Abstract
Pseudomonas aeruginosa expresses two lectins which are implicated in adhesion and biofilm formation. In this study, we demonstrate that P. aeruginosa LecB is involved in pilus biogenesis and proteolytic activity. Moreover, neither lectin was involved in adhesion to human tracheobronchial mucin. We infer that some of the ascribed functions are secondary effects on other systems rather than effects of the lectins themselves.
Lectins are sugar-binding proteins that presumably bind to carbohydrates located on cell surfaces and possibly to secreted glycoproteins. They are commonly found in plants, animals, and bacteria and are thought to play roles in bacterial infections. For example, some bacterial lectins have been suggested to mediate bacterial adhesion to mammalian cells and to respiratory mucins in cystic fibrosis (CF) and to play a role in cell-cell interactions (5, 9, 10, 29). The opportunistic pathogen Pseudomonas aeruginosa synthesizes two lectins, LecA and LecB (also known as PA-IL and PA-IIL), with specificity for galactose and fucose, respectively. Many roles have been suggested for these lectins, including adhesion of the bacterium to airway epithelial cells and injury to cells (1, 15). Furthermore, LecB was recently reported to be involved in biofilm formation (27). The latter phenomenon has been ascribed to surface-exposed LecB protein. These observations, combined with the properties of lectins, led us to hypothesize that the lectins in P. aeruginosa may exert some effect on surface-exposed proteins. Although both P. aeruginosa lectins were initially thought to be located in the cytoplasm (11), later results suggested that LecB may be present on the surface of sessile Pseudomonas cells (20).
The data on the role of these lectins in bacterial adhesion are, however, not entirely clear. To date, there have not been any studies with mutants of these lectins that demonstrate a role for these proteins in bacterial adhesion. Given that LecA is intracellular and there is some question concerning the amount of LecB on the surface of the cell, we initially approached the issue of the functions of these lectins with the hypothesis that they may function in posttranslational modification of bacterial proteins, perhaps by acting as sugar carriers. However, our investigations have led us to some striking new conclusions on the function of LecB, the best-studied of these two lectins.
Comparative membrane proteome analysis.
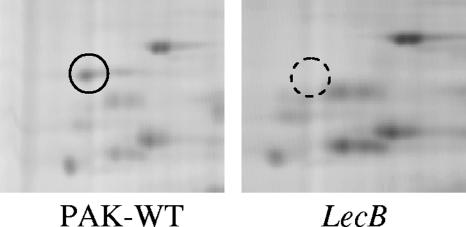
lecA and lecB mutants were constructed by insertional inactivation of the P. aeruginosa PAK lecA and lecB genes with gentamicin resistance gene cassettes. The insertions were verified by sequencing of the genes and Southern blotting. The bacterial strains and plasmids used in this study are listed in Table 1. This study was initially designed to ascertain whether there were any differences in the expression of any of the membrane proteins of the mutants as a result of the mutations. Membrane proteins were chosen because the implied actions of the lectins were all surface-related phenomena, e.g., adhesion and biofilm formation. Membrane proteins were isolated from the wild-type strain PAK and its isogenic lec mutants as described previously (12) and were subjected to two-dimensional (2-D) differential fluorescence gel electrophoresis. The preparations were labeled with fluorescent Cy3 and Cy5 dyes according to the supplier's instructions (Amersham Biosciences). Labeled samples were mixed together and run on the same isoelectric focusing and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels so that the same protein labeled with a Cy dye migrates to the same position on the 2-D gel. Image analysis of 2-D gels by Phoretix software (Nonlinear Dynamics) revealed that one protein spot identified as PilJ by QSTAR mass spectrometry was sixfold less in the lecB mutant than in the wild-type strain (Fig. 1); however, in the lecA mutant no difference in the protein expression profile was observed. pilJ is a member of an operon containing pilG to -L. To determine if the observed differential expression of PilJ is regulated at the transcriptional level, we generated a transcriptional pilG::lacZ promoter fusion construct and inserted it into P. aeruginosa strain PAK and its lecB mutant. β-Galactosidase assays were performed as described by Miller (17). No significant difference in the transcriptional activity was observed (data not shown), suggesting that the effect on the expression of PilJ seen in the 2-D studies takes place at the posttranscriptional level. Neither lectin was detected in the wild-type membrane preparations used for proteomic studies, suggesting that they may be expressed at very low levels or are not found in significant amounts in the outer membranes. Since there was a difference in the expression of the PilJ protein in the lecB mutant, the wild-type protein was subjected to gas chromatography-mass spectrometry and liquid chromatography-mass spectrometry at the Complex Carbohydrate Research Center (Athens, Ga.) to examine whether it was glycosylated. No carbohydrates were found attached to this protein.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli DH5α | hsdR recA lacZYA φ80 lacZΔM15 | GIBCO-BRL |
| P. aeruginosa | ||
| PAK | Wild type | Steve Lory |
| LecA strain | PAK LecA::Gmr | This study |
| LecB strain | PAK LecB::Gmr | This study |
| PAKpilG::lacZ | PAK harboring pDN19lacΩ with pilGHIJK promoter fused to lac | This study |
| PAKlecB::lacZ strain | LecB strain harboring pDN19lacΩ with pilGHIJK promoter fused to lacZ | This study |
| lecB+ strain | lecB complemented | This study |
| PAK-NP | PAK pilA::Tcr | 25 |
| Plasmids | ||
| pUC19 | Cloning vector, Ampr | New England Biolabs |
| pUC7G | Plasmid with a gentamicin cassette | |
| pUClecA::Gm | pUC19 containing a 2.0-kb PAK lecA gene with gentamicin cassette | This study |
| pUClecB::Gm | pUC19 containing a 2.0-kb PAK lecB gene with gentamicin cassette | This study |
| pMMB67EH | Broad-host-range cloning vector, tac promoter, Carbr | 7 |
| pDN19lacΩ | Promoterless lacZ oriV oriT Tcr Strr Ω | 28 |
| Primers | ||
| LecAFor | 5′-cccaaaggatccGCCTGCCGTCGCAGATCAACC-3′ | BamHI site inserted |
| LecARev | 5′-cccaaaaagcttGCGGGCGGCAGACCGTCC-3′ | HindIII site inserted |
| LecBFor | 5′-cccaaaggatccGCTGCACACATGGTGGCGC-3′ | BamHI site inserted |
| LecBRev | 5′-cccaaaaagcttGGAACGCATCGCCGGCCAGG-3′ | HindIII site inserted |
| LecA1 | 5′-GGGCAGGTAACGTCGATTATCTGCAGTCCGGGCGATGTCATTACCAT-3′ | Two bases incorporated to create PstI site |
| LecA2 | 5′-ATGGTAATGACATCGCCCGGACTGCAGATAATCGACGTTACCTGCCC-3′ | Two bases incorporated to create PstI site |
| LecB1 | 5′-CGAGACGGCCGCGACCTGCAGCGGGCAAAGCACCA-3′ | One base incorporated to create PstI site |
| LecB2 | 5′-TGGTGCTTTGCCCGCTGCAGGTCGCGGCCGTCTCG-3′ | One base incorporated to create PstI site |
Gmr, gentamicin resistance; Ampr, ampicillin resistance; Carbr, carbenicillin resistance; Tcr, tetracycline resistance; Strr, streptomycin resistance. In primer sequences, lowercase denotes nucleotides added to facilitate restriction digestion at the marked sites in bold. Nucleotides added to create a restriction site are underlined.
FIG. 1.
Proteomic profile of the outer membranes of P. aeruginosa PAK and its lecB mutant using a 2-D differential fluorescence gel electrophoresis system. Membrane extracts from the wild-type PAK and the lecB mutant were subjected to two-dimensional gel electrophoresis. The normal protein spot is indicated by a circle, whereas the corresponding down-regulated protein in the lecB mutant is indicated by a dashed circle.
The LecB mutant was defective in twitching motility.
In P. aeruginosa, twitching motility is mediated by type IV pili and is controlled by a complex chemosensory pathway comprised of the proteins PilG, PilH, PilI, PilJ, PilK, ChpA, ChpB, and ChpC (30). Kearns et al. (13) had observed that a P. aeruginosa pilJ mutant was deficient in twitching motility and expressed no type IV pili. Therefore, the twitching motility phenotype of the mutants was examined via the subsurface stab assay. Twitching assays demonstrated that PAK and the lecA mutant twitched normally, whereas the lecB mutant was nontwitching. This defect could be complemented by providing an intact copy of the lecB gene cloned into the low-copy-number broad-host-range plasmid pMMB67EH (8), indicating that inactivation of lecB was responsible for the loss of twitching motility.
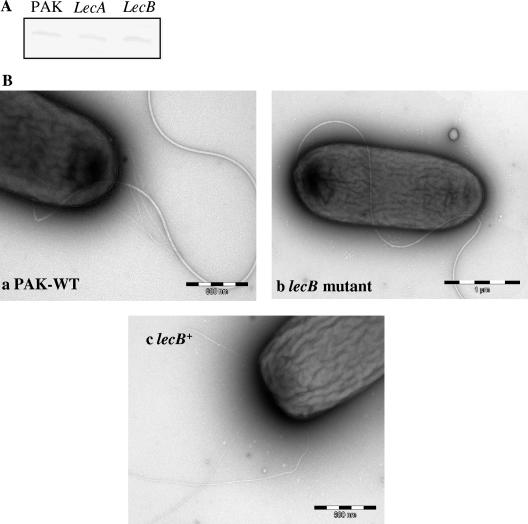
To further characterize the defect in twitching motility, Western blot analysis was performed to determine the levels of PilA, the major pilus subunit, in the lecB mutant. The mutant produced wild-type levels of intracellular PilA (Fig. 2A). However, transmission electron microscopic examination of the lecB mutant and the complemented lecB mutant demonstrated that lecB was involved in pilus biogenesis (Fig. 2B) in the mutant lacking pili. Thus, the twitching defect in the lecB mutant is not due to a transcriptional change in PilJ or PilA synthesis but is most likely due to an inability to assemble surface pili.
FIG. 2.
(A) Western blot analysis of PilA production by PAK and by lecA and lecB mutants. PilA was detected with anti-PilA antibody and is indicative of intracellular pilin in these strains. (B) Transmission electron microscopy of PAK, the lecB mutant, and the complemented lecB mutant (lecB+). Cells were used to coat carbon grids, stained with 1% phosphotungstic acid, washed, and dried. The mounted bacteria were viewed with a Hitachi H-7000 transmission electron microscope.
Impairment in biofilm formation is due to lack of pilus synthesis.
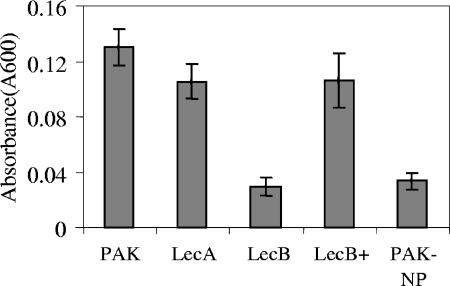
lecB, flagellar, and type IV pilus mutants of P. aeruginosa have been shown to be impaired in biofilm formation (14, 21). We therefore compared the ability of the P. aeruginosa wild-type strain, lecA and lecB mutants, and a nonpiliated PAK strain (PAK-NP) to form biofilms (22). The behavior of the lecB mutant was identical to that of the pilA mutant. As shown in Fig. 3, more than a 50% defect in biofilm formation was observed compared to the wild-type strain. The complemented lecB mutant formed a biofilm comparable to that of the wild-type strain. Student's t test was used to compare means from three experiments. Differences were considered significant when P values were <0.05. It was proposed that LecB may facilitate cell-cell interaction, since purified LecB binds to the surface of P. aeruginosa (27). The LecB protein was recently reported to be present in the outer membrane fraction (27), which would support such a role, but whether the protein is localized on the periplasmic face or the external surface of the outer membrane is unclear. With this new information that pili are not assembled in the lecB mutant, the most likely explanation for the defect in biofilm formation may be the absence of pili in this mutant.
FIG. 3.
Quantification of biofilm formation by the PAK, lecA mutant, lecB mutant, complemented lecB mutant (lecB+), and PAK-NP strains in a crystal violet microtiter plate assay after 24 h. Absorbance was measured at 600 nm. The means ± standard deviation (error bars) were determined from at least three independent experiments. The difference between the parent strain and lecB mutant was statistically significant (P < 0.05).
Role of LecB in protease activity.
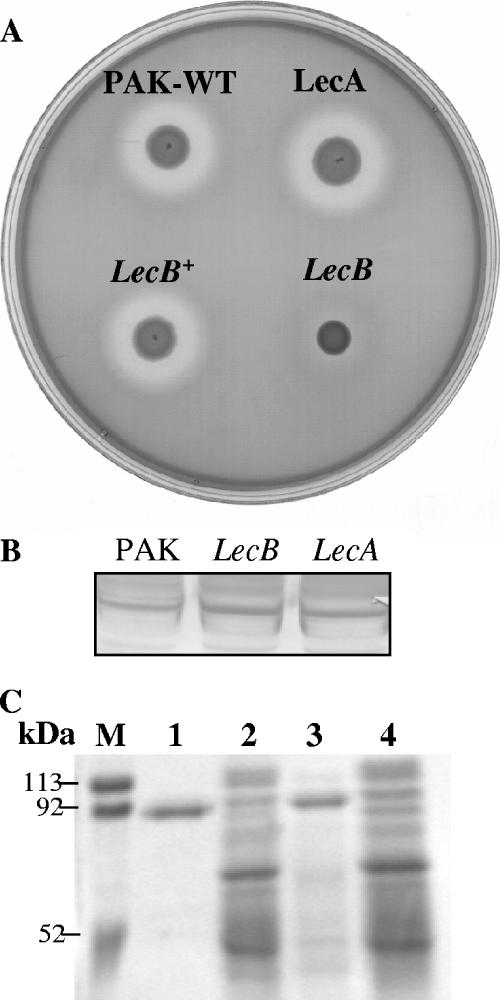
P. aeruginosa is a prolific exporter of virulence factors. Its genome harbors genes for the six known protein secretion systems that have been observed in gram-negative bacteria (26). Since the lecB mutant was defective in type IV secretion, we examined whether the mutants also had defects in other secretion systems. In P. aeruginosa, the type II secretory pathway is used to export the largest number of proteins, including phospholipase C, alkaline protease, exotoxin A, LasA, and LasB (elastase) (3, 4, 16, 19, 23). We examined the proteolytic activities of lecA and lecB mutants by growing them on casein-milk agar plates. In P. aeruginosa, caseinolytic activity is due mainly to the actions of LasB, alkaline protease, and protease IV (6). After 36 h, the wild-type strain, the lecA mutant, and the complemented lecB mutant produced a proteolytic zone, while a very weak zone was produced by the lecB mutant (Fig. 4A). We therefore examined the levels of LasB in the LecB mutant by immunoblot using antibody against LasB protein. Both the intracellular pool and secreted LasB were unaltered in the mutant (Fig. 4B), suggesting that the reduced caseinolytic activity is due not to a LasB defect but to either alkaline protease or protease IV. Alkaline protease is a metalloprotease whose activity is inhibited by EDTA (6). Therefore, to eliminate alkaline protease, we took advantage of the specific ability of protease IV to cleave lactoferrin in the presence of EDTA (31). Supernatants from overnight cultures of the wild type, the lecB mutant, and the complemented lecB mutant were concentrated 125-fold, and lactoferrin degradation by these supernatants was examined as described by Wilderman et al. (31) in the presence of EDTA. Degradation products were then analyzed by SDS-PAGE. As shown in Fig. 4C, the lecB mutant did not exhibit any lactoferrin-degrading activity, whereas hydrolysis of the substrate was observed with the wild type and the complemented lecB mutant. Since neither the secretion nor the size of LasB is affected in the lecB mutant, we presume that the defect in caseinolytic activity is probably due to modulation of protease IV activity. Notably, SDS-PAGE analysis of the secreted proteins showed that many extracellular proteins were absent from the lecB mutant.
FIG. 4.
(A) Caseinolytic activity of PAK, the lecA mutant, the lecB mutant, and the complemented lecB mutant (lecB+). Strains were grown on milk-casein agar plates. The appearance of a clear zone was observed after 36 h. (B) Western blot analysis of elastase (LasB) production by wild-type PAK and the lecA and lecB mutants. LasB was detected with anti-LasB antibody and is indicative of levels of elastase in these strains. (C) Lactoferrin degradation by wild-type PAK, the lecB mutant, and the complemented lecB mutant. Lactoferrin (1 μg) was incubated with concentrated supernatants of the bacteria at 37°C for 1 h. Metalloproteases were inhibited by adding EDTA to the supernatants. Digestion products were analyzed by SDS-PAGE, and gels were stained with Coomassie blue. Molecular mass markers are shown. Lane 1, lactoferrin (∼90 kDa); lane 2, wild-type PAK; lane 3, lecB mutant; lane 4, complemented lecB strain.
In addition, the mutants were assayed for phospholipase and exotoxin A secretion but found to be unaffected. Furthermore, no effect on the secretion of ExoS and ExoT, which are secreted through the type III secretion system (7, 32), was observed in the lectin mutants.
P. aeruginosa lectins are not involved in adhesion to respiratory mucins.
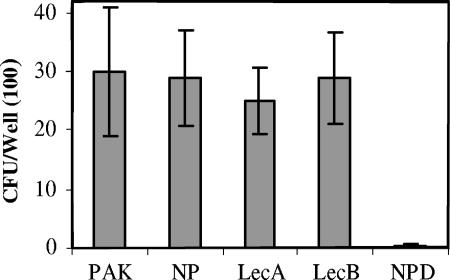
Pseudomonas lectins have been suggested but not demonstrated to be involved in adhesion to respiratory mucins in cystic fibrosis (18). In order to examine this, we performed assays of binding to CF mucins as described previously (24). A P. aeruginosa mutant lacking pili (PAK-NP) (25) attaches to mucins as efficiently as the wild-type strain (24), whereas a PAK-NP mutant lacking functional fliD (PAK-NPD), which encodes the flagellar cap protein, was severely impaired in mucin adhesion (2). These strains were used as controls. As shown in Fig. 5, both lectin mutants were unimpaired in their ability to bind to CF mucin compared to the wild-type strain, whereas the PAK-NPD mutant showed a marked defect. Respiratory mucins contain galactose and fucose, among a number of sugars; therefore, one would predict that if there is surface expression of these lectins, then mucin binding may be a prominent function. This was not demonstrable in these studies.
FIG. 5.
Adhesion of PAK, the lecA mutant, the lecB mutant, PAK-NP, and PAK-NPD to human tracheobronchial mucin. The means ± standard deviation (error bars) were determined from at least three independent experiments. Only the NPD mutant was significantly different from the wild-type strain (P < 0.001).
In summary, our results indicate that P. aeruginosa LecB is involved in multiple functions. It affects the expression of PilJ, a member of the pilus biogenesis pathway, which in turn affects pilus formation. Its role in pilus biogenesis would explain its role in biofilm formation in vitro. The studies also demonstrate a role for LecB in the proteolytic activity of P. aeruginosa. Given that this protein affects both type II and type IV secretion processes, one can assume that the action may be at the level where these two secretion pathways intersect. In addition, we have shown that neither lectin is involved in mucin adhesion. Thus, the exact place and role of these interesting lectins in cell physiology remain unsolved.
Acknowledgments
We thank Dennis Ohman, Barbara Iglewski, S. Jin, and M. Vasil for providing the antibodies used in this study.
This work was supported by NIH grant AI 47852 to R.R.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 2 October 2006.
REFERENCES
- 1.Alverdy, J., C. Holbrook, F. Rocha, L. Seiden, R. L. Wu, M. Musch, E. Chang, D. Ohman, and S. Suh. 2000. Gut-derived sepsis occurs when the right pathogen with the right virulence genes meets the right host: evidence for in vivo virulence expression in Pseudomonas aeruginosa. Ann. Surg. 232:480-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1998. The Pseudomonas aeruginosa flagellar cap protein, FliD, is responsible for mucin adhesion. Infect. Immun. 66:1000-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball, G., E. Durand, A. Lazdunski, and A. Filloux. 2002. A novel type II secretion system in Pseudomonas aeruginosa. Mol. Microbiol. 43:475-485. [DOI] [PubMed] [Google Scholar]
- 4.Bally, M., A. Filloux, M. Akrim, G. Ball, A. Lazdunski, and J. Tommassen. 1992. Protein secretion in Pseudomonas aeruginosa: characterization of seven xcp genes and processing of secretory apparatus components by prepilin peptidase. Mol. Microbiol. 6:1121-1131. [DOI] [PubMed] [Google Scholar]
- 5.Beachey, E. H. 1981. Bacterial adherence: adhesin-receptor interactions mediating the attachment of bacteria to mucosal surface. J Infect. Dis. 143:325-345. [DOI] [PubMed] [Google Scholar]
- 6.Caballero, A. R., J. M. Moreau, L. S. Engel, M. E. Marquart, J. M. Hill, and R. J. O'Callaghan. 2001. Pseudomonas aeruginosa protease IV enzyme assays and comparison to other Pseudomonas proteases. Anal. Biochem. 290:330-337. [DOI] [PubMed] [Google Scholar]
- 7.Frank, D. W. 1997. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol. Microbiol. 26:621-629. [DOI] [PubMed] [Google Scholar]
- 8.Fürste, J. P., W. Pansegrau, R. Frank, H. Blocker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 9.Gabius, H. J., S. Andre, H. Kaltner, and H. C. Siebert. 2002. The sugar code: functional lectinomics. Biochim. Biophys. Acta 1572:165-177. [DOI] [PubMed] [Google Scholar]
- 10.Gilboa-Garber, N., D. Avichezer, and N. C. Garber. 1997. Bacterial lectins: properties, structure, effects, function, and applications, p. 369-398. In H.-J. Gabius and S. Gabius (ed.), Glycosciences: status and perspectives. Chapman and Hall, Weinheim, Germany.
- 11.Glick, J., and N. C. Garber. 1983. The intracellular localization of Pseudomonas aeruginosa lectins. J. Gen. Microbiol. 129:3085-3090. [DOI] [PubMed] [Google Scholar]
- 12.Henningsen, R., B. L. Gale, K. M. Straub, and D. C. DeNagel. 2002. Application of zwitterionic detergents to the solubilization of integral membrane proteins for two-dimensional gel electrophoresis and mass spectrometry. Proteomics 2:1479-1488. [DOI] [PubMed] [Google Scholar]
- 13.Kearns, D. B., J. Robinson, and L. J. Shimkets. 2001. Pseudomonas aeruginosa exhibits directed twitching motility up phosphatidylethanolamine gradients. J. Bacteriol. 183:763-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48:1511-1524. [DOI] [PubMed] [Google Scholar]
- 15.Laughlin, R. S., M. W. Musch, C. J. Hollbrook, F. M. Rocha, E. B. Chang, and J. C. Alverdy. 2000. The key role of Pseudomonas aeruginosa PA-I lectin on experimental gut-derived sepsis. Ann. Surg. 232:133-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, P. V. 1966. The roles of various fractions of Pseudomonas aeruginosa in its pathogenesis. II. Effects of lecithinase and protease. J. Infect. Dis. 116:112-116. [DOI] [PubMed] [Google Scholar]
- 17.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Mitchell, E., C. Houles, D. Sudakevitz, M. Wimmerova, C. Gautier, S. Perez, A. M. Wu, N. Gilboa-Garber, and A. Imberty. 2002. Structural basis for oligosaccharide-mediated adhesion of Pseudomonas aeruginosa in the lungs of cystic fibrosis patients. Nat. Struct. Biol. 9:918-921. [DOI] [PubMed] [Google Scholar]
- 19.Morihara, K. 1964. Production of elastase and proteinase by Pseudomonas aeruginosa. J. Bacteriol. 88:745-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morimoto, M., H. Saimoto, H. Usui, Y. Okamoto, S. Minami, and Y. Shigemasa. 2001. Biological activities of carbohydrate-branched chitosan derivatives. Biomacromolecules 2:1133-1136. [DOI] [PubMed] [Google Scholar]
- 21.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 22.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 23.Pavlovskis, O. R., and F. B. Gordon. 1972. Pseudomonas aeruginosa exotoxin: effect on cell cultures. J. Infect. Dis. 125:631-636. [DOI] [PubMed] [Google Scholar]
- 24.Ramphal, R., L. Koo, K. S. Ishimoto, P. A. Totten, J. C. Lara, and S. Lory. 1991. Adhesion of Pseudomonas aeruginosa pilin-deficient mutants to mucin. Infect. Immun. 59:1307-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saiman, L., K. Ishimoto, S. Lory, and A. Prince. 1990. The effect of piliation and exoproduct expression on the adherence of Pseudomonas aeruginosa to respiratory epithelial monolayers. J. Infect. Dis. 161:541-548. [DOI] [PubMed] [Google Scholar]
- 26.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 27.Tielker, D., S. Hacker, R. Loris, M. Strathman, J. Wingender, S. Wilhelm, F. Rosenau, and K.-E. Jaeger. 2005. Pseudomonas aeruginosa lectin LecB is located in the outer membrane and is involved in biofilm formation. Microbiology 151:1313-1323. [DOI] [PubMed] [Google Scholar]
- 28.Totten, P. A., and S. Lory. 1990. Characterization of the type a flagellin gene from Pseudomonas aeruginosa PAK. J. Bacteriol. 172:7188-7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wentworth, J. S., F. E. Austin, N. C. Garber, N. Gilboa-Garber, C. A. Paterson, and R. J. Doyle. 1991. Cytoplasmic lectins contribute to the adhesion of P. aeruginosa. Biofouling 4:99-104. [Google Scholar]
- 30.Whitchurch, C. B., A. J. Leech, M. D. Young, D. Kennedy, J. L. Sargent, J. J. Bertrand, A. B. Semmler, A. S. Mellick, P. R. Martin, R. A. Alm, M. Hobbs, S. A. Beatson, B. Huang, L. Nguyen, J. C. Commolli, J. N. Engel, A. Darzins, and J. S. Mattick. 2004. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 52:873-893. [DOI] [PubMed] [Google Scholar]
- 31.Wilderman, P. J., A. I. Vasil, Z. Johnson, M. J. Wilson, H. E. Cunliffe, I. L. Lamont, and M. L. Vasil. 2001. Characterization of an endoprotease (PrpL) encoded by a PvdS-regulated gene in Pseudomonas aeruginosa. Infect. Immun. 69:5385-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yahr, T. L., J. Goranson, and D. W. Frank. 1996. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol. Microbiol. 22:991-1003. [DOI] [PubMed] [Google Scholar]