Abstract
The Schizosaccharomyces pombe global corepressors Tup11 and Tup12, which are orthologs of Saccharomyces cerevisiae Tup1, are involved in glucose-dependent transcriptional repression and chromatin alteration of the fbp1+ gene. The fbp1+ promoter contains two regulatory elements, UAS1 and UAS2, one of which (UAS2) serves as a binding site for two antagonizing C2H2 Zn finger transcription factors, the Rst2 activator and the Scr1 repressor. In this study, we analyzed the role of Tup proteins and Scr1 in chromatin remodeling at fbp1+ during glucose repression. We found that Scr1, cooperating with Tup11 and Tup12, functions to maintain the chromatin of the fbp1+ promoter in a transcriptionally inactive state under glucose-rich conditions. Consistent with this notion, Scr1 is quickly exported from the nucleus to the cytoplasm at the initial stage of derepression, immediately after glucose starvation, at which time Rst2 is known to be imported into the nucleus. In addition, chromatin immunoprecipitation assays revealed a switching of Scr1 to Rst2 bound at UAS2 during glucose derepression. On the other hand, Tup11 and Tup12 persist in the nucleus and bind to the fbp1+ promoter under both derepressed and repressed conditions. These observations suggest that Tup1-like proteins recruited to the fbp1+ promoter are controlled by either of two antagonizing C2H2 Zn finger proteins. We propose that the actions of Tup11 and Tup12 are regulated by reciprocal nuclear shuttling of the two antagonizing Zn finger proteins in response to the extracellular glucose concentration. This notion provides new insights into the molecular mechanisms of the Tup family corepressors in gene regulation.
A proper response to extracellular stresses is vital for the homeostasis of biological systems. Therefore, transcriptional regulation in response to stress signaling must be rigorously controlled. Transcription preferentially occurs in accessible chromatin domains, where acetylation of histones and local chromatin remodeling are induced to facilitate the recruitment of transcriptional regulators and RNA polymerases onto DNA. Such local chromatin accessibility is under the regulation of transcription activators and repressors that can bind specifically to cis-acting regulatory elements and subsequently recruit coactivators and corepressors, respectively (26, 34, 41).
The Saccharomyces cerevisiae Tup1 protein is a global corepressor with WD40 repeats that can interact with Ssn6 (35, 47) and has been suggested to be a potential yeast ortholog of Groucho (reviewed in reference 4) family corepressors (7). The Ssn6-Tup1 complex regulates the expression of numerous genes controlled by a variety of DNA binding proteins involved in transcriptional repression under the control of cell type, glucose, DNA damage, and other cellular stress signals (36, 49). Tup1 binds to histones, histone deacetylases, transcription regulators, and RNA polymerase II (5, 12, 35, 51, 53). This suggests potential roles of the Ssn6-Tup1 complex in transcriptional regulation by altering chromatin or the stability of the transcription machinery. In fact, the Ssn6-Tup1 complex has been shown to establish repressive chromatin structures around promoters (3, 8, 9) and to inhibit the function of the basal transcription machinery (24, 35, 56). Tup1 is recruited to the promoters of target genes via interactions with various sequence-specific DNA binding repressors. For example, Tup1 is recruited by the Mig1 repressor to glucose-repressed genes (46).
The Schizosaccharomyces pombe (fission yeast) Tup11 and Tup12 proteins are redundant counterparts of Tup1 which are involved in transcriptional glucose repression of the fbp1+ gene, encoding fructose-1,6-bisphosphatase (20, 30). Furthermore, Tup11 and Tup12 are required for the proper induction of chromatin alteration and later activation of transcription for specific environmental stresses at the fbp1+ and cta3+ promoters (15). The closest S. pombe homolog of Mig1 is Scr1, a C2H2 Zn finger protein that represses the transcription of inv1+ (44) and fbp1+ (31). Note that Mig1 and Scr1 are highly conserved around their C2H2 Zn finger domains (Fig. 1A). In addition, an scr1+ deletion displays genetic interactions with deletion of either tup11+ or tup12+ (20).
FIG. 1.
Tup11-Tup12 and Scr1 act in the same pathway to regulate fbp1+ expression. (A) Alignment of C2H2 Zn finger domains in S. pombe Scr1 and Rst2 and S. cerevisiae Mig1 by ClustalW (45; http://www.ebi.ac.uk/clustalw/). Identical amino acid residues are shaded. (B) Cells of the wild-type strain (K131) were cultured in YER (containing 6% glucose), and the density of the culture was monitored by the optical density at 600 nm (OD600). Cells were harvested at the times indicated by “M” and “S.” (C) Results of Northern analysis. Cells of the wild-type strain (WT; K131) and the tup11Δ tup12Δ (tupΔΔ; PKH40), scr1Δ (PKH164), and scr1Δ tup11Δ tup12Δ (PKH186) mutant strains were cultured in YER (M, glucose +) and collected at the densities indicated in panel B. Glucose starvation was carried out by transferring a portion of the mid-log-phase cells to YED (containing 0.1% glucose and 3% glycerol) and culturing them for 3 h (M, glucose −). Expression of fbp1+ was analyzed by Northern blotting. Expression of cam1+ (43) was also analyzed and used as an internal control to normalize the expression levels of fbp1+.
Transcription of the fbp1+ gene is regulated in response to environmental glucose (17-19, 48). Exposure of fission yeast cells to a high concentration of extracellular glucose results in an intracellular cyclic AMP (cAMP) signal (2, 25, 29) to activate the cAMP-dependent kinase protein kinase A (PKA) by dissociation of the regulatory subunit Cgs1 from the catalytic subunit Pka1 (reviewed in reference 55). The activated PKA signal then represses the transcription of genes, including fbp1+ (2, 17, 21), by inhibiting the C2H2 Zn finger transcription activator Rst2 (13, 23). Thus, the Rst2 C2H2 Zn finger protein is assumed to have an antagonizing role to that of the C2H2 Zn finger repressor protein Scr1. Interestingly, Rst2 has significant homology to Mig1 and Scr1 at the C2H2 Zn finger domain (Fig. 1A).
PKA activation is also antagonistic to a pathway of stress-activated protein kinases (SAPKs), i.e., Spc1/Sty1. Glucose starvation stimulates the SAPK pathway, leading to the derepression of fbp1+ transcription (22, 40, 42). The SAPK signal is mediated by the CREB/ATF-type transcription factor Atf1 (22, 39, 42, 52), a basic leucine zipper (bZIP) protein forming a heterodimer with another bZIP protein, Pcr1 (50).
Gene regulation of fbp1+ requires two upstream cis-acting elements, called UAS1 and UAS2, which include cAMP response element-like and stress response element (STRE)-like DNA sequences (31). Aft1-Pcr1 and Rst2 can bind to the UAS1 and UAS2 sequences, respectively (13, 31). A protein complex is formed on UAS2 in an Scr1-dependent manner, suggesting a direct or indirect interaction of the Scr1 repressor with UAS2 (31). Thus, UAS2 appears to serve as a common binding site for the antagonizing C2H2 Zn finger Rst2 activator and Scr1 repressor.
We previously reported that Tup11 and Tup12, together with Rst2, are involved in the chromatin opening at fbp1+ during glucose derepression (15). It has been reported that scr1+ displays a genetic interaction with tup11+ and tup12+ in glucose repression at fbp1+. Thus, the Tup11-Tup12 corepressors may repress chromatin alteration at fbp1+ with Scr1. However, cooperative mechanisms of Scr1 and Tup11-Tup12 in the chromatin regulation at fbp1+ remain to be elucidated.
In this study, we present evidence for reciprocal nuclear translocation of the two counteracting Zn finger proteins Scr1 and Rst2, with which the Tup11-Tup12 proteins function to induce proper chromatin responses of the fbp1+ promoter during glucose repression and derepression. We discuss a model for the regulation of transcriptional repression by Tup family corepressors and C2H2 Zn finger proteins.
MATERIALS AND METHODS
Fission yeast strains, genetic methods, and media.
The S. pombe strains used in this study are listed in Table 1. General genetic procedures were carried out as described previously (11). Minimal medium (SD) (38) was used for the culture of S. pombe unless stated otherwise. Strain construction was carried out by mating haploids on sporulation medium (SPA) (11), followed by tetrad dissection. The standard rich yeast extract medium YEL (with 2% glucose) (11) was used for culturing cells. YER medium (yeast extract with 6% glucose) and YED medium (yeast extract with 0.1% glucose plus 3% glycerol) were used for glucose repression and derepression, respectively. Transformation was performed by the lithium acetate method as previously described (16). All strains were grown in 200 ml of YEL in 2-liter flasks at 30°C. To select kanamycin-resistant (Kanr) colonies, culture suspensions were inoculated onto YE plates, incubated for 16 h, and then replica plated onto YE plates containing 100 μg/ml of Geneticin (Sigma). For the construction of strains expressing proteins with epitope tags, we employed a PCR-based integration method (1). These strains expressing fusion proteins (Scr1-FLAG, Scr1-green fluorescent protein [Scr1-GFP], Tup11-FLAG, etc.) can properly repress and induce fbp1+ transcription, indicating that the fusion proteins are functional.
TABLE 1.
S. pombe strains used in this study
| Strain | Genotypea |
|---|---|
| K131 | h− ade6-M26 leu1-32 |
| PKH40 | h− ade6-M26 tup11::ura4 tup12::ura4 leu1-32 ura4-D18 |
| PKH164 | h− ade6-M26 scr1::ura4 ura4-D18 |
| PKH166 | h− ade6-M26 tup11-3flag≪kanMX6 leu1-32 |
| PKH167 | h− ade6-M26 tup12-3flag≪kanMX6 leu1-32 |
| PKH168 | h− ade6-M26 tup11-GFP≪kanMX6 leu1-32 |
| PKH169 | h− ade6-M26 scr1::ura4 tup11-3flag≪kanMX6 ura4-D18 |
| PKH170 | h− ade6-M26 scr1::ura4 tup12-3flag≪kanMX6 ura4-D18 |
| PKH171 | h− ade6-M26 scr1-3flag≪kanMX6 ura4-D18 |
| PKH186 | h− ade6-M26 tup11::ura4 tup12::ura4 scr1::ura4 leu1-32 ura4-D18 |
| PKH187 | h− ade6-M26 scr1-GFP≪kanMX6 leu1-32 |
| PKH188 | h− ade6-M26 tup12-GFP≪kanMX6 leu1-32 |
| PKH241 | h− ade6-M26 scr1-GFP≪kanMX6 spc1::ura4 ura4-D18 |
| PKH243 | h− ade6-M26 scr1-GFP≪kanMX6 pka1::ura4 ura4-D18 leu1-32 his7-366 |
| PKH246 | h− ade6-M26 scr1-GFP≪kanMX6 cgs1::ura4 ura4-D18 leu1-32 |
| PKH251 | h+ ade6-M26 scr1-GFP≪kanMX6 atf1::ura4 ura4-D18 leu1-32 his3-D1 |
| PKH418 | h+ ade6-M26 scr1-GFP≪kanMX6 tor1::ura4 ura4-D18 |
| PKH453 | h− ade6-M26 rst2-3flag≪kanMX6 leu1-32 |
≪ indicates linking of the marker gene to the inserted gene.
Northern blot analysis.
The probes to detect transcripts of fbp1+ and cam1+ were prepared from PCR-amplified DNA fragments, and the DNA fragments were further labeled with 32P, using a random priming kit (Amersham Pharmacia, Piscataway, NJ). The nucleotide sequences of the primers used for fbp1+ and cam1+ probes were as described previously (15). Total RNA was prepared from S. pombe cells according to a method described elsewhere (6). For Northern blot analysis, 10 μg of total RNA was denatured with formamide, electrophoresed in 1.5% agarose gels containing formaldehyde (37), and blotted onto a charged nylon membrane (Biodyne B membrane; Pall).
Chromatin analysis.
Analysis of chromatin structure by indirect end labeling was done according to the method of Mizuno et al. (28). The DNA samples were analyzed by Southern analysis as described below. To analyze chromatin around the fbp1+ promoter, the micrococcal nuclease (MNase)-treated DNA was digested with ClaI and separated by electrophoresis in a 1.5% agarose gel (40-cm long) containing Tris-acetate-EDTA buffer. The separated DNA fragments were alkali transferred to charged nylon membranes (Biodyne B membrane; Pall). The probe used for indirect end labeling of the fbp1+ region was the same probe used for Northern analysis, as described above.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation (ChIP) was performed as described previously (54), with slight modifications, as briefly described below. Fifty milliliters of culture was incubated with 1.4 ml of a 37% formaldehyde solution for 15 min at room temperature, and 2.5 ml of 2.5 M glycine was added and incubated for 5 min. After centrifugation, collected cells were washed twice with cold Tris-buffered saline. The cells were mixed with 400 μl of lysis 140 buffer (0.1% sodium deoxycholate, 1 mM EDTA, 50 mM HEPES-KOH [pH 7.5], 140 mM NaCl, 1% Triton X-100), and 0.6 ml of zirconia beads was added. After disruption of the cells using a multibead shocker (Yasuikikai, Japan), the suspension was sonicated five times for 30 s each and centrifuged at 4°C for 10 min, and the supernatant was collected as a whole-cell extract. Three hundred microliters of whole-cell extract was mixed with 4 μl of anti-FLAG antibody M2 (Sigma) and 40 μl of DYNA protein A beads (DYNAL) and allowed to immunoprecipitate at 4°C overnight. The precipitates were washed with lysis 140 buffer twice and with lysis 500 buffer (0.1% sodium deoxycholate,1 mM EDTA, 50 mM HEPES-KOH [pH 7.5], 500 mM NaCl, 1% Triton X-100) once and then further washed with wash buffer (0.5% sodium deoxycholate, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 10 mM Tris-HCl [pH 8.0]) twice and with TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) once. The well-washed precipitates were mixed with 100 μl of elution buffer (10 mM EDTA, 1% sodium dodecyl sulfate, 50 mM Tris-HCl [pH 8.0]), and the immunoprecipitated protein-DNA complexes were eluted at 65°C for 15 min (IP sample). The IP sample or 3 μl of whole-cell extract was mixed with 150 μl or 250 μl of 1% sodium dodecyl sulfate-containing TE buffer and incubated at 37°C for 8 h. After incubation, the temperature was shifted to 65°C, and the sample was further incubated overnight. After incubation, DNA was phenol-chloroform extracted from each of the samples and slot blotted onto a charged nylon membrane (Biodyne B membrane; Pall), followed by Southern blot analysis to quantify the DNA content. Quantification was conducted using a Fuji BAS2000 image analyzer by obtaining a calibration curve with various concentrations of input genome sample. The probes were amplified from S. pombe genomic DNA by PCR, using primer sets fbp1-1 (ACGATCTAACGAAACAGGAA and CCCTTTGTGGACATTTAGAC), fbp1-2 (GAAAATTCCACGGGACATTAG and CCCTTCCTATTAGCAATAAGG), fbp1-3 (GGGATGAAAACAATCAACCTC and GGAATGCAGCAACGAAAATC), fbp1-4 (GATTTTCGTTGCTGCATTCC and CCTATGATTTGATGTCTAGC), fbp1-5 (GCTAGACATGAAATGATACC and CATTCCACCCTATTCATC), fbp1-6 (GGGTGGAATGAGTCCGC and GTTCCGCGAATCATAAGCC), fbp1-7 (CGCGGAACTAAACATAGCG and GCTAGAAACCGAGTGGTG), fbp1-8 (GCCCAACTTAACTCAGCTC and GCTTCTGATTGTATCGGCG), and fbp1-9 (CGCCGATACAATCAGAAGC and CGATGAGTTTGCAGCATCC).
To investigate whether Scr1-3Flag and Rst2-3Flag bind to UAS2 in the fbp1+ promoter, the primer set fbp1-6 was used together with the control primer set ade10 (ACGTAGCAAACAAAGCAG and CTAATTCCTACAGAACTG).
Fluorescence microscopy.
The cells were collected by filtration and suspended with 5 μl of culture on a slide glass. Fluorescence images of living cells were taken with a cooled charge-coupled device camera and stored digitally using MetaMorph software (Universal Imaging, Downingtown, PA). For fixed samples, the culture was centrifuged for 5 s, and the pelleted cells were suspended in 70% ethanol, washed with phosphate-buffered saline, and suspended in 5 μl of phosphate-buffered saline containing Hoechst 33342 dye (0.1 mg/ml).
RESULTS
Tup11-Tup12 corepressors and the Scr1 repressor function in the same pathway to repress chromatin remodeling around the fbp1+ promoter.
The fission yeast Tup11-Tup12 corepressors are required for glucose repression of fbp1+ transcription, possibly collaborating with the C2H2 Zn finger Scr1 repressor (20, 30, 31). To investigate their cooperative roles in chromatin regulation, we first investigated the genetic relationship between the tup11Δ tup12Δ double deletion and scr1Δ deletion strains. Both the tup11Δ tup12Δ and scr1Δ strains were cultured to mid-log phase (Fig. 1C, lanes M) or early stationary phase (lanes S) in YER medium containing 6% glucose (for glucose-rich conditions). The cells in mid-log phase were cultured further for 3 h in YED medium containing 0.1% glucose (for glucose-starved conditions). As previously reported (15, 20), a robust transcriptional activation of fbp1+ was detected by Northern analysis after glucose starvation of wild-type cells. The tup11Δ tup12Δ strain exhibited a slight derepression of fbp1+ transcription in glucose-fed cells at early stationary phase.
The scr1Δ mutation also conferred a slight induction of fbp1+ transcription, even at early stationary phase (relative ratios compared to the derepressed level in the wild type, 0.5 and 0.1 for the tup11Δ tup12Δ and scr1Δ strains, respectively), whereas no induction was observed in wild-type cells under the same conditions (Fig. 1C, lanes S and +). The expression levels of fbp1+ in the tup11Δ tup12Δ and scr1Δ strains were 2.2 and 1.4 times higher, respectively, than that in the wild type at mid-log phase under glucose-starved conditions (Fig. 1C, lanes M and −). Thus, the effects of the tup11Δ tup12Δ double deletion on fbp1+ glucose repression are much more severe than those of the scr1Δ single deletion. More importantly, the phenotype of a triple deletion mutant deleted for tup11+, tup12+, and scr1+ was similar to the case for the tup11Δ tup12Δ mutant at the mid-log and early stationary phases. Thus, Scr1 appears to act in the same pathway as Tup11 and Tup12.
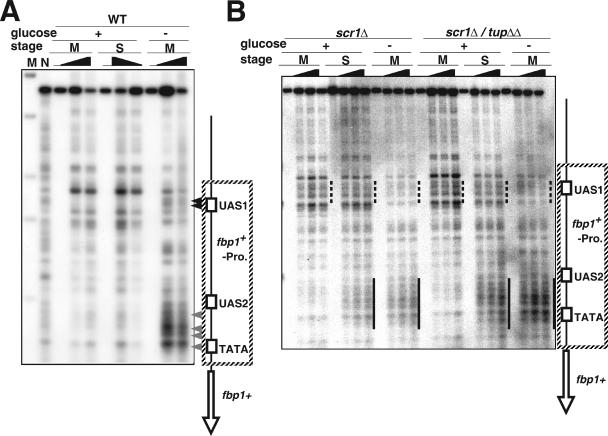
We further analyzed the chromatin structure around the fbp1+ promoter under the conditions described above. In the wild-type strain grown under glucose-rich conditions as previously reported (15), chromatin around the UAS1 and UAS2-TATA sites was relatively resistant to MNase digestion, presenting only a couple of bands around the UAS1 site (Fig. 2A). Under glucose-starved conditions at mid-log phase, some faint bands appeared in the UAS1 region (Fig. 2A, filled arrowheads), and several hypersensitive sites were generated around the UAS2-TATA region (Fig. 2A, gray arrowheads).
FIG. 2.
Tup11-Tup12 and Scr1 act together to regulate the chromatin structure in the fbp1+ promoter. (A and B) Chromatin structures around the fbp1+ promoter in wild-type (WT), scr1Δ, and scr1Δ tup11Δ tup12Δ (scr1Δ tupΔΔ) strains, using the same strains as those described in the legend to Fig. 1C. Lanes contain chromatin from mid-log-phase (M) and early-stationary-phase (S) cells. The isolated chromatin was digested with 0, 20, 30, or 50 units/ml of MNase at 37°C for 5 min. Purified DNA was digested with ClaI and analyzed by Southern blot analysis as described in Materials and Methods. Black and gray arrowheads indicate regions with MNase-sensitive sites within UAS1 (the open square labeled UAS1, positions −1566 to −1573 from the first A of the fbp1+ open reading frame) and around UAS2 (the open square labeled UAS2, positions −926 to −938), respectively.
On the other hand, in the scr1Δ and scr1Δ tup11Δ tup12Δ strains, some sensitive sites appeared constitutively in the UAS1 region from the mid-log to early stationary stages, even when the cells were cultured in a glucose-rich medium (Fig. 2B, dashed lines). For the chromatin from scr1Δ tup11Δ tup12Δ cells grown to early stationary phase under glucose-rich conditions, several hypersensitive sites appeared around the UAS2-TATA region (Fig. 2B, thick lines), similar to those for the glucose-starved wild-type cells. Chromatin from the scr1Δ mutant cells exhibited a similar but weaker pattern of hypersensitive sites around the UAS2-TATA region, especially under glucose-rich conditions at early stationary phase (Fig. 2B, thick lines). Under the same conditions, the scr1Δ mutant and scr1Δ tup11Δ tup12Δ triple mutant exhibited a chromatin configuration very similar to that of glucose-starved wild-type cells. These results indicate that the Scr1 repressor and Tup11-Tup12 corepressors function in the same pathway to establish or maintain transcriptionally repressed chromatin in the fbp1+ region.
Exchange of Scr1/Rst2 on UAS2 in response to glucose starvation.
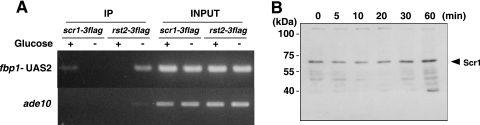
Both the Scr1 repressor and the Rst2 activator are C2H2 Zn finger proteins that appear to bind UAS2 in the fbp1+ promoter (13, 31). However, Scr1 and Rst2 have opposite roles in regulating fbp1+ transcription, and the Rst2 activator is thought to act to create a more open chromatin structure in the fbp1+ promoter region, in opposition to the action of Tup11-Tup12 (15). To address how these related proteins function antagonistically while sharing the same binding site, we first conducted ChIP analysis and examined the binding of the Scr1 repressor and the Rst2 activator to UAS2 in the fbp1+ promoter. We found that Rst2 binds to UAS2 under glucose-starved conditions, while Scr1 binds to UAS2 under glucose-rich conditions (Fig. 3A). This result indicates that Rst2 replaces Scr1 at UAS2 in response to glucose starvation.
FIG. 3.
Scr1 and Rst2 bind to UAS2 in the fbp1+ promoter under repressing and derepressing conditions, respectively. (A) Cells of scr1-3flag (PKH171) and rst2-3flag (PKH453) strains were cultured in YER to mid-log phase, and portions were shifted to YED medium and cultured for 60 min. ChIP experiments were conducted as described in Materials and Methods. The binding of Scr1-3FLAG and Rst2-3FLAG to UAS2 in the fbp1+ promoter was detected by PCR. Whole genomic DNA (1% input) was amplified at the same time. The primer set amplifying the ade10 locus was used as a control. (B) Cells of the scr1-3flag (PKH171) strain were cultured in YER to mid-log phase, and portions were shifted to YED medium and harvested at the indicated times after the medium shift. Protein samples were prepared from the same amount of cells (5 × 107 cells). The arrowhead indicates the band corresponding to Scr1-3Flag.
To examine whether the Scr1-Rst2 switch in response to glucose starvation is regulated by posttranslational modifications (e.g., phosphorylation) or degradation of the Scr1 protein, we examined the level of Scr1 protein in the course of glucose starvation by Western blotting. As shown in Fig. 3B, no visible changes in protein level or mobility, which would be suggestive of a posttranslational modification, were detected. However, we cannot exclude the possibility of posttranslational modifications without mobility shifts, and hence it still remains unsolved whether Scr1-Rst2 switching is regulated via posttranslational protein modification.
Nuclear export of the Scr1 repressor.
Higuchi et al. reported that the function of Rst2 is regulated by its nuclear localization (13). Rst2 is exported from the nucleus in response to glucose/cAMP signaling in a PKA pathway-dependent manner. These observations led us to speculate that switching between Scr1 and Rst2 on UAS2 may be controlled by nuclear import and export.
To test this possibility, we constructed a strain expressing Scr1-GFP and analyzed the protein's subcellular localization. We found that Scr1-GFP localizes to the nucleus under glucose-rich conditions and to the cytoplasm under glucose-starved conditions (Fig. 4A). A time-lapse examination of Scr1-GFP revealed that Scr1-GFP in the nucleus under glucose-rich conditions is exported quickly to the cytoplasm within 5 min of exposure to glucose-starved conditions (Fig. 4B). We further confirmed that nuclear import of Scr1-GFP occurs immediately after the reexposure of cells to glucose (Fig. 4B). Moreover, we examined the behavior of Tup11-GFP and Tup12-GFP. As shown in Fig. 4C, Tup11-GFP and Tup12-GFP were found in the nucleus under either glucose-rich or glucose starvation conditions, indicating that Tup11 and Tup12 are not regulated by changes in localization (Fig. 4C).
FIG. 4.
Scr1 translocates from the nucleus to the cytoplasm upon glucose starvation. (A) Cells of scr1-GFP strain PKH187 were cultured in YER to mid-log phase, and a portion was transferred to YED. The cells were fixed and observed as described in Materials and Methods. Bar, 10 μm. (B) Live observation of Scr1-GFP. Cells of the scr1-GFP strain (PKH 187) were transferred from YER to YED (glucose positive to glucose negative) or from YED to YER (glucose negative to glucose positive). The amount of time after the medium change is indicated. Bars, 10 μm. (C) Tup11 and Tup12 persist in the nucleus under repressing and derepressing conditions. Cells of the tup11-GFP (PKH168) and tup12-GFP (PKH188) strains were cultured, fixed, and observed as described for panel A. Bars, 10 μm.
Since the nuclear localization of Rst2 is regulated by PKA phosphorylation (13), we further tested the localization of Scr1-GFP in strains lacking a functioning mitogen-activated protein (MAP) kinase pathway (spc1Δ and atf1Δ strains), PKA pathway (cgs1Δ and pka1Δ strains) (reviewed in reference 55), or TOR pathway (tor1Δ strain) (27). We detected proper localization patterns of Scr1-GFP in all mutant strains (Fig. 5), indicating that these intracellular signaling pathways do not regulate the nuclear localization of Scr1.
FIG. 5.
scr11-GFP strains lacking pka1 (PKH243), cgs1 (PKH246), atf1 (PKH251), spc1 (PKH241), or tor1 (PKH418) were cultured, fixed, and observed as described in the legend to Fig. 4A. Bar, 10 μm.
Tup11 and Tup12 occupy UAS2 in the fbp1+ promoter even under derepressed conditions.
The observations described above led us to speculate that under glucose-repressing conditions, Tup11 and Tup12 are recruited to UAS2 by Scr1, which is preloaded on UAS2 in place of Rst2. To test this notion, we analyzed the binding of Tup11 and Tup12 to the fbp1+ promoter by ChIP analysis. For quantitative analysis, the 5′ promoter region of fbp1+ was divided into segments of ∼250 bp (Fig. 6A), and the probe for each region was used in slot blot analysis to measure the ChIP efficiency at that segment.
FIG. 6.
Tup11-Tup12 binding to the UAS2 region of the fbp1+ promoter. (A) Schematic drawing of the probes used to quantify DNA precipitated with Tup11 or Tup12. Probes 3, 6, and 7 contain UAS1, UAS2, and the TATA box, respectively. (B) Chromatin immunoprecipitation of Tup11 and Tup12. Cells of the tup11-3flag (PKH166) and tup12-3flag (PKH167) strains were cultured in YER to mid-log phase for the glucose-positive samples. One-half of each culture was transferred to YED and grown for 3 h for the glucose-negative samples. DNAs from 1% input and IP samples were quantified by slot blotting followed by hybridization with the above probes. IP efficiencies are presented (% IP). (C) Scr1 is dispensable for loading of Tup11-Tup12 onto the fbp1+ promoter UAS2. The tup11-3flag strain lacking scr1 (PKH169) and the tup12-3flag strain lacking scr1 (PKH 170) were cultured as described for panel B. To quantify the binding of Tup11 and Tup12 to UAS2, probe 6 was used for hybridization.
We found that the FLAG-tagged versions of Tup11 and Tup12 were enriched in the UAS2 region. Their occupancy on DNA was significantly higher under derepressed conditions (Fig. 6B and C). Considering the amount of whole genomic DNA in the input control, the IP efficiencies of Tup11 and Tup12 around UAS2 were estimated to be ∼1.0% and ∼0.2% under derepressed and repressed conditions, respectively. We further examined the requirement of Scr1 in the binding of Tup11 and Tup12 to UAS2 (Fig. 6C) and found that the Scr1 repressor was dispensable for their binding. Therefore, Scr1 is not required for the recruitment of Tup11 and Tup12 to the fbp1+ promoter. Tup1 and Groucho cannot bind to DNA directly (reviewed in reference 4), so other DNA binding proteins may be required for the recruitment of Tup11and Tup12 to DNA. Several proteins with zinc fingers related to those of Rst2 and Scr1 are encoded by the S. pombe genome and may contribute to the multiple band shifts observed with the UAS2 probe (31).
DISCUSSION
In the present study, we characterized the roles of the Scr1 repressor and the Tup1-like corepressors Tup11 and Tup12 during glucose repression of fbp1+. We found that Tup proteins function in concert with the Zn finger Scr1 repressor and Rst2 activator proteins, which undergo reciprocal nuclear shuttling and the resultant exchange of their occupancies at UAS2. Importantly, it turned out that Tup proteins persist at UAS2 under both repressed and derepressed conditions.
Switching of Zn finger proteins Scr1 and Rst2 to regulate the function of Tup11-Tup12 complexes.
We showed that Scr1, a Zn finger repressor, and the Tup11-Tup12 corepressors function in concert to repress chromatin remodeling in the fbp1+ promoter and the transcription of fbp1+ (Fig. 1 and 2). Since we previously reported that Rst2, a Zn finger activator, acts to antagonize the function of Tup11-Tup12 in chromatin repression, it is supposed that Scr1 and Rst2 act toward Tup proteins in a mutually antagonizing manner (15). Interestingly, the Zn finger domains of S. pombe Rst2 and Scr1 and S. cerevisiae Mig1 are highly conserved (Fig. 1A), and they can bind to a common consensus sequence called STRE. Rst2 and Scr1 are supposed to share the binding site on the STRE-containing UAS2 regulatory element in the fbp1+ promoter (13, 31). Thus, it is supposed that these Zn finger proteins compete with each other for the common binding site. In fact, our ChIP data demonstrate that the Scr1 repressor and Rst2 activator bind to UAS2 under repressing and derepressing conditions, respectively (Fig. 3A).
Interestingly, Scr1 rapidly translocates from the nucleus to the cytoplasm in response to glucose starvation (Fig. 4). This localization pattern is reciprocal to that of Rst2 (13). Therefore, such reciprocal nuclear shuttling can avoid conflicts between the Rst2 activator and the Scr1 repressor at an STRE in the fbp1+ promoter. These results suggest that Tup proteins are regulated to be repressive or nonrepressive by exchanging the Zn finger proteins Scr1 and Rst2 (Fig. 7). This explains why the Rst2 activator is dispensable for fbp1+ transcription per se in tup11Δ tup12Δ mutants (15). Presumably, Scr1 facilitates the ability of Tup proteins to repress fbp1+ transcription under glucose-rich conditions, and once cells encounter glucose starvation conditions, Scr1 is replaced by Rst2 on UAS2. This may be the first step toward the activation of transcription, which might involve the subsequent action of the Aft1-Pcr1 activator at UAS1. It will be intriguing to learn whether or not similar reciprocal shuttling of antagonizing Zn finger proteins occurs to control transcriptional regulation in higher eukaryotes.
FIG. 7.
Model of chromatin remodeling regulation around the fbp1+ promoter region by switching between two antagonizing Zn finger proteins. See Discussion for details.
Tup11 and Tup12 persist at the fbp1+ promoter under derepressing conditions.
We have shown here that Tup11 and Tup12 bind persistently to UAS2 (Fig. 6). These results suggest that the major function of Tup proteins is to regulate chromatin configurations in repressive or nonrepressive states by replacing the Zn finger proteins Scr1 and Rst2 (Fig. 7) rather than by simply inducing transcriptional repression. This idea seems reasonable, since S. cerevisiae Tup1 also resides at promoters to activate chromatin alteration by recruiting a histone acetyltransferase complex (32), and more importantly, Tup1 occupancy at the SUC2 promoter also increases under derepressing conditions (32).
The persistent binding of Tup proteins at the fbp1+ promoter during derepression suggests that the binding of Tup proteins to a promoter does not necessarily result in transcriptional repression. Tup proteins may function as transcriptional activators and stay at the promoter to promote chromatin remodeling by recruiting SAGA and SWI/SNF remodeling complexes under derepressing conditions, as proposed in the case of S. cerevisiae (32, 33). This seems unlikely, though, since a robust transcriptional activation of fbp1+ was observed in the tup11Δ tup12Δ strain. Alternatively, Tup proteins may function as transcriptional regulators that allow specific chromatin responses to distinct extracellular environments, as suggested previously (10). In fact, we previously demonstrated that the tup11Δ tup12Δ mutations conferred chromatin remodeling and unusual transcription activation at fbp1+ in response to stresses that do not normally induce fbp1+ transcription (14). We speculate that Tup proteins bound to the fbp1+ UAS2 may prevent nonspecific chromatin remodeling that is not coupled to specific changes in extracellular circumstances. The exchange of the Zn finger proteins Scr1 and Rst2 may regulate the activity of Tup proteins to establish appropriate and specific transcriptional activation in the fbp1+ promoter. As such, transcriptional activity does not simply correlate with the amount of Tup proteins bound to individual promoters. However, more studies will be needed to understand how the Tup proteins act to repress transcription under glucose-rich conditions yet allow high-level transcription under derepressing conditions, at which time they display increased occupancy at the fbp1+ promoter.
Proper responses to extracellular stresses are vital for the survival of all eukaryotes. For the appropriate responses, biological systems have developed many signaling pathways to activate specific genes depending on the distinct environmental stresses. As discussed above, Tup proteins may act as a regulation center, interacting with both positive and negative regulators under distinct conditions to ensure the specificity of transcriptional activation in response to particular stresses. Such sophisticated gene regulation by Tup proteins, rather than their simple action as repressors, could also be important in higher eukaryotes, because the system consisting of MAP kinase pathways, PKA pathways, Tup proteins, and Zn finger proteins is highly conserved. More precise molecular investigations of Tup proteins and their Zn finger partners will provide further understanding of genetic responses to environmental stresses.
Acknowledgments
K.H. thanks all members of the Genetic System Regulation Laboratory and the Cellular & Molecular Biology Laboratory at RIKEN for helpful discussions and Takehiko Shibata for valuable comments. We thank Takatomi Yamada for critical advice regarding chromatin immunoprecipitation analysis. We also thank Y. Ichikawa and R. Nakazawa for DNA sequencing and Y. Sakuma for technical assistance.
This work was supported by grants from the following sources: a basic research grant from the Bio-Oriented Technology Research Advancement Institution (to T. Shibata and K. Ohta) and grants-in-aid for scientific research on priority areas from the Ministry of Education, Science, Culture, and Sports of Japan (to K. Ohta).
Footnotes
Published ahead of print on 6 October 2006.
REFERENCES
- 1.Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 2.Byrne, S. M., and C. S. Hoffman. 1993. Six git genes encode a glucose-induced adenylate cyclase activation pathway in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 105:1095-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper, J. P., S. Y. Roth, and R. T. Simpson. 1994. The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes Dev. 8:1400-1410. [DOI] [PubMed] [Google Scholar]
- 4.Courey, A. J., and S. Jia. 2001. Transcriptional repression: the long and the short of it. Genes Dev. 15:2786-2796. [DOI] [PubMed] [Google Scholar]
- 5.Edmondson, D. G., M. M. Smith, and S. Y. Roth. 1996. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 10:1247-1259. [DOI] [PubMed] [Google Scholar]
- 6.Elder, R. T., E. Y. Loh, and R. W. Davis. 1983. RNA from the yeast transposable element Ty1 has both ends in the direct repeats, a structure similar to retrovirus RNA. Proc. Natl. Acad. Sci. USA 80:2432-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flores-Saaib, R. D., and A. J. Courey. 2000. Analysis of Groucho-histone interactions suggests mechanistic similarities between Groucho- and Tup1-mediated repression. Nucleic Acids Res. 28:4189-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gavin, I. M., M. P. Kladde, and R. T. Simpson. 2000. Tup1p represses Mcm1p transcriptional activation and chromatin remodeling of an a-cell-specific gene. EMBO J. 19:5875-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gavin, I. M., and R. T. Simpson. 1997. Interplay of yeast global transcriptional regulators Ssn6p-Tup1p and Swi-Snf and their effect on chromatin structure. EMBO J. 16:6263-6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenall, A., A. P. Hadcroft, P. Malakasi, N. Jones, B. A. Morgan, C. S. Hoffman, and S. K. Whitehall. 2002. Role of fission yeast Tup1-like repressors and Prr1 transcription factor in response to salt stress. Mol. Biol. Cell 13:2977-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutz, H., H. Heslot, U. Leupold, and N. Loprieno. 1974. Schizosaccharomyces pombe, p. 395-446. In R. D. King (ed.), Handbook of genetics, vol. 1. Plenum, New York, N.Y. [Google Scholar]
- 12.Herschbach, B. M., M. B. Arnaud, and A. D. Johnson. 1994. Transcriptional repression directed by the yeast α2 protein in vitro. Nature 370:309-311. [DOI] [PubMed] [Google Scholar]
- 13.Higuchi, T., Y. Watanabe, and M. Yamamoto. 2002. Protein kinase A regulates sexual development and gluconeogenesis through phosphorylation of the Zn finger transcriptional activator Rst2p in fission yeast. Mol. Cell. Biol. 22:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirota, K., T. Hasemi, T. Yamada, K. I. Mizuno, C. S. Hoffman, T. Shibata, and K. Ohta. 2004. Fission yeast global repressors regulate the specificity of chromatin alteration in response to distinct environmental stresses. Nucleic Acids Res. 32:855-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirota, K., C. S. Hoffman, T. Shibata, and K. Ohta. 2003. Fission yeast Tup1-like repressors repress chromatin remodeling at the fbp1(+) promoter and the ade6-M26 recombination hotspot. Genetics 165:505-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirota, K., K. Tanaka, Y. Watanabe, and M. Yamamoto. 2001. Functional analysis of the C-terminal cytoplasmic region of the M-factor receptor in fission yeast. Genes Cells 6:201-214. [DOI] [PubMed] [Google Scholar]
- 17.Hoffman, C. S., and F. Winston. 1991. Glucose repression of transcription of the Schizosaccharomyces pombe fbp1 gene occurs by a cAMP signaling pathway. Genes Dev. 5:561-571. [DOI] [PubMed] [Google Scholar]
- 18.Hoffman, C. S., and F. Winston. 1990. Isolation and characterization of mutants constitutive for expression of the fbp1 gene of Schizosaccharomyces pombe. Genetics 124:807-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffman, C. S., and F. Winston. 1989. A transcriptionally regulated expression vector for the fission yeast Schizosaccharomyces pombe. Gene 84:473-479. [DOI] [PubMed] [Google Scholar]
- 20.Janoo, R. T., L. A. Neely, B. R. Braun, S. K. Whitehall, and C. S. Hoffman. 2001. Transcriptional regulators of the Schizosaccharomyces pombe fbp1 gene include two redundant Tup1p-like corepressors and the CCAAT binding factor activation complex. Genetics 157:1205-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin, M., M. Fujita, B. M. Culley, E. Apolinario, M. Yamamoto, K. Maundrell, and C. S. Hoffman. 1995. sck1, a high copy number suppressor of defects in the cAMP-dependent protein kinase pathway in fission yeast, encodes a protein homologous to the Saccharomyces cerevisiae SCH9 kinase. Genetics 140:457-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanoh, J., Y. Watanabe, M. Ohsugi, Y. Iino, and M. Yamamoto. 1996. Schizosaccharomyces pombe gad7+ encodes a phosphoprotein with a bZIP domain, which is required for proper G1 arrest and gene expression under nitrogen starvation. Genes Cells 1:391-408. [DOI] [PubMed] [Google Scholar]
- 23.Kunitomo, H., T. Higuchi, Y. Iino, and M. Yamamoto. 2000. A zinc-finger protein, Rst2p, regulates transcription of the fission yeast ste11(+) gene, which encodes a pivotal transcription factor for sexual development. Mol. Biol. Cell 11:3205-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, M., S. Chatterjee, and K. Struhl. 2000. Genetic analysis of the role of Pol II holoenzyme components in repression by the Cyc8-Tup1 corepressor in yeast. Genetics 155:1535-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maeda, T., N. Mochizuki, and M. Yamamoto. 1990. Adenylyl cyclase is dispensable for vegetative cell growth in the fission yeast Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 87:7814-7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mannervik, M., Y. Nibu, H. Zhang, and M. Levine. 1999. Transcriptional coregulators in development. Science 284:606-609. [DOI] [PubMed] [Google Scholar]
- 27.Matsuo, T., Y. Kubo, Y. Watanabe, and M. Yamamoto. 2003. Schizosaccharomyces pombe AGC family kinase Gad8p forms a conserved signaling module with TOR and PDK1-like kinases. EMBO J. 22:3073-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizuno, K., Y. Emura, M. Baur, J. Kohli, K. Ohta, and T. Shibata. 1997. The meiotic recombination hot spot created by the single-base substitution ade6-M26 results in remodeling of chromatin structure in fission yeast. Genes Dev. 11:876-886. [DOI] [PubMed] [Google Scholar]
- 29.Mochizuki, N., and M. Yamamoto. 1992. Reduction in the intracellular cAMP level triggers initiation of sexual development in fission yeast. Mol. Gen. Genet. 233:17-24. [DOI] [PubMed] [Google Scholar]
- 30.Mukai, Y., E. Matsuo, S. Y. Roth, and S. Harashima. 1999. Conservation of histone binding and transcriptional repressor functions in a Schizosaccharomyces pombe Tup1p homolog. Mol. Cell. Biol. 19:8461-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neely, L. A., and C. S. Hoffman. 2000. Protein kinase A and mitogen-activated protein kinase pathways antagonistically regulate fission yeast fbp1 transcription by employing different modes of action at two upstream activation sites. Mol. Cell. Biol. 20:6426-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papamichos-Chronakis, M., T. Petrakis, E. Ktistaki, I. Topalidou, and D. Tzamarias. 2002. Cti6, a PHD domain protein, bridges the Cyc8-Tup1 corepressor and the SAGA coactivator to overcome repression at GAL1. Mol. Cell 9:1297-1305. [DOI] [PubMed] [Google Scholar]
- 33.Proft, M., and K. Struhl. 2002. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell 9:1307-1317. [DOI] [PubMed] [Google Scholar]
- 34.Ptashne, M., and A. Gann. 1997. Transcriptional activation by recruitment. Nature 386:569-577. [DOI] [PubMed] [Google Scholar]
- 35.Redd, M. J., M. B. Arnaud, and A. D. Johnson. 1997. A complex composed of tup1 and ssn6 represses transcription in vitro. J. Biol. Chem. 272:11193-11197. [DOI] [PubMed] [Google Scholar]
- 36.Roth, S. Y. 1995. Chromatin-mediated transcriptional repression in yeast. Curr. Opin. Genet. Dev. 5:168-173. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Sherman, F., G. Fink, and J. Hicks. 1986. Methods in yeast genetics: laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Shiozaki, K., and P. Russell. 1996. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 10:2276-2288. [DOI] [PubMed] [Google Scholar]
- 40.Stettler, S., E. Warbrick, S. Prochnik, S. Mackie, and P. Fantes. 1996. The wis1 signal transduction pathway is required for expression of cAMP-repressed genes in fission yeast. J. Cell Sci. 109:1927-1935. [DOI] [PubMed] [Google Scholar]
- 41.Struhl, K. 1995. Yeast transcriptional regulatory mechanisms. Annu. Rev. Genet. 29:651-674. [DOI] [PubMed] [Google Scholar]
- 42.Takeda, T., T. Toda, K. Kominami, A. Kohnosu, M. Yanagida, and N. Jones. 1995. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 14:6193-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeda, T., and M. Yamamoto. 1987. Analysis and in vivo disruption of the gene coding for calmodulin in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 84:3580-3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka, N., N. Ohuchi, Y. Mukai, Y. Osaka, Y. Ohtani, M. Tabuchi, M. S. Bhuiyan, H. Fukui, S. Harashima, and K. Takegawa. 1998. Isolation and characterization of an invertase and its repressor genes from Schizosaccharomyces pombe. Biochem. Biophys. Res. Commun. 245:246-253. [DOI] [PubMed] [Google Scholar]
- 45.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Treitel, M. A., and M. Carlson. 1995. Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc. Natl. Acad. Sci. USA 92:3132-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varanasi, U. S., M. Klis, P. B. Mikesell, and R. J. Trumbly. 1996. The Cyc8 (Ssn6)-Tup1 corepressor complex is composed of one Cyc8 and four Tup1 subunits. Mol. Cell. Biol. 16:6707-6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vassarotti, A., and J. D. Friesen. 1985. Isolation of the fructose-1,6-bisphosphatase gene of the yeast Schizosaccharomyces pombe. Evidence for transcriptional regulation. J. Biol. Chem. 260:6348-6353. [PubMed] [Google Scholar]
- 49.Wahi, M., K. Komachi, and A. D. Johnson. 1998. Gene regulation by the yeast Ssn6-Tup1 corepressor. Cold Spring Harb. Symp. Quant. Biol. 63:447-457. [DOI] [PubMed] [Google Scholar]
- 50.Watanabe, Y., and M. Yamamoto. 1996. Schizosaccharomyces pombe pcr1+ encodes a CREB/ATF protein involved in regulation of gene expression for sexual development. Mol. Cell. Biol. 16:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watson, A. D., D. G. Edmondson, J. R. Bone, Y. Mukai, Y. Yu, W. Du, D. J. Stillman, and S. Y. Roth. 2000. Ssn6-Tup1 interacts with class I histone deacetylases required for repression. Genes Dev. 14:2737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilkinson, M. G., M. Samuels, T. Takeda, W. M. Toone, J. C. Shieh, T. Toda, J. B. Millar, and N. Jones. 1996. The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev. 10:2289-2301. [DOI] [PubMed] [Google Scholar]
- 53.Wu, J., N. Suka, M. Carlson, and M. Grunstein. 2001. TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell 7:117-126. [DOI] [PubMed] [Google Scholar]
- 54.Yamada, T., K. I. Mizuno, K. Hirota, N. Kon, W. P. Wahls, E. Hartsuiker, H. Murofushi, T. Shibata, and K. Ohta. 2004. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J. 23:1792-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamamoto, M., Y. Imai, and Y. Watanabe. 1997. Mating and sporulation in Schizosaccharomyces pombe, p. 1037-1106. In J. R. Prigle, J. R. Broach, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 56.Zaman, Z., A. Z. Ansari, S. S. Koh, R. Young, and M. Ptashne. 2001. Interaction of a transcriptional repressor with the RNA polymerase II holoenzyme plays a crucial role in repression. Proc. Natl. Acad. Sci. USA 98:2550-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]