Abstract
A 3,463-bp plasmid, pSCM201, was isolated from a halophilic archaeon, Haloarcula sp. strain AS7094. The minimal replicon that is essential and sufficient for autonomous replication and stable maintenance in Haloarcula hispanica was determined by deletion analysis of the plasmid. This minimal replicon (∼1.8 kb) consisted of only two functionally related segments: (i) a putative origin (ori201) containing an AT-rich region and sets of repeats and (ii) an adjacent gene encoding a putative replication initiation protein (Rep201). Electron microscopic observation and Southern blotting analysis demonstrated that pSCM201 replicates via a theta mechanism. Precise mapping of the putative origin suggested that the replication initiated from a fixed site close to the AT-rich region and proceeded unidirectionally toward the downstream rep201 gene, which was further confirmed by electron microscopic analysis of the ClaI-digested replication intermediates. To our knowledge, this is the first unidirectional theta replication plasmid experimentally identified in the domain of archaea. It provides a novel plasmid system to conduct research on archaeal DNA replication.
Archaea are phylogenetically distinct from bacteria and exhibit significant similarities to eukaryotes in their genetic information processing (1, 3, 23, 40). Therefore, they have the potential of serving as simple models for understanding eukaryotic biology, particularly in the field of DNA replication (1, 40, 41). Although most of the archaeal replication proteins are eukaryote-like, others are more similar to the counterparts of bacteria or are simply archaeon-specific factors; thus, future work on archaeal genetics may provide insights into the replication mechanism and the evolution of that process (23).
Among the domain of the archaea, haloarchaea are well suited to the study of archaeal genetics. They are easy to culture and are the first archaea that could be efficiently transformed (46, 47). Haloarchaea are rich in plasmids. Plasmids represent more than 25% of the genetic material of the cells in some haloarchaeal strains (20) and could provide versatile replicons for investigations of archaeal-DNA replication. For instance, Haloferax volcanii contains four smaller replicons (plasmids and minichromosomes) ranging in size from 6.4 to 690 kb (9, 10), and Haloarcula marismortui has eight smaller replicons ranging from 33 to 410 kb (2).
Several haloarchaeal plasmids have been isolated and sequenced, some of which have been used to create haloarchaeal vectors (6, 9, 18-22, 27, 30, 36, 38, 49, 50). Interestingly, most of these plasmids (e.g., pGRB1, pHGN1, pHSB1, pHK2, and pNB101) are grouped into the family of rolling-circle-replicating (RCR) plasmids, as they encode homologous replication initiation proteins containing conserved motifs (20, 50). The single-stranded-DNA (ssDNA) forms, a hallmark of RCR plasmids, are also detected in the host strains of the plasmids pNB101 and pGRB1 (45, 50). In contrast, the plasmid pHV2 (9, 27) and megaplasmids such as pHH1 (6, 39) and pNRC100 (36, 38) are suggested to be theta replication plasmids based on their sequence information (20). The minimal replicons of pNRC100 and pHH1, which contain a large AT-rich region and at least one gene encoding the putative replication protein, have been identified (36, 39). However, the molecular details of the replication of the haloarchaeal plasmids, especially for the theta replication plasmids that may extensively use the host eukaryote-like replication machinery, have yet to be established.
pSCM201 is a novel plasmid isolated from the halophilic archaeon Haloarcula sp. strain AS7094, which has little homology with those haloarchaeal plasmids reported so far. Recently, it has been used to develop a stable vector for the expression of a halocin immunity protein in Haloarcula hispanica (48). In this study, we report the sequence features of pSCM201 and the investigation of its replication mechanism. The minimal replicon containing a replication origin (ori201) and an adjacent gene (rep201) was determined. The replication initiation point (RIP) at the vicinity of an AT-rich region in ori201 was identified, and the unidirectional theta replication model of pSCM201 was established. To our knowledge, this is the first unidirectional theta-type replication plasmid experimentally identified in the domain of archaea. It provides a novel system for archaeal genetic research.
MATERIALS AND METHODS
Strains, plasmids, and primers.
The halophilic archaeon Haloarcula sp. strain AS7094 is the natural host of pSCM201. The H. hispanica strain ATCC 33960 was used as recipient for pSCM201 derivatives. These haloarchaeal strains were cultivated at 37°C in AS-168 medium as described previously (29). When necessary, mevinolin was added to a final concentration of 3 to 5 μg/ml. Escherichia coli strain JM109 was used as the host for all cloning and sequencing experiments and cultured in Luria-Bertani medium (42). When needed, ampicillin was added to a final concentration of 100 μg/ml for selection. The cloning vector pUCm-T (Sangon, Shanghai, China) was used to clone PCR products. All primers used in this study are listed in Table 1, and the strains and plasmids are listed in Table 2.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′ to 3′) |
|---|---|
| 16sF | ATTCCGGTTGATCCTGCCGGA |
| 16sR | AGGAGGTGATCCAGCCGCAGA |
| orf1F | AAGCGACTAAGCCACATC |
| orf1R | CCTTCTCTGCCTCTATCT |
| cRF | GCGACAATACTCATCACG |
| cRR | GCAGGATGATACCGAAGC |
| 204F | TCCCCATCACTGTGCTGC |
| 204R | GCGGGGGAACATAAAACC |
| 205F | ACGAAGTGACCGAGAAGG |
| 209R | CCAGACGGAACCACCATC |
| 212R | GGCTACGGGGAAATGCTT |
| 213R | GCCTATTCACCCATCAGC |
| 214R | ACCGCCTTCTCTGCCTCT |
| 216F | GACCGCTACACCCACGAG |
| P66 | CGCGAGAAAGGCGCTACAC |
| P67 | TCACGCCACTGGTCAAACG |
| RIPF | ACCTATGACAGCAAGCAAGCCTAC |
| RIPR | GACGAACGAACAGCGATAAACCAG |
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| Haloarcula sp. strain AS7094 | Natural host of pSCM201 | IMCAS |
| Haloarcula hispanica strain ATCC 33960 | Recipient of pSCM201 derivatives | ATCC |
| Escherichia coli JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) F′ [traD36 proAB+lacIq lacZΔM15] | 42 |
| Plasmids | ||
| pUCm-T | 2.77-kb vector for cloning PCR products; Ampr | Sangon, China |
| pUCmT | 2.77-kb replication type of pUCm-T | This work |
| pBluescript II KS− | 2.96-kb cloning vector; Ampr | Stratagene |
| pUBP2 | 12.3-kb shuttle vector; Ampr Mevr | 6 |
| pBR322 | 4,361-bp plasmid; Ampr Tetr | New England Biolabs |
| pSCM201 | 3,463-bp natural plasmid of Haloarcula sp. strain AS7094; GenBank accession no. AY443099 | This work |
| pSCM2011 | The 3,463-bp HindIII fragment from pSCM201 was cloned into pBluescript II KS−; Ampr | This work |
| pSCM2012 | The 3,463-bp ClaI fragment from pSCM201 was cloned into pBluescript II KS−; Ampr | This work |
| pSCM202 | The 3,463-bp NcoI fragment from pSCM201 was cloned into pUCmT and then the 3.5-kb Mevr fragment was inserted; Ampr Mevr | This work |
| pSCM203 | The 3.5-kb Mevr fragment was cloned into pSCM2012; Ampr Mevr | This work |
| pSCM204 | The 3,389-bp PCR product (204F-204R) was cloned into pUCm-T and then the 3.5-kb Mevr fragment was inserted; Ampr Mevr | This work |
| pSCM212 | The 2,143-bp PCR product (204F-212R) was cloned into pUCm-T and then the 3.5-kb Mevr fragment was inserted; Ampr Mevr | This work |
| pSCM213 | The 2,069-bp PCR product (204F-213R) was cloned into pUCm-T and then the 3.5-kb Mevr fragment was inserted; Ampr Mevr | This work |
| pSCM214 | The 1,730-bp PCR product (204F-214R) was cloned into pUCm-T and then the 3.5-kb Mevr fragment was inserted; Ampr Mevr | This work |
| pSCM216 | The 1,999-bp PCR product (216F-209R) was cloned into pUCm-T and then the 3.5-kb Mevr fragment was inserted; Ampr Mevr | This work |
| pSCM217 | The 1,885-bp PCR product (205F-209R) was cloned into pUCm-T and then the 3.5-kb Mevr fragment was inserted; Ampr Mevr | This work |
Isolation of the cryptic plasmid pSCM201 and DNA sequencing.
The plasmid pSCM201 was isolated from haloarchaeal strain AS7094 by the alkaline lysis method (37). For DNA sequencing, pSCM201 DNA was digested at either the HindIII or the ClaI site to generate two kinds of linear-plasmid DNA molecules, which were inserted into pBluescript II KS− (Stratagene) at the corresponding sites to yield two different recombinant clones and sequenced by primer walking with the dideoxynucleotide termination cycle sequencing method.
The Genetics Computer Group (GCG) software package (University of Wisconsin, Madison, WI) was used to assemble and analyze the sequences. The FASTA and BLAST programs of the GCG package were used for homology searches in the GenBank and Swiss-Prot protein databases. The helix-turn-helix (HTH) motif was identified by GYM 2.0 software (http://www.cs.fiu.edu/∼giri/bioinf/GYM2/prog.html).
Determination of the minimal replicon.
In order to identify the minimal replicon of pSCM201, different regions of this plasmid were amplified by PCR and ligated into the cloning vector pUCm-T to generate a series of derivative plasmids. Then the mevinolin-resistance (Mevr) gene was cut as an EcoRI-KpnI fragment from pUBP2 (6) and introduced into the same sites of the above-named plasmids. The resulting plasmids were then transformed into H. hispanica ATCC 33960 as described by Cline et al. (12).
Copy number determination and plasmid stability assays.
To measure the copy number of pSCM201 per chromosome equivalent, a comparison was made between the hybridization signals from the single-copy, chromosome-borne cop1 gene (the GenBank accession number is D31880) and the pSCM201-borne rep201 gene in total DNA extracts of AS7094. Restriction enzyme SalI was selected to digest the total DNA of AS7094 and to linearize pSCM201, resulting in a clear discrimination between chromosome- and pSCM201-borne signals. A rep201-specific probe (562 bp) was amplified by PCR using the primers orf1F and orf1R, and a cop1-specific probe (562 bp) was amplified with the primers cRF and cRR (Table 1). The two PCR products had the same G+C content of 60.5%. The hybridized-signal levels of the rep201 gene and cop1 gene were quantified by using a phosphorimager (Amersham Biosciences) with ImageQuant 5.2 software. The plasmid copy number was estimated by comparing the signals generated by the plasmid and the chromosomal cop1 genes. The relative copy numbers of other pSCM201 derivatives were evaluated by comparing the relative intensities of the plasmid bands as described by Emond et al. (14), and their stability was assayed by plasmid maintenance in the absence of selection pressure as described by Ng and DasSarma (36). Experiments were done in triplicate.
Isolation of RIs.
The plasmid replication intermediates (RIs) of pSCM201 from AS7094 and those of pSCM2011 from E. coli were isolated as described by Mojica et al. (33) and Santamaria et al. (43), respectively, except that alkaline and RNase treatments were avoided. For the RIP mapping assay, the RIs of plasmids containing single-stranded DNA were enriched by BND cellulose (Sigma) chromatography as described elsewhere (26). For electron microscopy, the RIs of pSCM201 were further separated by electrophoresis in a low-melting-temperature agarose gel. We then isolated the DNA from the gel region between the supercoiled plasmid and high-molecular-weight chromosomal bands, which included all possible plasmid RIs.
Detection of RIs by electron microscopy.
Samples for electron microscopy were prepared as described by Burkardt et al. (8). Micrographs were recorded using a Hitachi H-700A electron microscope. pBR322 was treated in a similar way and was used as the size marker.
RIP mapping.
RIP mapping was performed as described by Gerbi and Bielinsky (17). Briefly, Vent (exo-)polymerase was used to extend from a labeled primer to the DNA-RNA junctions of the nascent-strand templates in the RIs, in which the nicked contaminating DNA had been removed by λ exonuclease treatment. For the primer extension experiments of both of the pSCM201 RIs from Haloarcula sp. strain AS7094 and pSCM2011 RIs from E. coli, the primers RIPF (located about 250 bp upstream of the AT-rich region) and RIPR (located about 170 bp downstream of the AT-rich region) (Table 1 and see Fig. 5) were applied by using the following program: 3 min at 94°C and then 30 cycles of 1 min at 94°C, 1 min at 70°C, and 1.5 min at 72°C. Sequencing reactions were performed in parallel with the fmol DNA cycle sequencing system (Promega) according to the protocols of the manufacturer with the same primers, and all samples were electrophoresed and analyzed on a 6% acrylamide gel containing 8 M urea and 1× Tris-borate-EDTA. The primers used in RIP mapping and DNA sequencing were labeled with T4 polynucleotide kinase with [γ-32P]ATP (5,000 Ci/mmol).
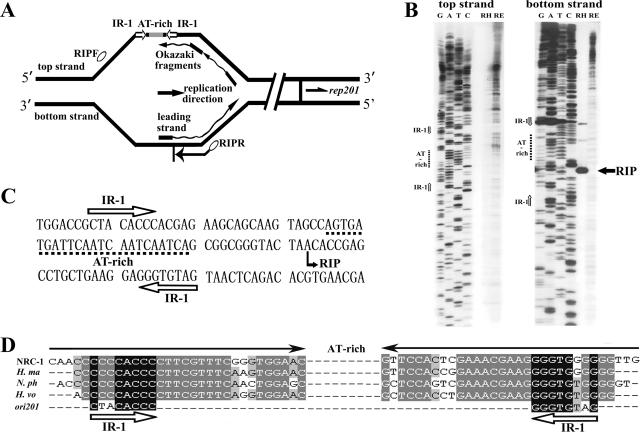
FIG. 5.
Mapping of the RIP of pSCM201. (A) Outline of RIP mapping for the unidirectional theta replication of pSCM201 (not to scale). Nascent DNAs (leading strand and Okazaki fragments) initiated by a small RNA primer (black rectangles) were used as the templates in the primer extension reaction to determine the RIP. Ovals indicate the primers used in RIP mapping. The directions of pSCM201 replication, rep201 transcription, and primer extension are indicated by arrows. (B) Identification of the replication initiation point by RIP mapping. Primer extension was performed with the oligonucleotides RIPF for the top strand (left panel) and RIPR for the bottom strand (right panel). Lanes contain dideoxy sequencing reaction mixtures (GATC) and the products of the primer extension of pSCM201 RIs from Haloarcula sp. strain AS7094 (RH) and pSCM2011 RIs from E. coli (RE). The filled arrow indicates the RIP position of pSCM201. The open arrows indicate the positions of IR-1. The dashed lines indicate the positions of the AT-rich region. (C) Nucleotide sequence of a region containing IR-1, the AT-rich region, and the RIP of pSCM201. (D) Conserved inverted repeats in ori201 and the chromosomal replication origins of the four other haloarchaea. The filled arrows indicate the long inverted repeat flanking the AT-rich region in the four haloarchaeal chromosomal replication origins. The open arrows indicate IR-1 of ori201, which shares high homology with the inner sequence of those chromosomal long inverted repeats. NRC-1, Halobacterium sp. strain NRC-1; H. ma, Haloarcula marismotui; N. ph, Natronomonas pharaonis; H. vo, Haloferax volcanii.
Nucleotide sequence accession numbers.
The nucleotide sequences of pSCM201 and the 16S rRNA gene of Haloarcula sp. strain AS7094 were deposited in GenBank under the accession numbers AY443099 and DQ334803, respectively.
RESULTS
Isolation of plasmid pSCM201 from the archaeon Haloarcula sp. strain AS7094.
To find novel plasmids in the extremely halophilic archaea, we have screened more than 20 haloarchaeal strains isolated from salt lakes or marine salterns in the People's Republic of China. An endogenous plasmid about 3.5 kb in size, named pSCM201, was isolated from a halophilic archaeon strain, AS7094. To identify the host strain, the 16S rRNA gene of AS7094 was amplified with the specific haloarchaeal primers (16sF and 16sR in Table 1) and the sequence was determined. The BLAST sequence in the NCBI database showed that AS7094 was closely related to Haloarcula argentinensis (the homology of the 16S rRNA gene is about 99%); thus, the host strain of pSCM201 was named Haloarcula sp. strain AS7094.
Sequence analysis of pSCM201.
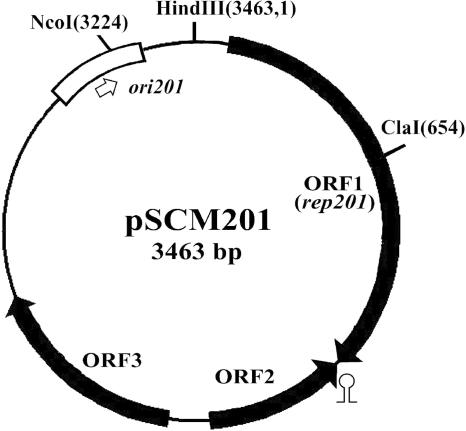
DNA sequence analysis showed that pSCM201 is a circular molecule of 3,463 bp (Fig. 1) with a G+C content of 59.9%, which is in accordance with the high G+C content of the genomes or plasmids of haloarchaea (6, 18, 19, 22, 27, 36, 38, 50). Examination of the six possible phases of the plasmid sequence revealed the presence of three open reading frames (ORFs) (ORF1, ORF2, and ORF3) in the two strands, which cover about 70% of the entire sequence (Fig. 1). These ORFs were supposed to encode three polypeptides with 399, 135, and 212 amino acids. No significant homology was found in the NCBI protein databases for the deduced proteins encoded by ORF2 and ORF3. However, a putative leucine zipper (LZ) motif (residues 88 to 108), an HTH motif (residues 129 to 150), and an ATPase domain (residues 148 to 309) were detected in the ORF1-encoded protein (Fig. 2). Since most replication initiation proteins harbor DNA-binding motifs and possess ATPase activities (28), and the LZ motif was also found in Rep proteins of many iteron-controlled plasmids (11, 13), we presumed that the protein encoded by ORF1 was the replication initiation protein of pSCM201 named Rep201 (44.7 kDa). Rep201 is highly acidic, with a calculated pI of 4.74. This acidic feature has been suggested as a major adaptive mechanism of most haloarchaeal proteins to function in conditions of nearly saturating salinity (24).
FIG. 1.
Map of pSCM201. The three open reading frames, named ORF1 (rep201), ORF2, and ORF3, are indicated with filled arrows. ori201 is indicated by a box, and the replication direction is indicated by an open arrow. The stem-loop represents the putative terminator of rep201. The restriction sites for appropriate enzymes are indicated, with their positions appearing in parentheses. Base 1 is arbitrarily defined by the first base of the HindIII recognition site.
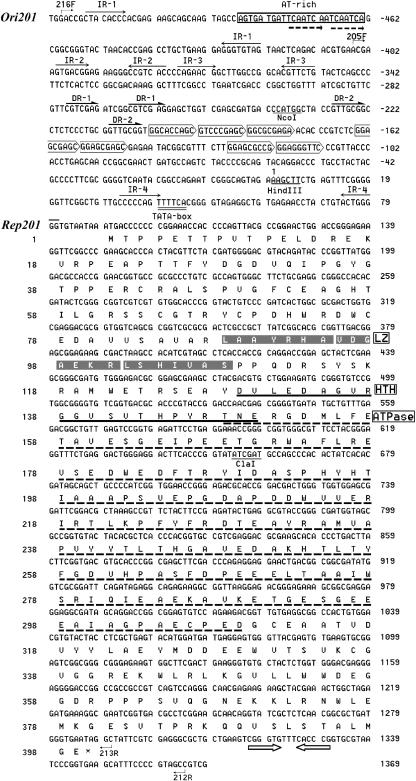
FIG. 2.
Sequence analysis of the replicon of pSCM201. The AT-rich region of ori201 is boxed, and direct repeats within this AT-rich region are indicated with dashed arrows. The inverted repeats (IR-1 to IR-4) are shown with arrows and the two sets of direct repeats (DR-1 and DR-2) with half arrows. The putative iterons are surrounded by pentagons. The putative TATA box for rep201 is double underlined. The putative transcriptional terminator of rep201 is indicated with open arrows. The restriction sites (HindIII, NcoI, ClaI) are underlined. Primers' positions are indicated with bent arrows. The amino acid sequence of Rep201 (399 amino acids), encoded by the rep201 gene, is indicated below the DNA sequence. The numbers on the right indicate the nucleotide numbers of pSCM201, while the numbers on the left indicate amino acid numbers of Rep201. The putative LZ motif, the HTH motif, and the ATPase domain of Rep201 are also indicated.
Upstream of the rep201 gene, we detected a 24-bp AT-rich region (70.9% A+T) located about 550 bp 5′ of the rep201 start codon (Fig. 2), which is flanked by perfect inverted repeat IR-1 (5′-CTACACCC-3′). Downstream of this region, there are sets of perfect inverted and direct repeats, followed by seven copies of a 9-bp iteron-like direct repeat with a consensus sequence of GGAGCGAGC (Fig. 2). Since these features resemble the replication origins of many theta replication plasmids, we presumed that the above-described region is the replication origin of pSCM201 and named it ori201 (Fig. 1 and 2).
Determination of the minimal replicon of pSCM201.
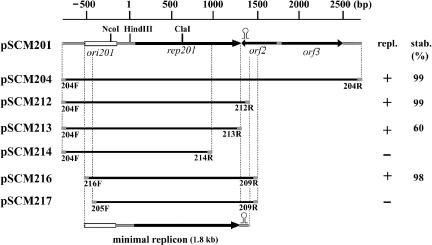
In order to determine the minimal replicon of pSCM201, the different regions of this plasmid plus the Mevr gene were cloned into the E. coli cloning vector pUCm-T without any haloarchaeal replicons (see Materials and Methods) and tested for their abilities to replicate and maintain themselves in H. hispanica ATCC 33960. The minimal replicon (∼1.8 kb) was demonstrated to contain only the putative replication origin region (ori201) and the adjacent rep201 gene, as the deletion of ORF2, ORF3, and the partial noncoding sequence downstream of ORF3 could not interfere with replication (Fig. 3). The pSCM201-derived vectors are very stable in transformed H. hispanica cells, and the minimal replicon is sufficient for stable maintenance of the derived plasmids. For instance, after 100 generations of growth, even without selection pressure, the pSCM201 derivatives (pSCM204, pSCM212, pSCM216) were sustained in more than 98% of cells of transformed H. hispanica (Fig. 3), and the copy numbers are equal to that of pSCM201 (about 10 ± 2 per chromosome).
FIG. 3.
Determination of the minimal replicon of pSCM201. A linear restriction map is shown on the top of the diagram. Putative ORFs (black arrows), ori201 (rectangle), and the terminator (stem-loop) shown on the restriction map were inferred from sequence analysis. Thick lines under the linear map represent the fragments cloned for autonomous-replication assays. The names of primers (indicated as gray block) are placed under the ends of each fragment. The names of recombinant plasmids containing these fragments are indicated on the left. The ability of the derivatives to replicate (repl.) in Haloarcula hispanica is indicated as “+” (yes) or “−” (no). Stability (stab.) indicates the ratio of cells (percentage) harboring the plasmids after 100 generations of growth without selective pressure.
The rep201 gene is necessary for this plasmid's replication. Interruption of the rep201 gene at the ClaI site resulted in the loss of the replication ability of the derivative plasmid pSCM203 (Table 2) in H. hispanica. Removing the last 112 amino acids from Rep201 also abolished replication (pSCM214) (Fig. 3). Moreover, when deletions extended into the inverted repeat (nucleotide 1317) downstream of rep201, which we considered to be the transcriptional terminator of rep201 (Fig. 2), the derivative pSCM213 became unstable. About 40% of H. hispanica cells transformed with pSCM213 lost the plasmids after 100 generations without selection pressure, and the copy number of pSCM213, if it existed, decreased to five ± two per chromosome. Likewise, the ori201 region was also required for plasmid replication. A shuttle vector named pSCM217, which was short only of the AT-rich region in comparison to pSCM216, lost its replication ability in H. hispanica (Fig. 3). Even with the interruption of this region at the NcoI site by insertion of an ∼6.2-kb fragment including the pUCm-T sequence and Mevr gene, the resultant plasmid, pSCM202 (Table 2), could not replicate in haloarchaea.
Taking these results together, we conclude that the putative replication origin ori201 combined with its adjacent gene (rep201), which constitutes the minimal replicon of pSCM201, is essential and sufficient for plasmid replication in halophilic archaea.
pSCM201 replicates via a theta replication mechanism.
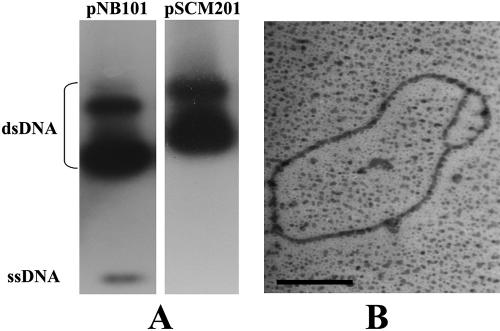
To determine whether pSCM201 replicates via an RCR or a theta replication mechanism, we first checked to determine if ssDNA intermediates of this plasmid were generated, as it is the hallmark of an RCR plasmid (25, 45, 50). The plasmid RIs were separated by agarose gel electrophoresis and blotted onto a nylon membrane. Then, the DNA of pSCM201 was detected with a rep201-derived probe that was amplified by primers orf1F and orf1R (Table 1) and labeled with [α-32P]dCTP, and the known RCR-type plasmid pNB101 (50) was used as the positive control, which was detected with a probe amplified by primers P66 and P67 (Table 1). No ssDNA of pSCM201 was detected in this assay even after prolonged exposure of the film, while under the same conditions, the ssDNA of pNB101 was detected (Fig. 4A). The absence of ssDNA RIs suggested that pSCM201 might not replicate via the RCR mechanism.
FIG. 4.
Analyses of RIs of pSCM201 by Southern blotting (A) and electron microscopy (B). (A) Detection of the ssDNA RIs of pNB101 (left panel) and pSCM201 (right panel). (B) Uncut replication bubble-shaped molecule of pSCM201. The bar corresponds to 0.5 kb. dsDNA, double-stranded DNA.
To explore whether pSCM201 replicates via a theta replication mechanism, we then enriched the RIs and subjected them to electron microscopic analysis. The characteristic bubble of theta-type replication was observed in the pSCM201 RIs (Fig. 4B). In addition, when the ClaI-cleaved RIs of pSCM201 were subjected to neutral/neutral two-dimension agarose gel electrophoresis, an arc supposed to be formed from the replication bubble-carrying molecules was also detected (data not shown). These results demonstrate that pSCM201 is indeed a theta replication plasmid.
RIP mapping of pSCM201.
To further characterize the plasmid, we determined the potential RIP within the ori201 by RIP mapping (17). This technique can map the replication initiation site with a nucleotide resolution and has recently been successfully applied to determine the precise RIPs of eukaryotic and archaeal chromosomes (5, 32, 41). The principle of RIP mapping is to extend a labeled primer to the DNA-RNA junctions of RNA-primed nascent strands with Vent (exo-)polymerase, and the RIP is usually defined as the transition point between the leading and lagging strands. This technique is capable of determining the RIP of a theta-type plasmid, in which replication also initiates with RNA primers.
To investigate if the RIP of pSCM201 is located within putative ori201 and to precisely identify its position, the vicinity of the AT-rich region in ori201 was primarily taken into account, and the oligonucleotides RIPF and RIPR (Table 1 and Fig. 5A) were subjected to primer extension reactions (see Materials and Methods). It was clearly revealed that the primer extension from RIPR, which is located downstream of the AT-rich region with a direction reverse to that of rep201 transcription, gave one prominent band with the nascent DNA templates of pSCM201 from Haloarcula sp. strain AS7094 (Fig. 5B, bottom strand panel, lane RH). In contrast, primer extension with the samples of pSCM2011 containing the complete sequence of pSCM201 isolated from E. coli did not give rise to such a specific extension product with the same primer (Fig. 5B, bottom strand panel, lane RE). This is consistent with the notion that ori201 would function in Haloarcula sp. strain AS7094 but not in E. coli. Furthermore, when the same nascent DNAs were used as the templates, no apparent and specific product was generated with RIPF, the primer located upstream of the AT-rich region with the rep201 direction of transcription (Fig. 5B, top strand panel).
These results are significantly different from those of bidirectional theta replication but support the argument that pSCM201 replicates unidirectionally. As to a unidirectional replication origin, only the primer orientated in the opposite direction of replication (RIPR in the case of pSCM201) can produce a prominent extension product, which points to the DNA-RNA junction of the leading strand and thus represents the RIP. Characteristically, the primer in the other direction upstream of the RIP could hardly give rise to any extension products, since almost no RNA-primed nascent DNA in that direction would be generated in such a unidirectional theta replication origin. Our results suggested that pSCM201 replicated from a fixed site 14 bp downstream of the AT-rich region and between the two half parts of a conserved inverted repeat, IR-1(Fig. 5B, C, and D), and progressed unidirectionally toward the downstream rep201 gene.
Electron microscopic analysis of the replication direction of pSCM201.
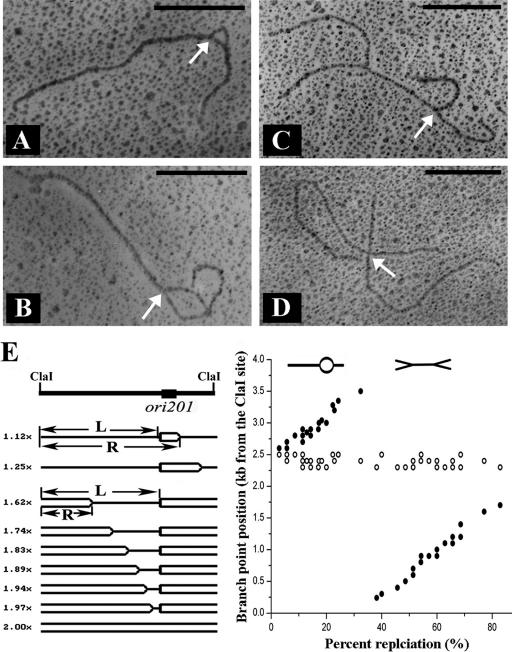
To further confirm the unidirectional replication of pSCM201, the single ClaI restriction site was used to linearize the plasmid, thus introducing a convenient reference point in electron microscopic analysis. Measurements of about 20 bubble-carrying molecules (Fig. 6A and B) revealed that the sizes of the long unreplicated arms were constant (2,400 ± 100 bp). Measurements of about 20 double-Y-shaped molecules showed that the size of one fork was constant (1,100 ± 100 bp) but that the other was changeable but not more than 2,400 ± 100 bp (Fig. 6C). For X-shaped molecules (Fig. 6D), the longer and shorter forks were usually about 2,400 and 1,100 bp, respectively. These data are summarized in Fig. 6E. In both the bubble-carrying and double-Y-shaped replication intermediates, the length from the ClaI site to the left-hand branch point or right-hand branch point was calculated. It was revealed that the position of the left-hand branch point did not move with an increasing extent of replication but that the position of the right-hand branch point depended linearly on the extent of replication (Fig. 6E), thus confirming the unidirectional replication mode.
FIG. 6.
Analysis of ClaI-cleaved RIs of pSCM201 by electron microscopy. (A and B) Replication bubble-shaped molecules of pSCM201. (C) Double-Y-shaped molecule of pSCM201. (D) X-shaped molecule of pSCM201. The bars correspond to 1.0 kb. The white arrows indicate the replication initiation point. (E) Analysis of the branch-point positions of the replication forks in the pSCM201 RIs. (Left) The pSCM201 RIs were schematically ordered by increasing extent of replication (one line indicates an unreplicated region; two lines indicate a replicated region). The positions of left-hand (L) and right-hand (R) branch points were measured relative to the same ClaI site. (Right) Positions of the left-hand (white circles) or right-hand (black circles) branch points are plotted versus the extent of replication.
This left-hand branch point (∼2,400 bp downstream or 1,100 bp upstream of the ClaI site (Fig. 6A to D) was in very good accordance with the RIP pinpointed by RIP mapping, as the distance from the designated RIP (Fig. 5C) to the downstream ClaI site was exactly 1,103 bp.
DISCUSSION
In this work, we characterize a novel haloarchaeal plasmid, pSCM201 (3,463 bp), and show that this small, naturally occurring plasmid replicates via a unidirectional theta-replicating mechanism, initiating in the vicinity of an AT-rich region within ori201. The evidence for this replication mechanism was based on the following observations: (i) the minimal replicon of pSCM201 containing the putative ori201 region and rep201 gene showed structural similarities with theta replication plasmids but not RCR plasmids; (ii) unlike RCR plasmids, pSCM201 did not generate ssDNA RIs; (iii) electron microscopy studies showed the presence of the replication bubble of RIs; and (iv) RIP mapping of pSCM201 determined that the location of RIP was in the vicinity of the AT-rich region in ori201 and revealed that the replication progressed in only one direction, which was further confirmed by electron microscopy. To our knowledge, pSCM201 is the first unidirectional theta replication plasmid experimentally identified in the domain of archaea.
pSCM201 is very stable. The vectors derived from the complete minimal replicon of pSCM201 had a constant copy number of about 10 for at least 100 generations in the absence of selective pressure. So we concluded that the most-efficient control elements were within this minimal replicon. In contrast to the stable plasmid pSCM212, the unstable and low-copy-number plasmid pSCM213 lacked only the putative terminator of rep201 (Fig. 3), indicating that Rep201 may participate in the control of plasmid replication initiation as well as its copy number. Rep201 contains a putative LZ motif and thus has the potential to form a dimer. Indeed, such a dimer was detected in the native polyacrylamide gel with the purified His-tagged Rep201 protein (data not shown), suggesting that Rep201 may be involved in the control of plasmid copy number via handcuffing, like many bacterial theta replication plasmids (11). Interestingly, Rep201 had an ATPase domain (residues 148 to 309) that is different from those of bacterial theta replication plasmids, which usually lack ATPase activities and might need the ATPase activities of DnaA to melt the origin (11). Therefore, Rep201 may have some novel functions not yet observed in its bacterial counterparts.
It is noteworthy that the inverted repeat IR-1 (5′-CTACACCC-3′) flanking the AT-rich region in ori201, is similar to the inner sequence (5′-CCCCACCC-3′) of putative origin recognition boxes of Halobacterium sp. strain NRC-1 (Fig. 5D). It has been speculated that this inner sequence is the binding site of the initiation proteins of archaea, Orc1/Cdc6, or other DNA replication initiation proteins (4, 41). Similar structures containing an inverted repeat flanking an AT-rich region were found to be highly conserved upstream of the orc7 gene (coding for the putative replication initiation protein) in the chromosomes of three other halophiles, Haloarcula marismortui, Haloferax volcanii (4), and Natronomonas pharaonis (15) (Fig. 5D). In addition, chromosomes of two other archaea, Pyrococcus abyssi (32, 34) and Sulfolobus solfataricus (41), are also reported to contain an inverted repeat/AT-rich region upstream of their orc7 homologs. This structure is reminiscent of a duplex unwinding element, a feature found in many bacteria and eukaryotic origins of replication and thought to facilitate the melting of DNA at the origin (35). Such a structure in pSCM201 would contribute to its theta replication, as the RIP of pSCM201 was located just between the inverted repeat (IR-1) and the nearby AT-rich region. Moreover, there are a pair of 7-bp direct repeats (TCAATCA) within the AT-rich region which might be the entry sites of helicase or its loader, as in E. coli plasmid pSC101 (7). Downstream of the AT-rich region, there are sets of inverted repeats and direct repeats, including seven putative iterons. These repeats might be the binding sites of Rep201 or other involved proteins.
The theta-type replication of plasmids, including many iteron-controlled bacterial plasmids (31), is usually unidirectional (13). In the well-studied unidirectional theta replication plasmid pBR322, it was found that the orientation of the DnaA boxes is critical for the efficiency of unidirectional replication (44). It is suggested that DnaA may interact more strongly with one of the DNA strands on which helicase loading would be more productive for unidirectional replication (16). For pSCM201, it will be interesting to investigate the possible correlation of replication direction and the structural asymmetry of ori201 in the future.
Acknowledgments
We thank Peijin Zhou for providing several haloarchaeal strains for plasmid screening and Juan Wang at the University of Pennsylvania for helpful comments during preparation of the manuscript.
This work was supported by grants from the National Basic Research Program (973 Program) of China (2004CB719603), the National 863 Program of China (2004AA626010), the National Natural Science Foundation of China (30570029), and the Chinese Academy of Sciences (KSCX-YW-G-023).
Footnotes
Published ahead of print on 22 September 2006.
REFERENCES
- 1.Allers, T., and M. Mevarech. 2005. Archaeal genetics—the third way. Nat. Rev. Genet. 6:58-73. [DOI] [PubMed] [Google Scholar]
- 2.Baliga, N. S., R. Bonneau, M. T. Facciotti, M. Pan, G. Glusman, E. W. Deutsch, P. Shannon, Y. Chiu, R. S. Weng, R. R. Gan, P. Hung, S. V. Date, E. Marcotte, L. Hood, and W. V. Ng. 2004. Genome sequence of Haloarcula marismortui: a halophilic archaeon from the Dead Sea. Genome Res. 14:2221-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernander, R. 2000. Chromosome replication, nucleoid segregation and cell division in archaea. Trends Microbiol. 8:278-283. [DOI] [PubMed] [Google Scholar]
- 4.Berquist, B. R., and S. DasSarma. 2003. An archaeal chromosomal autonomously replicating sequence element from an extreme halophile, Halobacterium sp. strain NRC-1. J. Bacteriol. 185:5959-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bielinsky, A. K., and S. A. Gerbi. 1998. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science 279:95-98. [DOI] [PubMed] [Google Scholar]
- 6.Blaseio, U., and F. Pfeifer. 1990. Transformation of Halobacterium halobium: development of vectors and investigation of gas vesicle synthesis. Proc. Natl. Acad. Sci. USA 87:6772-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bramhill, D., and A. Kornberg. 1988. A model for initiation at origins of DNA replication. Cell 54:915-918. [DOI] [PubMed] [Google Scholar]
- 8.Burkardt, H., and R. Lurz. 1984. Electron microscopy, p. 286-288. In A. Pühler and K. N. Timmis (ed.), Advanced molecular genetics. Springer Verlag, Berlin, Germany.
- 9.Charlebois, R. L., W. L. Lam, S. W. Cline, and W. F. Doolittle. 1987. Characterization of pHV2 from Halobacterium volcanii and its use in demonstrating transformation of an archaebacterium. Proc. Natl. Acad. Sci. USA 84:8530-8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charlebois, R. L., L. C. Schalkwyk, J. D. Hofman, and W. F. Doolittle. 1991. Detailed physical map and set of overlapping clones covering the genome of the archaebacterium Haloferax volcanii DS2. J. Mol. Biol. 222:509-524. [DOI] [PubMed] [Google Scholar]
- 11.Chattoraj, D. K. 2000. Control of plasmid DNA replication by iterons: no longer paradoxical. Mol. Microbiol. 37:467-476. [DOI] [PubMed] [Google Scholar]
- 12.Cline, S. W., W. L. Lam, R. L. Charlebois, L. C. Schalkwyk, and W. F. Doolittle. 1989. Transformation methods for halophilic archaebacteria. Can. J. Microbiol. 35:148-152. [DOI] [PubMed] [Google Scholar]
- 13.del Solar, G., R. Giraldo, M. J. Ruiz-Echevarria, M. Espinosa, and R. Diaz-Orejas. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62:434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emond, E., R. Lavallee, G. Drolet, S. Moineau, and G. LaPointe. 2001. Molecular characterization of a theta replication plasmid and its use for development of a two-component food-grade cloning system for Lactococcus lactis. Appl. Environ. Microbiol. 67:1700-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falb, M., F. Pfeiffer, P. Palm, K. Rodewald, V. Hickmann, J. Tittor, and D. Oesterhelt. 2005. Living with two extremes: conclusions from the genome sequence of Natronomonas pharaonis. Genome Res. 15:1336-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgopoulos, C. 1989. The E. coli dnaA initiation protein: a protein for all seasons. Trends Genet. 5:319-321. [DOI] [PubMed] [Google Scholar]
- 17.Gerbi, S. A., and A. K. Bielinsky. 1997. Replication initiation point mapping. Methods 13:271-280. [DOI] [PubMed] [Google Scholar]
- 18.Hackett, N. R., M. P. Krebs, S. DasSarma, W. Goebel, U. L. RajBhandary, and H. G. Khorana. 1990. Nucleotide sequence of a high copy number plasmid from Halobacterium strain GRB. Nucleic Acids Res. 18:3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall, M. J., and N. R. Hackett. 1989. DNA sequence of a small plasmid from Halobacterium strain GN101. Nucleic Acids Res. 17:10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmes, M. L., F. Pfeifer, and M. L. Dyall-Smith. 1995. Analysis of the halobacterial plasmid pHK2 minimal replicon. Gene 153:117-121. [DOI] [PubMed] [Google Scholar]
- 21.Holmes, M., F. Pfeifer, and M. Dyall-Smith. 1994. Improved shuttle vectors for Haloferax volcanii including a dual-resistance plasmid. Gene 146:117-121. [DOI] [PubMed] [Google Scholar]
- 22.Kagramanova, V. K., N. I. Derckacheva, and A. S. Mankin. 1988. The complete nucleotide sequence of the archaebacterial plasmid pHSB from Halobacterium, strain SB3. Nucleic Acids Res. 16:4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelman, L. M., and Z. Kelman. 2003. Archaea: an archetype for replication initiation studies? Mol. Microbiol. 48:605-615. [DOI] [PubMed] [Google Scholar]
- 24.Kennedy, S. P., W. V. Ng, S. L. Salzberg, L. Hood, and S. DasSarma. 2001. Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence. Genome Res. 11:1641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan, S. A. 2005. Plasmid rolling-circle replication: highlights of two decades of research. Plasmid 53:126-136. [DOI] [PubMed] [Google Scholar]
- 26.Kiger, J. A., Jr., and R. L. Sinsheimer. 1969. Vegetative lambda DNA. IV. Fractionation of replicating lambda DNA on benzoylated-naphthoylated DEAE cellulose. J. Mol. Biol. 40:467-490. [DOI] [PubMed] [Google Scholar]
- 27.Lam, W. L., and W. F. Doolittle. 1989. Shuttle vectors for the archaebacterium Halobacterium volcanii. Proc. Natl. Acad. Sci. USA 86:5478-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, D. G., and S. P. Bell. 2000. ATPase switches controlling DNA replication initiation. Curr. Opin. Cell Biol. 12:280-285. [DOI] [PubMed] [Google Scholar]
- 29.Li, Y., H. Xiang, J. Liu, M. Zhou, and H. Tan. 2003. Purification and biological characterization of halocin C8, a novel peptide antibiotic from Halobacterium strain AS7092. Extremophiles 7:401-407. [DOI] [PubMed] [Google Scholar]
- 30.Mankin, A. S., I. M. Zyrianova, V. K. Kagramanova, and R. A. Garrett. 1992. Introducing mutations into the single-copy chromosomal 23S rRNA gene of the archaeon Halobacterium halobium by using an rRNA operon-based transformation system. Proc. Natl. Acad. Sci. USA 89:6535-6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marszalek, J., and J. M. Kaguni. 1994. DnaA protein directs the binding of DnaB protein in initiation of DNA replication in Escherichia coli. J. Biol. Chem. 269:4883-4890. [PubMed] [Google Scholar]
- 32.Matsunaga, F., C. Norais, P. Forterre, and H. Myllykallio. 2003. Identification of short ‘eukaryotic’ Okazaki fragments synthesized from a prokaryotic replication origin. EMBO Rep. 4:154-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mojica, F. J., F. Charbonnier, G. Juez, F. Rodriguez-Valera, and P. Forterre. 1994. Effects of salt and temperature on plasmid topology in the halophilic archaeon Haloferax volcanii. J. Bacteriol. 176:4966-4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myllykallio, H., P. Lopez, P. Lopez-Garcia, R. Heilig, W. Saurin, Y. Zivanovic, H. Philippe, and P. Forterre. 2000. Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science 288:2212-2215. [DOI] [PubMed] [Google Scholar]
- 35.Natale, D. A., A. E. Schubert, and D. Kowalski. 1992. DNA helical stability accounts for mutational defects in a yeast replication origin. Proc. Natl. Acad. Sci. USA 89:2654-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng, W. L., and S. DasSarma. 1993. Minimal replication origin of the 200-kilobase Halobacterium plasmid pNRC100. J. Bacteriol. 175:4584-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng, W. L., C. F. Yang, J. T. Halladay, P. Arora, and S. DasSarma. 1995. Isolation of genomic and plasmid DNAs from Halobacterium halobium, p. 181-184. In S. DasSarma and E. M. Fleischmann (ed.), Archaea: a laboratory manual. Vol. 1. Halophiles. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 38.Ng, W. V., S. A. Ciufo, T. M. Smith, R. E. Bumgarner, D. Baskin, J. Faust, B. Hall, C. Loretz, J. Seto, J. Slagel, L. Hood, and S. DasSarma. 1998. Snapshot of a large dynamic replicon in a halophilic archaeon: megaplasmid or minichromosome? Genome Res. 8:1131-1141. [DOI] [PubMed] [Google Scholar]
- 39.Pfeifer, F., and P. Ghahraman. 1993. Plasmid pHH1 of Halobacterium salinarium: characterization of the replicon region, the gas vesicle gene cluster and insertion elements. Mol. Gen. Genet. 238:193-200. [DOI] [PubMed] [Google Scholar]
- 40.Robinson, N. P., and S. D. Bell. 2005. Origins of DNA replication in the three domains of life. FEBS J. 272:3757-3766. [DOI] [PubMed] [Google Scholar]
- 41.Robinson, N. P., I. Dionne, M. Lundgren, V. L. Marsh, R. Bernander, and S. D. Bell. 2004. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell 116:25-38. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Santamaria, D., E. Viguera, M. L. Martinez-Robles, O. Hyrien, P. Hernandez, D. B. Krimer, and J. B. Schvartzman. 2000. Bi-directional replication and random termination. Nucleic Acids Res. 28:2099-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seufert, W., B. Dobrinski, R. Lurz, and W. Messer. 1988. Functionality of the dnaA protein binding site in DNA replication is orientation-dependent. J. Biol. Chem. 263:2719-2723. [PubMed] [Google Scholar]
- 45.Sioud, M., G. Baldacci, P. Forterre, and A. M. de Recondo. 1988. Novobiocin induces accumulation of a single strand of plasmid pGRB-1 in the archaebacterium Halobacterium GRB. Nucleic Acids Res. 16:7833-7842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soppa, J. 2005. From replication to cultivation: hot news from Haloarchaea. Curr. Opin. Microbiol. 8:737-744. [DOI] [PubMed] [Google Scholar]
- 47.Soppa, J. 2006. From genomes to function: haloarchaea as model organisms. Microbiology 152:585-590. [DOI] [PubMed] [Google Scholar]
- 48.Sun, C., Y. Li, S. Mei, Q. Lu, L. Zhou, and H. Xiang. 2005. A single gene directs both production and immunity of halocin C8 in a haloarchaeal strain AS7092. Mol. Microbiol. 57:537-549. [DOI] [PubMed] [Google Scholar]
- 49.Zhou, M., H. Xiang, C. Sun, and H. Tan. 2004. Construction of a novel shuttle vector based on an RCR-plasmid from a haloalkaliphilic archaeon and transformation into other haloarchaea. Biotechnol. Lett. 26:1107-1113. [DOI] [PubMed] [Google Scholar]
- 50.Zhou, M., H. Xiang, C. Sun, Y. Li, J. Liu, and H. Tan. 2004. Complete sequence and molecular characterization of pNB101, a rolling-circle replication plasmid from the haloalkaliphilic archaeon Natronobacterium sp. strain AS7091. Extremophiles 8:91-98. [DOI] [PubMed] [Google Scholar]