proteomic analysis of Candida albicans yeast and hyphal cell wall and associated proteins. Proteomics 6:2147-2156. [DOI] [PubMed] [Google Scholar]
- 11.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ernst, J. F. 2000. Transcription factors in Candida albicans—environmental control of morphogenesis. Microbiology 146:1763-1774. [DOI] [PubMed] [Google Scholar]
- 13.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillum, A. M., E. Y. Tsay, and D. R. Kirsch. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179-182. [DOI] [PubMed] [Google Scholar]
- 15.Giusani, A. D., M. Vinces, and C. A. Kumamoto. 2002. Invasive filamentous growth of Candida albicans is promoted by Czf1p-dependent relief of Efg1p-mediated repression. Genetics 160:1749-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes, A. L., B. L. Todd, and P. J. Espenshade. 2005. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell 120:831-842. [DOI] [PubMed] [Google Scholar]
- 17.Ihmels, J., S. Bergmann, J. Berman, and N. Barkai. 2005. Comparative gene expression analysis by differential clustering approach: application to the Candida albicans transcription program. PLoS Genet. 1:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iyer, V. R., C. E. Horak, C. S. Scafe, D. Botstein, M. Snyder, and P. O. Brown. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409:533-538. [DOI] [PubMed] [Google Scholar]
- 19.Kadosh, D., and A. D. Johnson. 2001. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol. Cell. Biol. 21:2496-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamran, M., A. M. Calcagno, H. Findon, E. Bignell, M. D. Jones, P. Warn, P. Hopkins, D. W. Denning, G. Butler, T. Rogers, F. A. Muhlschlegel, and K. Haynes. 2004. Inactivation of the transcription factor gene ACE2 in the fungal pathogen Candida glabrata results in hypervirulence. Eukaryot. Cell 3:546-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly, M. T., D. M. MacCallum, S. D. Clancy, F. C. Odds, A. J. Brown, and G. Butler. 2004. The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence. Mol. Microbiol. 53:969-983. [DOI] [PubMed] [Google Scholar]
- 22.Khalaf, R. A., and R. S. Zitomer. 2001. The DNA binding protein Rfg1 is a repressor of filamentation in Candida albicans. Genetics 157:1503-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumamoto, C. A. 2005. A contact-activated kinase signals Candida albicans invasive growth and biofilm development. Proc. Natl. Acad. Sci. USA 102:5576-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwast, K. E., L. C. Lai, N. Menda, D. T. James III, S. Aref, and P. V. Burke. 2002. Genomic analyses of anaerobically induced genes in Saccharomyces cerevisiae: functional roles of Rox1 and other factors in mediating the anoxic response. J. Bacteriol. 184:250-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lan, C. Y., G. Newport, L. A. Murillo, T. Jones, S. Scherer, R. W. Davis, and N. Agabian. 2002. Metabolic specialization associated with phenotypic switching in Candida albicans. Proc. Natl. Acad. Sci. USA 99:14907-14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 27.Martin, D., C. Brun, E. Remy, P. Mouren, D. Thieffry, and B. Jacq. 2004. GOToolBox: functional analysis of gene datasets based on gene ontology. Genome Biol. 5:R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin-Cuadrado, A. B., E. Duenas, M. Sipiczki, C. R. De Aldana, and F. Del Rey. 2003. The endo-beta-1,3-glucanase Eng1p is required for dissolution of the primary septum during cell separation in Schizosaccharomyces pombe. J. Cell Sci. 116:1689-1698. [DOI] [PubMed] [Google Scholar]
- 29.McBride, H. J., Y. Yu, and D. J. Stillman. 1999. Distinct regions of the Swi5 and Ace2 transcription factors are required for specific gene activation. J. Biol. Chem. 274:21029-21036. [DOI] [PubMed] [Google Scholar]
- 30.McNemar, M. D., and W. A. Fonzi. 2002. Conserved serine/threonine kinase encoded by CBK1 regulates expression of several hypha-associated transcripts and genes encoding cell wall proteins in Candida albicans. J. Bacteriol. 184:2058-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukhopadhyay, K., T. Prasad, P. Saini, T. J. Pucadyil, A. Chattopadhyay, and R. Prasad. 2004. Membrane sphingolipid-ergosterol interactions are important determinants of multidrug resistance in Candida albicans. Antimicrob. Agents Chemother. 48:1778-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nantel, A., D. Dignard, C. Bachewich, D. Harcus, A. Marcil, A. P. Bouin, C. W. Sensen, H. Hogues, M. Van Het Hoog, P. Gordon, T. Rigby, F. Benoit, D. C. Tessier, D. Y. Thomas, and M. Whiteway. 2002. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 13:3452-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nasmyth, K., G. Adolf, D. Lydall, and A. Seddon. 1990. The identification of a second cell cycle control on the HO promoter in yeast: cell cycle regulation of SW15 nuclear entry. Cell 62:631-647. [DOI] [PubMed] [Google Scholar]
- 34.Nelson, B., C. Kurischko, J. Horecka, M. Mody, P. Nair, L. Pratt, A. Zougman, L. D. McBroom, T. R. Hughes, C. Boone, and F. C. Luca. 2003. RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol. Biol. Cell 14:3782-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Conallain, C., M. T. Doolin, C. Taggart, F. Thornton, and G. Butler. 1999. Regulated nuclear localisation of the yeast transcription factor Ace2p controls expression of chitinase (CTS1) in Saccharomyces cerevisiae. Mol. Gen. Genet. 262:275-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pasrija, R., S. Krishnamurthy, T. Prasad, J. F. Ernst, and R. Prasad. 2005. Squalene epoxidase encoded by ERG1 affects morphogenesis and drug susceptibilities of Candida albicans. J. Antimicrob. Chemother. 55:905-913. [DOI] [PubMed] [Google Scholar]
- 37.Pasrija, R., T. Prasad, and R. Prasad. 2005. Membrane raft lipid constituents affect drug susceptibilities of Candida albicans. Biochem. Soc. Trans. 33:1219-1223. [DOI] [PubMed] [Google Scholar]
- 38.Racki, W. J., A. M. Becam, F. Nasr, and C. J. Herbert. 2000. Cbk1p, a protein similar to the human myotonic dystrophy kinase, is essential for normal morphogenesis in Saccharomyces cerevisiae. EMBO J. 19:4524-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Setiadi, E. R., T. Doedt, F. Cottier, C. Noffz, and J. F. Ernst. 2006. Transcriptional response of Candida albicans to hypoxia: linkage of oxygen sensing and Efg1p-regulatory networks. J. Mol. Biol. 361:399-411. [DOI] [PubMed] [Google Scholar]
- 40.Simon, I., J. Barnett, N. Hannett, C. T. Harbison, N. J. Rinaldi, T. L. Volkert, J. J. Wyrick, J. Zeitlinger, D. K. Gifford, T. S. Jaakkola, and R. A. Young. 2001. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106:697-708. [DOI] [PubMed] [Google Scholar]
- 41.Sohn, K., C. Urban, H. Brunner, and S. Rupp. 2003. EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Mol. Microbiol. 47:89-102. [DOI] [PubMed] [Google Scholar]
- 42.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Todd, B. L., E. V. Stewart, J. S. Burg, A. L. Hughes, and P. J. Espenshade. 2006. Sterol regulatory element binding protein is a principal regulator of anaerobic gene expression in fission yeast. Mol. Cell. Biol. 26:2817-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vinces, M. D., C. Haas, and C. A. Kumamoto. 2006. Expression of the Candida albicans morphogenesis regulator gene CZF1 and its regulation by Efg1p and Czf1p. Eukaryot. Cell 5:825-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiss, E. L., C. Kurischko, C. Zhang, K. Shokat, D. G. Drubin, and F. C. Luca. 2002. The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J. Cell Biol. 158:885-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolfe, K. H., and D. C. Shields. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387:708-713. [DOI] [PubMed] [Google Scholar]
- 48.Wong, S., G. Butler, and K. H. Wolfe. 2002. Gene order evolution and paleopolyploidy in hemiascomycete yeasts. Proc. Natl. Acad. Sci. USA 99:9272-9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu, G., P. T. Spellman, T. Volpe, P. O. Brown, D. Botstein, T. N. Davis, and B. Futcher. 2000. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature 406:90-94. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.