Abstract
Clostridium difficile, a proteolytic strict anaerobe, has emerged as a clinically significant nosocomial pathogen in recent years. Pathogenesis is due to the production of lethal toxins, A and B, members of the large clostridial cytotoxin family. Although it has been established that alterations in the amino acid content of the growth medium affect toxin production, the molecular mechanism for this observed effect is not yet known. Since there is a paucity of information on the amino acid fermentation pathways used by this pathogen, we investigated whether Stickland reactions might be at the heart of its bioenergetic pathways. Growth of C. difficile on Stickland pairs yielded large increases in cell density in a limiting basal medium, demonstrating that these reactions are tied to ATP production. Selenium supplementation was required for this increase in cell yield. Analysis of genome sequence data reveals genes encoding the protein components of two key selenoenzyme reductases, glycine reductase and d-proline reductase (PR). These selenoenzymes were expressed upon the addition of the corresponding Stickland acceptor (glycine, proline, or hydroxyproline). Purification of the selenoenzyme d-proline reductase revealed a mixed complex of PrdA and PrdB (SeCys-containing) proteins. PR utilized only d-proline but not l-hydroxyproline, even in the presence of an expressed and purified proline racemase. PR was found to be independent of divalent cations, and zinc was a potent inhibitor of PR. These results show that Stickland reactions are key to the growth of C. difficile and that the mechanism of PR may differ significantly from that of previously studied PR from nonpathogenic species.
Clostridium difficile is the primary causative agent for antibiotic-induced diarrhea (termed C. difficile-associated disease) and is a rapidly emerging nosocomial pathogen in health care facilities around the world (14, 15, 20, 25). An outbreak of a more virulent epidemic strain of C. difficile in Quebec, Canada, was correlated with the death of more than 100 patients within a 6-month period in a single hospital (9-11, 27). A higher level of toxin produced by this toxigenic variant has been implicated in a higher death rate (10, 27, 47), and this strain has emerged not only in Canada but also in the United States (28, 32, 48). As concerning as these data are, they are likely a gross underestimate of the actual infection rate since there was not an active surveillance in place for reporting such infections and death rates due to C. difficile-associated disease during this time period.
Even more concerning is the dearth of information on the metabolic pathways of this emerging pathogen: few studies have addressed the means by which this anaerobe obtains energy. The only studies focused on physiology have determined that C. difficile requires five amino acids, leucine, isoleucine, proline, tryptophan, and valine, when grown in a defined medium (18, 22, 36). The addition of glycine to this minimal defined medium increases growth significantly (22). Toxin production (the primary means by which this organism causes disease) in C. difficile has been shown to increase upon addition or omission of certain amino acids to culture media (19, 22-24, 31, 49). Although the synthesis of toxins appears to be related to the level of certain amino acids in the culture medium, the molecular mechanism behind this link to fermentation pathways is still unexplained. Clearly, a better understanding of energy and carbon metabolism is needed since they may be tied directly to pathogenesis.
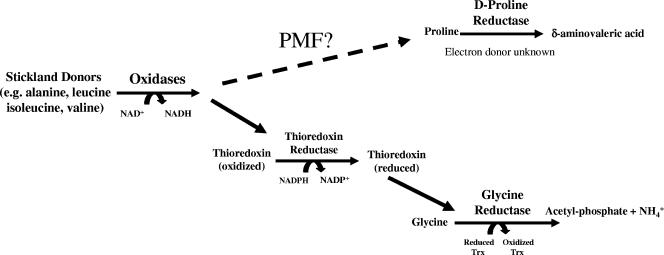
Using extracts of Clostridium sporogenes, Stickland (46) described the coupled fermentation of two amino acids in which one is oxidatively deaminated or decarboxylated (Stickland donor) and another amino acid (Stickland acceptor) is reductively deaminated or reduced (Fig. 1). This coupled amino acid fermentation has been demonstrated as a primary source of ATP generation in several model (nontoxigenic) organisms, including C. sporogenes, Clostridium sticklandii, and Eubacterium acidaminophilum (1, 4, 8, 42-44). Barker, also using cell extracts of C. sporogenes, established which amino acid combinations act as efficient Stickland donors and acceptors (2). The most efficient Stickland donors identified in these studies were leucine, isoleucine, and alanine, and the most efficient Stickland acceptors were glycine, proline, and hydroxyproline. Since these early reports on Stickland pairs, studies using C. sporogenes and C. sticklandii have demonstrated that both glycine and d-proline are efficient Stickland acceptors (2, 43-46). The two reductases that catalyze the reduction of the Stickland acceptors glycine and d-proline (glycine reductase [GR] and d-proline reductase [PR]) have best been characterized by Stadtman and her colleagues in C. sticklandii (6, 7, 42-45). Studies by Andreesen and his colleagues have also demonstrated that glycine derivatives (betaine and sarcosine) can also act as Stickland acceptors in E. acidaminophilum (1, 33), and the enzymes catalyzing the reduction of these amino acids are likely to contain the core subunits from the glycine reductase (1). Kabisch et al. also uncovered a selenoprotein subunit in the d-proline reductase (21). It should be noted that in the initial description of C. sticklandii, Stadtman and McClung reported that the closest related strain (based on biochemical characteristics) was the poorly understood C. difficile (45).
FIG. 1.
Overview of Stickland reactions. Based on previous studies with C. sticklandii, E. acidaminophilum, and C. sporogenes (1, 7, 42, 43, 46), a schematic overview is presented on the coupled oxidation and reduction of pairs of amino acids (Stickland reactions). In addition to oxidation of amino acids, Stickland reactions may also couple to the oxidation of purines and sugars, based on early work by Barker (2). The thioredoxin (Trx) and thioredoxin reductase (TrxR) systems have been suggested by Andreesen et al. to be directly linked to glycine reductase based on the colocalization of genes encoding Trx and TrxR with components of the glycine reductase in several organisms (1). The means by which reducing potential couples to d-proline reductase to produce proton motive force is unknown, although proline-dependent production of ΔpH has been demonstrated in C. sporogenes (29, 30).
d-Proline reductase catalyzes the reductive cleavage of the d-proline ring to yield δ-aminovaleric acid. Although this reaction does not result in the production of a molecule with high group transfer potential (compared to acetyl phosphate production by GR), it has been reported that PR is coupled to the generation of proton motive force (PMF) (30). This PMF generation likely represents the key energy-yielding pathway critical for anaerobes that specialize in Stickland fermentation of amino acids. The molecular mechanism by which PR couples to generate PMF has yet to be investigated. Although these selenoenzymes have been characterized in nontoxigenic strains, their putative role in the growth of the proteolytic toxigenic species of clostridia, such as C. difficile and C. botulinum, has not yet been evaluated.
In this report, we have begun to probe the metabolic pathways for energy metabolism in C. difficile. We have determined the nature of Stickland reactions in growth studies and purified and characterized both a d-proline reductase and a proline racemase to determine whether hydroxyproline could act directly as a Stickland acceptor. These studies are aimed at characterizing the bioenergetics of pathogenic clostridia and furthering our understanding of the biochemistry of Stickland reactions.
MATERIALS AND METHODS
Growth of C. difficile.
Two strains of C. difficile were used in this study, ATCC 9689 and strain 630. C. difficile 630 was kindly provided by Peter Mullany (Eastman Dental Institute, London, United Kingdom). The genome of strain 630 has been completely sequenced, and an initial analysis of the genome has been reported (40). Sequences utilized for annotation of genes were derived from data obtained by the Pathogen Sequencing Group at the Sanger Institute and can be obtained from ftp://ftp.sanger.ac.uk/pub/pathogens/cd. For routine growth and maintenance, cultures were grown in rich medium, TYPG (1.0% tryptone, 0.5% yeast extract, 10 mM KH2PO4, and 0.3% glucose), or in brain heart infusion (BHI; Oxoid).
In order to determine the need for selenium for growth of C. difficile on Stickland pairs, a basal medium was formulated to minimize contaminating selenium present in standard yeast extract. Torula yeast extract (Candida utilis; ICN) has been used in previous studies to establish selenium requirements for the production of the selenoenzyme nicotinic acid hydroxylase in Clostridium barkeri (16) as well as in the formulation of a mammalian diet demonstrated to cause selenium deficiency (5, 39). An autolysate of torula yeast extract (ICN) was generated by incubating yeast (250 g/liter in distilled water) at 50°C overnight, followed by clarification by centrifugation at 20,000 × g for 1 h at 4°C. The clarified autolysate was sterilized by autoclaving and stored at 4°C.
A basal medium, TTYP, formulated to minimize selenium contamination includes the following: 2 g/liter tryptone (Fisher Scientific), 0.1% torula yeast autolysate, 10 mM KH2PO4, and metal salts (10 μM CaCl2, 100 μM MgCl2, 100 μM FeSO4, 5 μM CoCl2, 5 μM NiCl2, 8 μM MnCl2, 3 μM Na2MoO42−). Selenium was added as selenite at a final concentration of 1 μM. After autoclaving, culture tubes (12 by 75 mm) were transferred to an anaerobic chamber with a 95% nitrogen-5% hydrogen atmosphere (Coy Laboratories, Grass Lake, Michigan) and Na2S was added to a final concentration of 0.03% to prereduce the medium. A 1% inoculum from a culture grown in the basal medium (TTYP) was used in all experiments to minimize the carryover of selenium from rich medium (brain heart infusion) stock cultures. Cultures were incubated in the anaerobic chamber in a model 2000 incubator (Coy Labs) at 37°C. Optical density measurements of cultures at 600 nm were determined using a Hewlett Packard 8453 diode array UV-visible spectrophotometer at the times indicated for each experiment.
Growth in defined medium was carried out as described by Karasawa et al. (22). l-4-Hydroxyproline was substituted for l-proline. In addition, phosphate (10 mM) was substituted for carbonate in the medium to maintain the pH of the culture, as CO2 is not a component of the gas mixture in the anaerobic chamber used in these studies.
75Se-labeling studies.
C. difficile strain 630 or ATCC 9689 was cultivated either in BHI (Oxoid Ltd., Basingstoke, England) or in TYPG medium. For the identification of selenoproteins, 10 μCi of 75Se (University of Missouri, Columbia), in the form of sodium selenite (50 nM), was added to each 9-ml culture (12- by 75-mm test tubes). Cells were harvested by centrifugation for 5 min at 5,000 × g, washed once with buffer A (0.1 M Tricine, pH 7.5, 0.1 mM phenylmethylsulfonyl fluoride, 0.5 mM EDTA), and resuspended in buffer A. Cells were lysed by sonication using a sonic dismembranator, model 100 (Fisher Scientific), for 10 seconds at a power output of 12 W, and the resultant crude cell extracts were clarified by centrifugation at 13,500 × g for 10 min at 4°C. Protein concentration was determined by the Bradford assay (3) using bovine serum albumin (Pierce) as a standard. Selenoproteins were identified by separating cell extracts using a 15% polyacrylamide gel, and radioisotope-labeled proteins were detected by PhosphorImager analysis (Molecular Dynamics).
Immunoblot analysis for glycine reductase selenoprotein A.
Polyclonal sheep antibodies to native selenoprotein A from C. sticklandii were a generous gift from Thressa Stadtman (NHLBI, NIH, Bethesda, Maryland). Cell extracts were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (15%), subsequently transferred to a polyvinylidene difluoride membrane, and blocked with Tris-buffered saline-Tween containing 2% bovine serum albumin for 1 h at 25°C. Membranes were incubated with primary antibody at 25°C for 4 h at a dilution of 1:500 in blocking buffer. After being washed with Tris-buffered saline-Tween, the membrane was incubated with secondary antibody (rabbit anti-sheep) conjugated with alkaline phosphatase (Sigma-Aldrich) at 25°C for 1 h. The blot was developed using BCIP (5-bromo-4-chloro-3-indolylphosphate) and Nitro Blue Tetrazolium as substrates for alkaline phosphatase in 100 mM Tris buffer, pH 9.0, 100 mM NaCl, and 5 mM MgCl2.
Purification of d-proline reductase.
Ten liters of C. difficile 630 was cultivated in a rich growth medium containing 20 g/liter tryptone, 10 g/liter yeast extract, 1.75 g/liter K2HPO4, 40 mM l-proline, 40 mM alanine, 1 μM selenite, and 0.05% Na2S to reduce the medium. The optimal concentrations of Stickland donor (alanine) and acceptor (proline) were determined experimentally and chosen based on cell yield determined by optical density at 24 to 36 h after inoculation (data not shown). Cells (harvested by centrifugation at 5,000 × g for 30 min) from 10 individual 1-liter cultures grown under identical conditions were pooled after resuspension in 25 ml of buffer A (50 mM Tris, pH 8.4, 1 mM EDTA, 1 mM dithiothreitol [DTT]), which contained 100 μM benzamidine to inhibit proteolysis. Cells were lysed by sonication at a power setting of 10 W using a Fisher Scientific model 100 sonifier in 30-second bursts until cell lysis was apparent. This crude cell lysate was clarified by centrifugation at 21,000 × g for 30 min at 4°C. Stepwise ammonium sulfate fractionation (25, 40, 60, and 85% saturation) was used as a first step to separate d-proline reductase-containing fractions from other cell components. The majority of the PR activity was obtained in the 40 to 60% ammonium sulfate pellet, and this fraction was subsequently applied to a phenyl-Sepharose (20 by 2.5 cm) column equilibrated in buffer A containing 2 M ammonium sulfate. Proteins were eluted using a 400-ml linear gradient of decreasing ammonium sulfate (2.0 M to 0.0 M) in buffer A. Active PR fractions (see description of assay below) were pooled and applied to EAH Sepharose (1.25 by 8 cm). After the column was washed in buffer A containing 0.1 M KCl to remove loosely bound proteins, PR was eluted by a 100-ml linear gradient of KCl (0.1 to 0.7 M) in buffer A. PR-containing fractions were then concentrated in an Amicon ultrafiltration membrane apparatus (YM-3 membrane) to a volume of 3.5 ml and applied to an S-200 gel filtration column (90 by 2.5 cm) equilibrated in buffer B (50 mM Tris, pH 8.4, 1 mM DTT, 250 mM KCl). PR-containing fractions eluted as a single sharp peak, and these fractions were aliquoted and quick-frozen in liquid nitrogen. This preparation was judged to be approximately 95% pure based on analysis by SDS-PAGE.
75Se-labeled PR was purified following essentially the same procedure for purification as that described above, with minor variations that did not affect the final protein preparation. The radiolabeled preparation of PR from 75Se-labeled cells was used for N-terminal sequence analysis and to confirm the presence of selenium within specific protein subunits of the purified enzyme complex.
Size exclusion chromatography.
Molecular mass standards used to calibrate the gel filtration column (Sephacryl S-200) were as follows: apoferritin, 480 kDa; gamma globulin, 160 kDa; bovine albumin, 67 kDa; chymotrypsinogen, 24 kDa; and cytochrome c, 13 kDa. Dextran blue was utilized to calculate the void volume of the column. From these data, a calibration curve was obtained to calculate the native molecular weight of the PR complex.
Fluorometric assay for d-proline reductase activity.
d-Proline reductase activity was assayed by following the DTT- and d-proline-dependent production of δ-aminovaleric acid, based on the method of Seto (41). The complete assay contained 50 mM Tricine, pH 8.4, 10 mM dithiothreitol, 1 μg of PR, and 1 mM d-proline. The detection was performed as previously described (41), with the only exception being the use of 96-well microplates for detection of the fluorometric product of the reaction of δ-aminovalerate with o-phthalaldehyde in a final volume of 200 μl. Initial rates were calculated based on the production of δ-aminovaleric acid per min at several time points (e.g., 2, 4, 6, and 8 min after the addition of enzyme). PR activities are reported as nanomoles per min per milligram of cell protein.
Expression and purification of proline racemase.
The gene encoding a putative proline racemase (prdF) was amplified by PCR from genomic DNA (strain 630) isolated using a Wizard genomic DNA kit (Promega, Madison, WI). A high-fidelity DNA polymerase enzyme mixture was utilized (DyNAzyme EXT; Finnzymes, Finland) for PCR. The sequences of the oligonucleotides utilized for amplification were as follows: 5′-GAATTTCATATGAAATTTAGCAGA-3′ and 5′-ATTGGATCCTTATTTAAGAATAAA-3′. Following digestion with NdeI and BamHI restriction enzymes, the resulting PCR product was directionally cloned into these corresponding sites in the expression vector pET15b (Novagen). The resultant plasmid, termed pSJ1, fuses the full-length PrdF protein with an N-terminal six-His tag to allow efficient purification by affinity chromatography.
For purification of the racemase, plasmid pSJ1 was transformed into Rosetta(DE3) and fresh transformants were used to inoculate 1 liter of modified Luria broth (10 g tryptone, 5 g yeast extract, 5 g NaCl) cultivated at 37°C in Fernbach flasks with shaking at 200 rpm. When the culture reached an optical density of 0.7 (600 nm), IPTG (isopropyl-β-d-thiogalactopyranoside) was added at 1 mM and the culture was incubated for an additional 16 h at 37°C before cells were harvested by centrifugation at 5,000 × g for 15 min. Cells were subsequently washed with buffer C (50 mM Tricine, pH 7.5, 150 mM NaCl), resuspended in 10 ml of buffer C, and lysed by sonication (Fisher Scientific Sonifier, model 100) at a power of 4 W in 30-second cycles until lysis was apparent. This crude cell lysate was clarified by centrifugation at 19,500 × g for 30 min at 4°C. The lysate was filtered through a 0.8 μM filter (Whatman), and NaCl was added to give a final concentration of 2 M. The lysate was applied to a 1-ml HiTrap chelating column (Amersham Pharmacia) and subsequently washed with buffer D (50 mM Tricine, pH 7.5, 2 M NaCl). Proline racemase was eluted by washing the column with buffer C containing 300 mM imidazole. This protein fraction was found to be at least 95% pure (based on SDS-PAGE analysis followed by Coomassie blue staining) and was subsequently concentrated 10-fold and stored at −80°C.
RESULTS AND DISCUSSION
Growth of C. difficile on Stickland amino acid pairs.
Both glycine and l-proline are required for optimal growth of C. difficile in defined media (18, 22, 36). Since the Stickland amino acid reductases (GR and PR) are selenoenzymes in nontoxigenic Clostridium sp., the metabolism of glycine and d-proline in the pathogenic C. difficile may also require selenium. To test this, we determined the growth of C. difficile in the presence of Stickland pairs in a basal culture medium with and without added selenium.
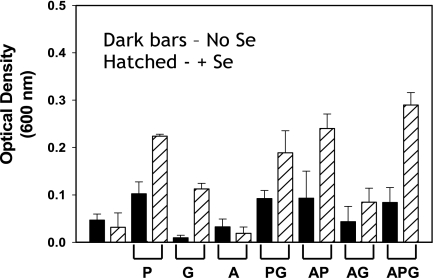
Complete removal of selenium from either defined or rich medium is made difficult by its presence as a trace contaminant in inorganic salts, yeast extract, and protein hydrolysates. The basal medium with torula yeast extract, termed TTYP, supported only a low level of growth of the C. difficile type strain (ATCC 9689) (Fig. 2). The addition of the Stickland donor alanine had no significant effect on growth. However, the addition of the Stickland acceptor d-proline increased growth significantly, especially when selenium was added (Fig. 2). Growth with glycine was increased only when selenium was also present in the medium. In the absence of selenium, glycine actually inhibited growth. This growth inhibition by glycine in the absence of selenium was partially alleviated by the addition of d-proline or by the Stickland donor alanine (Fig. 2). When the Stickland donor and both acceptors (d-proline and glycine) were added to the culture medium, overall growth increased in the presence of selenium. The growth yields (optical density) were obtained after 22 h, as this was determined to be the peak of optical density for growth in these culture media (data not shown). These results demonstrate that the growth of C. difficile in the presence of Stickland acceptors is dependent on selenium.
FIG. 2.
Growth of C. difficile requires selenium when cultured with Stickland pairs. Optical densities of cultures were measured at 22 h after inoculation (using a 1% inoculum), which was determined to be the peak density in growth curve studies (data not shown). Cultures were incubated at 37°C overnight in a 95% nitrogen-5% hydrogen atmosphere in a Coy anaerobic chamber. Selenium, when added, was in the form of selenite at a concentration of 1 μM. Torula yeast extract was used in the basal medium to reduce trace selenium normally present in TYPG medium (see Materials and Methods for details on TTYP medium composition). P, d-proline; G, glycine; A, alanine. The mean optical density from at least three individual cultures is plotted with the standard deviation plotted as error, and the experiment was reproduced at least three times.
Since several carbohydrates can be utilized as carbon sources by C. difficile (35), we determined the effect of added glucose on the stimulatory effect of Stickland amino acids and selenium on the growth of C. difficile. The addition of glucose to the culture medium only slightly increased the cell yield without changing the stimulation by Stickland acceptors and selenium (data not shown). This clearly demonstrates that although C. difficile can utilize glucose as a carbon source, the availability of amino acids as electron acceptors (Stickland fermentation) is a key limiting factor in growth yields of this pathogen.
We also tested the growth of a different strain, C. difficile 630, to confirm that increases in cell yield by the addition of Stickland pairs of amino acids are consistent between two C. difficile isolates. In the presence of equimolar (30 mM) alanine and glycine, C. difficile 630 cell yield was increased about 10-fold by the addition of selenium, reaching optical densities (measured at 600 nm) higher than 1.3 units in the TTYP basal medium. Growth on equimolar concentrations (30 mM) of alanine and d-proline was also enhanced by the addition of selenium but only about twofold (data not shown). Increasing the concentration of Stickland amino acid pairs to 100 mM did not increase the cell yield any higher than did the values with the 30 mM amino acid additions, likely due to limitations by other components in the basal growth medium. At all concentrations of amino acids tested, selenium was an integral requirement for higher growth rate and cell yield. These results show that for C. difficile, high cell yields can be obtained with either Stickland pair but only in the presence of selenium. Furthermore, these growth studies suggest that C. difficile likely uses the GR and PR selenoenzymes as a primary means of generating ATP, and this is reflected in the higher growth yields in selenium-containing media.
Hydroxyproline can substitute for proline in defined medium.
Hydroxyproline is a modified amino acid that is found in the host as a posttranslational modification of proline residues in collagen. To determine whether l-4-hydroxyproline can be utilized as a substitute for proline (as an electron acceptor) to support higher growth yield of C. difficile, we included l-4-hydroxyproline in a defined medium and omitted l-proline. Peak optical densities (22 h) obtained in the presence of l-4-hydroxyproline were similar (optical density of 0.97 ± 0.03) to readings for cultures with l-proline in the culture medium (optical density of 1.1 ± 0.10) (mean ± standard deviation). In the absence of either amino acid, no significant growth was observed, as expected (22). These results not only show that an amino acid derived solely from collagen (l-4-hydroxyproline) can serve to replace l-proline in defined medium but also demonstrate that the role of proline may not be for protein synthesis but as a needed electron acceptor, since hydroxyproline has been shown to be a Stickland acceptor in previous studies (2).
Analysis of genes encoding GR and PR.
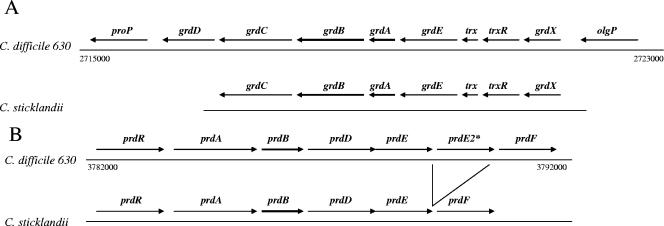
The genome sequence of C. difficile 630 was recently reported (40), and the sequence file is available from the Wellcome Trust Sanger Centre. Using established gene and protein sequences for glycine reductase and d-proline reductase from Clostridium sticklandii (21), we compared the genes encoding these enzymes in C. difficile 630. Figure 3 is a schematic of the organization of the genes that putatively encode the subunits of PR and GR on the chromosome of C. difficile 630. Genes encoding subunits of glycine reductase (denoted grd) are clustered together in an apparent operon, based on close linkage of the open reading frames in both C. difficile and C. sticklandii. Two of the genes, grdA and grdB, contain an in-frame UGA codon, suggesting that these two genes encode the two previously identified selenocysteine-containing subunits of GR (17). The locations of the SeCys residues in the C. difficile 630 GR amino acid sequences are conserved with respect to these residues in GRs from C. sticklandii and E. acidaminophilum. The gene order from grdX to grdC is also identical to that from C. sticklandii (17). It should be noted that a gene predicted to encode proline aminopeptidase (proP) is located at the end of the grd genes (Fig. 3A). This protein has been used as a biochemical confirmation for positive clinical isolates of C. difficile (12, 13), and its close linkage to the grd genes in C. difficile suggests a metabolic link of proline and glycine metabolism in C. difficile.
FIG. 3.
Identification of putative genes encoding glycine reductase and d-proline reductase in C. difficile 630. The sequence data used for annotation were produced by the Pathogen Sequencing Group at the Wellcome Trust Sanger Centre and were obtained from ftp://ftp.sanger.ac.uk/pub/pathogens/cd. A schematic representation of the genes encoded within two separate regions of the genomic DNA of Clostridium difficile 630, compared directly with similar operons in C. sticklandii, is shown. Numbers indicate the locations of the DNA coding regions within the linear genomic DNA sequence (Sanger Institute). (A) Alignment of the glycine reductase region from C. difficile 630 and C. sticklandii. (17). Primary putative assignment of genes in C. difficile was made by comparison with genes encoded by C. sticklandii (accession no. GI:11065682). The assignment of olgP (oligopeptidase) and proP (Xaa-Pro aminopeptidase) genes was made based on their homology to similar genes in Clostridium perfringens (accession no. GI:18145988) and Clostridium tetani (accession no. NP:782200), respectively. (B) Alignment of the proline reductase region from C. difficile 630 and C. sticklandii. Primary putative assignment of genes in C. difficile was made by comparison with genes encoded by C. sticklandii (accession no. GI:6899992).
The genes predicted to encode subunits of the PR, including one gene containing an in-frame UGA codon, are located within an apparent operon at a location distant from the genes for GR (Fig. 3B). The organization of this operon is also identical to that found in C. sticklandii, with the exception of a duplication of prdE in C. difficile (21). Located downstream, and potentially within the same operon of the prd genes, is an open reading frame predicted to encode the proline racemase (prdF). Proline racemase is critical for converting available l-proline to d-proline to generate the substrate for the d-proline reductase. In all of our growth studies, l-proline was interchangeable with d-proline, demonstrating the presence of an active proline racemase in the cell to convert l-proline to d-proline in C. difficile.
Selenium incorporation into glycine reductase and proline reductase.
Given the efficient growth of C. difficile strains on Stickland pairs of amino acids (including l-4-hydroxyproline) and the requirement for selenium, we sought to confirm the production of the predicted selenoproteins of GR and PR by labeling the cells with 75Se. Cells were cultured in the presence of 50 nM selenite (10 μCi 75Se) for 48 h in TYPG. The selenium-labeled proteins were identified by autoradiography of protein bands after separation on reducing, denaturing, 12% PAGE (Fig. 4A).
FIG. 4.
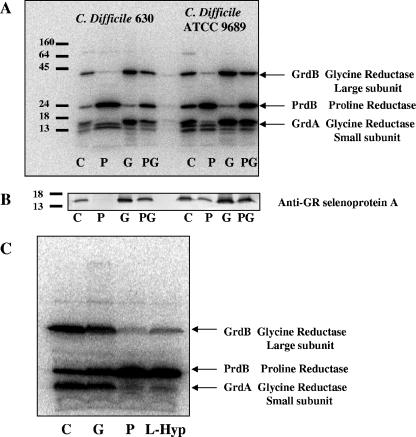
Radiolabeling (75Se) of C. difficile reveals increased proline reductase and glycine reductase upon the addition of glycine, d-proline, or l-4-hydroxyproline. (A) Cultures were grown in TYPG medium (see text) with 75Se (10 μCi) for 24 h, harvested, lysed by sonication, and separated by 12% SDS-PAGE. The locations of molecular mass markers are indicated at the left (in kDa). Arrows indicate the selenoprotein present in cell extracts grown under the following conditions: lanes 1 and 5, TYPG alone (C); lanes 2 and 6, TYGP plus d-proline (10 mM) (P); lanes 3 and 7, TYGP plus glycine (10 mM) (G); lanes 4 and 8, TYGP plus d-proline and glycine (10 mM each) (PG). (B) Immunoblot to detect the presence of glycine reductase selenoprotein A (small subunit). After SDS-PAGE, the same extracts from panel A were transferred to a polyvinylidene difluoride membrane and probed with polyclonal antibodies raised against GR selenoprotein A from C. sticklandii (kindly provided by T. C. Stadtman, NHLBI, NIH). (C) l-Proline and l-4-hydroxyproline also induce the production of a selenoprotein in C. difficile (strain 9689). Lane 1, TYPG medium (no addition) (C); lane 2, TYPG plus glycine (10 mM) (G); lane 3, TYPG plus l-proline (10 mM) (P); lane 4, TYPG plus l-4-hydroxyproline (10 mM) (L-Hyp). The predicted selenoproteins are indicated by arrows based on the molecular weights of the genes annotated in Fig. 3.
In both C. difficile 630 and type strain ATCC 9689, three major selenoproteins were identified upon labeling with 75Se (Fig. 4A). All three selenoproteins were expressed in the rich medium, with no additional amino acids, demonstrating that Stickland reactions are utilized during growth in protein hydrolysates. The addition of d-proline increased the 75Se-labeled protein band at 27 kDa, and the addition of glycine increased the levels of both a 17-kDa and a 47-kDa protein. These sizes correspond to the selenoprotein subunits predicted by genes encoding subunits of PR and GR (Fig. 3). Similar results were obtained when using BHI medium (data not shown). Additional minor radioactive bands may be attributed to other unknown selenoproteins or degradation products of selenoproteins.
The 17-kDa selenoprotein is GR selenoprotein A.
Polyclonal antibodies raised against the selenoprotein A from C. sticklandii were used to probe whether the small 17-kDa radiolabeled protein corresponds to GR selenoprotein A. These results are shown in Fig. 4B. An immunoreactive protein was detected at the same location as the radiolabeled protein (based on a calibrated protein marker). In addition, the intensity of the immunoreactive protein agrees with the level of the radiolabeled protein band from Fig. 4A. This confirms the identification of selenoprotein A and indicates that when glycine is present at higher concentrations in the culture medium, the use of glycine reductase is preferred and synthesis of the selenoprotein subunits of d-proline reductase is apparently repressed, since equal protein amounts from cell extracts were present on the membrane.
Hydroxyproline induces a selenoprotein similar in size to PrdB.
Barker originally reported the use of l-hydroxyproline as a Stickland acceptor in studies using cell extracts and reduced redox-active dyes (or hydrogen) as electron donors (2). Little is known about the use of hydroxyproline as a Stickland acceptor. Upon the addition of l-4-hydroxyproline to the culture medium, a selenoprotein similar in molecular weight to one produced when d-proline was added to the medium was produced at higher levels (Fig. 4C). This suggests that an enzyme similar to the previously studied PR from C. sticklandii (21, 43) is produced. The addition of either d-proline or hydroxyproline to the culture medium resulted in a substantial decrease in the level of glycine reductase selenoproteins A and B. This raised the question of whether the d-proline reductase from C. difficile reduces both d-proline and l-4-hydroxyproline (or a downstream metabolite of l-4-hydroxyproline) as substrates. Thus, we purified the radiolabeled selenoprotein induced by d-proline to determine the substrate specificity of this enzyme from C. difficile strain 630.
Purification and initial biochemical characterization of d-proline reductase.
Several preparations of PR were obtained in our studies, either by following radioisotope labeling (75Se) or by d-proline-dependent production of δ-aminovalerate in the presence of dithiothreitol as the electron donor as previously described (41). A purification scheme for PR that utilized enzyme activity for protein purification (as opposed to following radioisotope-labeled protein) is presented in Fig. 5 and Table 1. As described for PR from C. sticklandii (42), the enzyme complex did bind to EAH Sepharose, but some loss of activity was observed at this purification step (Table 1). A similar preparation (based on specific activity and SDS-PAGE analysis) was obtained by following 75Se-labeled fractions using the same chromatographic steps (data not shown) but with a slightly higher specific activity (875 units).
FIG. 5.
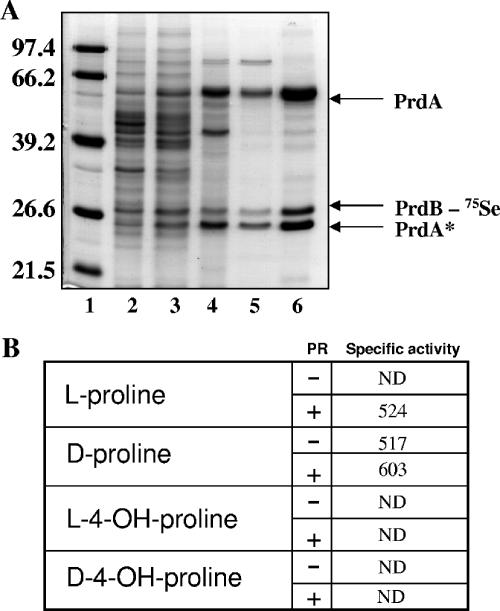
Purification and substrate specificity of C. difficile d-proline reductase. (A) Fractions from sequential steps of purification of d-proline reductase are separated by SDS-PAGE (15%) after being stained with Coomassie blue. Lane 1, molecular mass marker (size in kDa indicated at left); lane 2, crude cell extract; lane 3, 60% ammonium sulfate fraction; lane 4, phenyl-Sepharose; lane 5, EAH Sepharose; lane 6, Sephacryl S-200. Five micrograms of protein was loaded in each lane. The identification of protein subunits was accomplished by Edman degradation (see text for details). PrdA* is a proteolytically cleaved product of the precursor subunit PrdA (21). (B) Substrate specificity of d-proline reductase. Proline racemase (PrdF; Fig. 3) was purified by affinity chromatography (see Materials and Methods) and used to elucidate the substrate specificity of d-proline reductase from C. difficile. Specific activity was measured by assaying for δ-aminovaleric acid production as previously described (41). Proline racemase (1 μg) was preincubated with proline substrate for 30 min (30°C) prior to the addition of d-proline reductase. Each proline substrate was present in the reaction at a concentration of 1 mM. ND indicates no detectable activity. Activity is expressed as nmol min−1 mg−1 protein.
TABLE 1.
Purification of C. difficile d-proline reductase
| Sample | Protein amt (mg) | Sp act (nmol min−1 mg−1) | % Yield | Purification (fold) |
|---|---|---|---|---|
| Crude extract | 742 | 89.5 | 100 | |
| 60% (NH4)2SO4 | 125 | 173 | 33 | 1.9 |
| Phenyl-Sepharose | 24.9 | 314 | 11 | 3.5 |
| EAH Sepharose | 5.3 | 203 | 1.7 | 2.3 |
| Sephacryl S-200 | 1.4 | 513.6 | 1.1 | 5.7 |
Three separate protein subunits were observed in the purified preparation (Fig. 5). Edman degradation of these subunits confirmed that the largest subunit was indeed PrdA (SITLEAQ, initial methionine not present) and that the 27-kDa subunit was PrdB (SLTTVQGL). Excision of the protein bands from SDS-PAGE revealed that 75Se was present only in the PrdB subunit. The smallest subunit was initially blocked at the N terminus. Treatment of the protein with 0.1 N trifluoroacetic acid (which resulted in cleavage into smaller peptides) and subsequent analysis by Edman degradation revealed an internal sequence of PrdA (VRIM). Since no other gene within the prd or grd operon encodes a protein containing this amino acid sequence, it strongly suggests this is a processed PrdA subunit (PrdA*). The proteolysis of PrdA has been described in previous studies that demonstrate that posttranslational modification of this protein subunit is required for activation of the enzyme complex (1, 21, 42).
d-Proline reductase does not utilize either l- or d-hydroxyproline as a substrate.
Since it appears that PrdB selenoprotein is induced in radiolabeled cultures by the addition of either d-proline (l-proline) or l-4-hydroxyproline, we determined the substrate specificity of our preparation of PR. For this analysis, we also expressed and purified by affinity chromatography the proline racemase enzyme (see Materials and Methods for details). d-Proline reductase utilized d-proline as a substrate, but in the presence of l-proline, no δ-aminovaleric acid was produced (Fig. 5B). The addition of proline racemase and l-proline did result in comparable activities (Fig. 5B), confirming the activity of the expressed and purified proline racemase. We analyzed the kinetics of PR (higher activity preparation) with d-proline and found the apparent Km to be 200 μM, with a Vmax of 1,000 nmol min−1 mg−1. We also determined the optimum pH, which increased steadily upon increasing the pH above pH 7.0 and up to pH 8.6. We chose to use pH 8.4 for our assays since increasing pH values at or above 8.4 did not significantly increase PR activity. We also determined the native molecular weight of the enzyme complex by chromatography on Sephacryl S-200. The estimated native molecular mass was approximately 280 kDa, indicating that several subunits of PrdA and PrdB are present in the purified enzyme complex, although the composition cannot yet be determined. This is made difficult by the presence of a significant amount of unprocessed PrdA protein in the preparation, as this likely cannot contribute to enzyme activity (21, 42). To our knowledge, this is the first determination of the kinetic properties of PR from any organism.
Although the presence of proline racemase supported the conversion of l-proline to d-proline, neither l-4-hydroxyproline nor d-4-hydroxyproline was utilized as a substrate by the purified PR complex, even when present in concentrations of up to 5 mM. The addition of proline racemase also had no effect on the enzyme activity in the presence of these substrates (Fig. 5B). This would indicate that the d-proline reductase, in conjunction with the proline racemase, cannot directly convert hydroxyproline derived from collagen for this enzyme complex. However, the possibility remains that the product of the reduction of hydroxyproline to a hydroxyl derivative of δ-aminovaleric acid is not detected with o-phthalaldehyde (see Materials and Methods for details).
We also cultivated C. difficile 630 in the presence of 75Se-selenite and l-4-hydroxyproline as the Stickland acceptor and partially purified the radiolabeled protein induced under these growth conditions. This partially purified preparation also did not utilize l-4-hydroxyproline or d-4-hydroxyproline as a substrate in a DTT-dependent manner but did have good activity in the presence of d-proline. These results indicate that the selenoprotein induced upon the addition of l-4-hydroxyproline is in fact PR.
Divalent cations are not required for d-proline reductase activity.
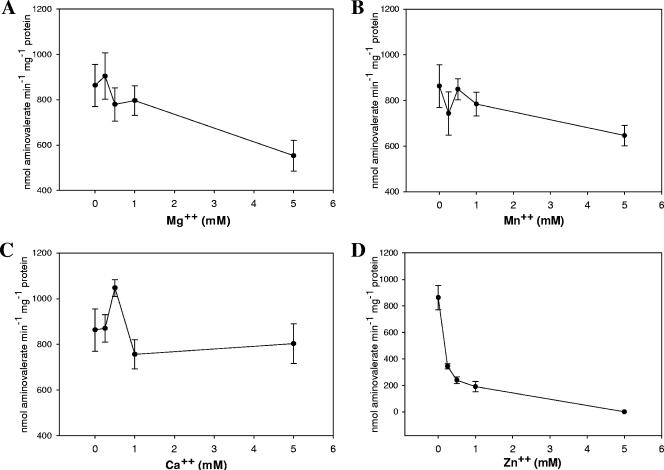
All previous characterization of d-proline reductases, whether in cell extracts or using purified preparations, have reported an absolute requirement for divalent cations, such as Mg2+. Our analysis of the required components for PR from C. difficile by omission of individual components of the enzyme assay revealed no requirement for any divalent cation (data not shown). The addition of EDTA or EGTA also did not significantly affect the activity of the enzyme. This is in contrast to previous reports (21, 43, 44). To further study this apparent discrepancy, we determined the activity of PR in the presence of various concentrations of magnesium, manganese, calcium, and zinc, and these results are summarized in Fig. 6.
FIG. 6.
Analysis of the requirement for divalent cations for d-proline reductase activity. The requirement for divalent cations for d-proline reductase activity was tested in the presence of several cations: (A) Mg2+, (B) Mn2+, (C) Ca2+, and (D) Zn2+. Specific activity is plotted versus cation concentration. The mean of these activities is plotted with the standard deviation shown as error. At least three independent enzyme assays are represented. Activity was determined by following the production of δ-aminovalerate as described in Materials and Methods.
The activity of PR increases slightly with magnesium added at 250 μM; however, this increase is not statistically significant. The addition of higher concentrations of magnesium reduced activity, as much as 30% when magnesium was present at 5 mM. It should be noted that this was the concentration typically used in previous reports of PR enzyme assays (21, 43, 44). No significant change in activity occurred in the presence of manganese, except for a decrease again at 5 mM. In contrast, the addition of calcium at 500 μM significantly increased PR activity. Nonetheless, this increase was not consistent when calcium was increased to millimolar levels. The addition of zinc dramatically reduced PR activity. This likely indicates the binding of zinc to the active-site selenocysteine and/or cysteine residue, which prevents catalytic reduction of the proline ring by the selenoprotein subunit.
Biochemical analysis of d-proline reductase revealed that hydroxyproline derivatives could not be used directly as electron acceptors. This suggests that a novel pathway may exist in C. difficile to interconvert hydroxyproline to proline so that it can be used to drive PMF via Stickland fermentations, since growth studies clearly show that hydroxyproline can be used as a Stickland acceptor. The lack of a requirement for divalent cations for PR catalysis and resistance to chelating agents was unexpected. This suggests that the PR from C. difficile may reduce the proline ring by a slightly different mechanism than PR isolated from other Stickland fermentors. The need for divalent cations is not well understood since the reaction mechanism for this enzyme has yet to be explored. The inhibition by zinc may indeed prove to be useful in probing this mechanism in future studies.
Based on the available nutrients in the host, it is tantalizing to suggest that collagen could serve as a primary source of these amino acids to act as electron acceptors. The structural domain of collagen is Gly-X-Y, where X is often proline and Y is often hydroxyproline (38). It has been reported that large clostridial toxin B from C. difficile can induce the production of matrix metalloproteinases (MMPs) (26, 34). MMP-2 could generate the needed oligopeptides for growth of C. difficile (glycine and proline rich) and gives a rationale to the type of noninvasive infection that is characteristic of this pathogen. This would also shed light on the apparent lack of collagenase activity exhibited by clinical isolates and explain a linkage between fermentation pathways and toxin regulation (37). Once small peptides are released by MMP-2, the oligopeptidase and proline aminopeptidase encoded by genes located adjacent to the grd operon (Fig. 3) would generate free proline, glycine, and hydroxyproline for use as Stickland acceptors. These free amino acids would then be used as Stickland acceptors for the synthesis of ATP by substrate-level phosphorylation (GR) as well as the production of PMF (PR). This potential linkage to toxin regulation and the use of collagen as a source of Stickland acceptors will be the focus of future studies.
Acknowledgments
We thank Thressa Stadtman from the National Institutes of Health for the generous gift of antibodies to the glycine reductase selenoprotein A from C. sticklandii. We also thank Peter Mullany (Dental Institute, London, United Kingdom) for the gift of C. difficile 630 and Rodney Levine for the Edman degradation analysis of d-proline reductase protein subunits.
Footnotes
Published ahead of print on 13 October 2006.
REFERENCES
- 1.Andreesen, J. R., M. Wagner, D. Sonntag, M. Kohlstock, C. Harms, T. Gursinsky, J. Jager, T. Parther, U. Kabisch, A. Grantzdorffer, A. Pich, and B. Sohling. 1999. Various functions of selenols and thiols in anaerobic gram-positive, amino acids-utilizing bacteria. Biofactors 10:263-270. [DOI] [PubMed] [Google Scholar]
- 2.Barker, H. A. 1961. Fermentation of nitrogenous organic compounds, p. 151-188. In I. C. Gunsalus and R. Y. Stanier (ed.), The bacteria, vol. 2. Academic Press, New York, N.Y. [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Britz, M. L., and R. G. Wilkinson. 1982. Leucine dissimilation to isovaleric and isocaproic acids by cell suspensions of amino acid fermenting anaerobes: the Stickland reaction revisited. Can. J. Microbiol. 28:291-300. [DOI] [PubMed] [Google Scholar]
- 5.Cases, J., V. Vacchina, A. Napolitano, B. Caporiccio, P. Besancon, R. Lobinski, and J. M. Rouanet. 2001. Selenium from selenium-rich Spirulina is less bioavailable than selenium from sodium selenite and selenomethionine in selenium-deficient rats. J. Nutr. 131:2343-2350. [DOI] [PubMed] [Google Scholar]
- 6.Cone, J. E., R. M. Del Rio, J. N. Davis, and T. C. Stadtman. 1976. Chemical characterization of the selenoprotein component of clostridial glycine reductase: identification of selenocysteine as the organoselenium moiety. Proc. Natl. Acad. Sci. USA 73:2659-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cone, J. E., R. M. del Rio, and T. C. Stadtman. 1977. Clostridial glycine reductase complex. Purification and characterization of the selenoprotein component. J. Biol. Chem. 252:5337-5344. [PubMed] [Google Scholar]
- 8.Costilow, R. N. 1977. Selenium requirement for the growth of Clostridium sporogenes with glycine as the oxidant in Stickland reaction systems. J. Bacteriol. 131:366-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eggertson, L. 2004. C. difficile hits Sherbrooke, Que., hospital: 100 deaths. Can. Med. Assoc. J. 171:436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eggertson, L. 2004. C. difficile: by the numbers. Can. Med. Assoc. J. 171:1331-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eggertson, L., and B. Sibbald. 2004. Hospitals battling outbreaks of C. difficile. Can. Med. Assoc. J. 171:19-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fedorko, D. P., and E. C. Williams. 1997. Use of cycloserine-cefoxitin-fructose agar and l-proline-aminopeptidase (PRO Discs) in the rapid identification of Clostridium difficile. J. Clin. Microbiol. 35:1258-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia, A., T. Garcia, and J. L. Perez. 1997. Proline-aminopeptidase test for rapid screening of Clostridium difficile. J. Clin. Microbiol. 35:3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.George, W. L., R. D. Rolfe, G. K. Harding, R. Klein, C. W. Putnam, and S. M. Finegold. 1982. Clostridium difficile and cytotoxin in feces of patients with antimicrobial agent-associated pseudomembranous colitis. Infection 10:205-208. [DOI] [PubMed] [Google Scholar]
- 15.George, W. L., V. L. Sutter, D. Citron, and S. M. Finegold. 1979. Selective and differential medium for isolation of Clostridium difficile. J. Clin. Microbiol. 9:214-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gladyshev, V. N., S. V. Khangulov, and T. C. Stadtman. 1994. Nicotinic acid hydroxylase from Clostridium barkeri: electron paramagnetic resonance studies show that selenium is coordinated with molybdenum in the catalytically active selenium-dependent enzyme. Proc. Natl. Acad. Sci. USA 91:232-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graentzdoerffer, A., A. Pich, and J. R. Andreesen. 2001. Molecular analysis of the grd operon coding for genes of the glycine reductase and of the thioredoxin system from Clostridium sticklandii. Arch. Microbiol. 175:8-18. [DOI] [PubMed] [Google Scholar]
- 18.Haslam, S. C., J. M. Ketley, T. J. Mitchell, J. Stephen, D. W. Burdon, and D. C. Candy. 1986. Growth of Clostridium difficile and production of toxins A and B in complex and defined media. J. Med. Microbiol. 21:293-297. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda, D., T. Karasawa, K. Yamakawa, R. Tanaka, M. Namiki, and S. Nakamura. 1998. Effect of isoleucine on toxin production by Clostridium difficile in a defined medium. Zentbl. Bakteriol. 287:375-386. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, S., C. R. Clabots, F. V. Linn, M. M. Olson, L. R. Peterson, and D. N. Gerding. 1990. Nosocomial Clostridium difficile colonisation and disease. Lancet 336:97-100. [DOI] [PubMed] [Google Scholar]
- 21.Kabisch, U. C., A. Grantzdorffer, A. Schierhorn, K. P. Rucknagel, J. R. Andreesen, and A. Pich. 1999. Identification of D-proline reductase from Clostridium sticklandii as a selenoenzyme and indications for a catalytically active pyruvoyl group derived from a cysteine residue by cleavage of a proprotein. J. Biol. Chem. 274:8445-8454. [DOI] [PubMed] [Google Scholar]
- 22.Karasawa, T., S. Ikoma, K. Yamakawa, and S. Nakamura. 1995. A defined growth medium for Clostridium difficile. Microbiology 141:371-375. [DOI] [PubMed] [Google Scholar]
- 23.Karasawa, T., T. Maegawa, T. Nojiri, K. Yamakawa, and S. Nakamura. 1997. Effect of arginine on toxin production by Clostridium difficile in defined medium. Microbiol. Immunol. 41:581-585. [DOI] [PubMed] [Google Scholar]
- 24.Karlsson, S., L. G. Burman, and T. Akerlund. 1999. Suppression of toxin production in Clostridium difficile VPI 10463 by amino acids. Microbiology 145:1683-1693. [DOI] [PubMed] [Google Scholar]
- 25.Kelly, C. P., and J. T. LaMont. 1998. Clostridium difficile infection. Annu. Rev. Med. 49:375-390. [DOI] [PubMed] [Google Scholar]
- 26.Koike, T., M. Kuzuya, T. Asai, S. Kanda, X. W. Cheng, K. Watanabe, Y. Banno, Y. Nozawa, and A. Iguchi. 2000. Activation of MMP-2 by Clostridium difficile toxin B in bovine smooth muscle cells. Biochem. Biophys. Res. Commun. 277:43-46. [DOI] [PubMed] [Google Scholar]
- 27.Loo, V. G., M. D. Libman, M. A. Miller, A. M. Bourgault, C. H. Frenette, M. Kelly, S. Michaud, T. Nguyen, L. Poirier, A. Vibien, R. Horn, P. J. Laflamme, and P. Rene. 2004. Clostridium difficile: a formidable foe. Can. Med. Assoc. J. 171:47-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loo, V. G., L. Poirier, M. A. Miller, M. Oughton, M. D. Libman, S. Michaud, A. M. Bourgault, T. Nguyen, C. Frenette, M. Kelly, A. Vibien, P. Brassard, S. Fenn, K. Dewar, T. J. Hudson, R. Horn, P. Rene, Y. Monczak, and A. Dascal. 2005. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 353:2442-2449. [DOI] [PubMed] [Google Scholar]
- 29.Lovitt, R. W., D. B. Kell, and J. G. Morris. 1987. The physiology of Clostridium sporogenes NCIB 8053 growing in defined media. J. Appl. Bacteriol. 62:81-92. [DOI] [PubMed] [Google Scholar]
- 30.Lovitt, R. W., D. B. Kell, and J. G. Morris. 1986. Proline reduction by Clostridium sporogenes is coupled to vectorial proton ejection. FEMS Microbiol. Lett. 36:269-273. [Google Scholar]
- 31.Maegawa, T., T. Karasawa, T. Ohta, X. Wang, H. Kato, H. Hayashi, and S. Nakamura. 2002. Linkage between toxin production and purine biosynthesis in Clostridium difficile. J. Med. Microbiol. 51:34-41. [DOI] [PubMed] [Google Scholar]
- 32.McDonald, L. C., G. E. Killgore, A. Thompson, R. C. Owens, Jr., S. V. Kazakova, S. P. Sambol, S. Johnson, and D. N. Gerding. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 353:2433-2441. [DOI] [PubMed] [Google Scholar]
- 33.Meyer, M., K. Granderath, and J. R. Andreesen. 1995. Purification and characterization of protein PB of betaine reductase and its relationship to the corresponding proteins glycine reductase and sarcosine reductase from Eubacterium acidaminophilum. Eur. J. Biochem. 234:184-191. [DOI] [PubMed] [Google Scholar]
- 34.Na, X., D. Zhao, H. W. Koon, H. Kim, J. Husmark, M. P. Moyer, C. Pothoulakis, and J. T. LaMont. 2005. Clostridium difficile toxin B activates the EGF receptor and the ERK/MAP kinase pathway in human colonocytes. Gastroenterology 128:1002-1011. [DOI] [PubMed] [Google Scholar]
- 35.Nakamura, S., S. Nakashio, K. Yamakawa, N. Tanabe, and S. Nishida. 1982. Carbohydrate fermentation by Clostridium difficile. Microbiol. Immunol. 26:107-111. [DOI] [PubMed] [Google Scholar]
- 36.Osgood, D. P., N. P. Wood, and J. F. Sperry. 1993. Nutritional aspects of cytotoxin production by Clostridium difficile. Appl. Environ. Microbiol. 59:3985-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poilane, I., T. Karjalainen, M. C. Barc, P. Bourlioux, and A. Collignon. 1998. Protease activity of Clostridium difficile strains. Can. J. Microbiol. 44:157-161. [PubMed] [Google Scholar]
- 38.Ramshaw, J. A., N. K. Shah, and B. Brodsky. 1998. Gly-X-Y tripeptide frequencies in collagen: a context for host-guest triple-helical peptides. J. Struct. Biol. 122:86-91. [DOI] [PubMed] [Google Scholar]
- 39.Rao, L., B. Puschner, and T. A. Prolla. 2001. Gene expression profiling of low selenium status in the mouse intestine: transcriptional activation of genes linked to DNA damage, cell cycle control and oxidative stress. J. Nutr. 131:3175-3181. [DOI] [PubMed] [Google Scholar]
- 40.Sebaihia, M., B. W. Wren, P. Mullany, N. F. Fairweather, N. Minton, R. Stabler, N. R. Thomson, A. P. Roberts, A. M. Cerdeno-Tarraga, H. Wang, M. T. Holden, A. Wright, C. Churcher, M. A. Quail, S. Baker, N. Bason, K. Brooks, T. Chillingworth, A. Cronin, P. Davis, L. Dowd, A. Fraser, T. Feltwell, Z. Hance, S. Holroyd, K. Jagels, S. Moule, K. Mungall, C. Price, E. Rabbinowitsch, S. Sharp, M. Simmonds, K. Stevens, L. Unwin, S. Whithead, B. Dupuy, G. Dougan, B. Barrell, and J. Parkhill. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779-786. [DOI] [PubMed] [Google Scholar]
- 41.Seto, B. 1979. Proline reductase: a sensitive fluorometric assay with o-phthalaldehyde. Anal. Biochem. 95:44-47. [DOI] [PubMed] [Google Scholar]
- 42.Seto, B., and T. C. Stadtman. 1976. Purification and properties of proline reductase from Clostridium sticklandii. J. Biol. Chem. 251:2435-2439. [PubMed] [Google Scholar]
- 43.Stadtman, T. C. 1956. Studies on the enzymic reduction of amino acids: a proline reductase of an amino acid-fermenting Clostridium, strain HF. Biochem. J. 62:614-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stadtman, T. C., and P. Elliott. 1957. Studies on the enzymic reduction of amino acids. II. Purification and properties of D-proline reductase and a proline racemase from Clostridium sticklandii. J. Biol. Chem. 228:983-997. [PubMed] [Google Scholar]
- 45.Stadtman, T. C., and L. S. McClung. 1957. Clostridium sticklandii nov. spec. J. Bacteriol. 73:218-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stickland, H. S. 1935. The chemical reactions by which Cl. sporogenes obtains its energy. Biochem. J. 28:1746-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valiquette, L., D. E. Low, J. Pepin, and A. McGeer. 2004. Clostridium difficile infection in hospitals: a brewing storm. Can. Med. Assoc. J. 171:27-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warny, M., J. Pepin, A. Fang, G. Killgore, A. Thompson, J. Brazier, E. Frost, and L. C. McDonald. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079-1084. [DOI] [PubMed] [Google Scholar]
- 49.Yamakawa, K., S. Kamiya, X. Q. Meng, T. Karasawa, and S. Nakamura. 1994. Toxin production by Clostridium difficile in a defined medium with limited amino acids. J. Med. Microbiol. 41:319-323. [DOI] [PubMed] [Google Scholar]