Abstract
The membrane topologies of the six subunits of Na+-translocating NADH:quinone oxidoreductase (Na+-NQR) from Vibrio cholerae were determined by a combination of topology prediction algorithms and the construction of C-terminal fusions. Fusion expression vectors contained either bacterial alkaline phosphatase (phoA) or green fluorescent protein (gfp) genes as reporters of periplasmic and cytoplasmic localization, respectively. A majority of the topology prediction algorithms did not predict any transmembrane helices for NqrA. A lack of PhoA activity when fused to the C terminus of NqrA and the observed fluorescence of the green fluorescent protein C-terminal fusion confirm that this subunit is localized to the cytoplasmic side of the membrane. Analysis of four PhoA fusions for NqrB indicates that this subunit has nine transmembrane helices and that residue T236, the binding site for flavin mononucleotide (FMN), resides in the cytoplasm. Three fusions confirm that the topology of NqrC consists of two transmembrane helices with the FMN binding site at residue T225 on the cytoplasmic side. Fusion analysis of NqrD and NqrE showed almost mirror image topologies, each consisting of six transmembrane helices; the results for NqrD and NqrE are consistent with the topologies of Escherichia coli homologs YdgQ and YdgL, respectively. The NADH, flavin adenine dinucleotide, and Fe-S center binding sites of NqrF were localized to the cytoplasm. The determination of the topologies of the subunits of Na+-NQR provides valuable insights into the location of cofactors and identifies targets for mutagenesis to characterize this enzyme in more detail. The finding that all the redox cofactors are localized to the cytoplasmic side of the membrane is discussed.
The Na+-pumping NADH:quinone oxidoreductase (Na+-NQR) is the main entry point for electrons into the aerobic respiratory chain of many marine and pathogenic bacteria. The enzyme oxidizes NADH, reduces quinone, and uses the free energy released in this redox reaction to generate a sodium motive force that can be used for motility and metabolic work (2, 5, 6, 8, 14, 19, 35, 43).
The Na+-NQR complex is made up of six subunits and accommodates a number of cofactors including several flavins (flavin adenine dinucleotide [FAD], flavin mononucleotide [FMN], and riboflavin) and a 2Fe-2S cluster (3, 4, 7). Two FMNs are bound covalently to subunits B and C of the enzyme, and a noncovalently bound FAD resides in subunit F (1, 15, 28). A molecule of ubiquinone-8 is believed to be bound near Gly-141 (Vibrio cholerae numbering) of NqrB on the basis of inhibitor studies (16). NqrF includes a motif typical of NADH binding sites. This evidence together with mutant studies indicate that this subunit is the entry point of electrons into the enzyme (29, 31).
Na+-NQR is an integral membrane enzyme. Five of the six subunits that make up the complex (all but NqrA) apparently include membrane-spanning segments. In order to elucidate the mechanism that operates in this enzyme, it is very important to know where a given point in the amino acid sequence is situated with respect to the membrane, whether it is on the cytoplasmic side, within the membrane itself, or on the periplasmic side. Topological information of this kind for the subunits NqrB, NqrC, and NqrF is important to elucidate where, in relation to the membrane plane, the different cofactors are located, particularly the covalently bound FMNs in subunits B and C. This information is essential for studying the mechanism that couples the redox reaction of Na+-NQR with the pumping of sodium. To function correctly, an ion pump must move ions across the membrane in a specific direction. In Na+-NQR, sodium ions are taken up from the cytoplasmic side of the membrane and are released on the extracellular side, resulting in the outer side of the cell membrane becoming positively charged with respect to the inner side. The directional (vectorial) nature of the ion-pumping process must be linked to an oriented placement of the protein with respect to the sides of the membrane. The localization of the redox cofactors and other putative pump-related sites with respect to the sides of the membrane is important for understanding the pump mechanism because it can reveal whether the work involved in moving Na+ up the membrane potential gradient is done during Na+ uptake or release. Furthermore, topological models of subunits NqrB, NqrD, and NqrE can help to identify conserved amino acid residues located within the membrane-spanning regions that are likely to be involved in sodium pathways.
Computer prediction programs can be used to generate topological maps of membrane proteins. We employed Web-based topology prediction algorithms to create a set of seven models for each subunit. The predictions generated in this way disagree in important respects. Importantly, the prediction of membrane-spanning helices is more accurate than predictions of absolute sidedness, i.e., whether the N terminus is on the cytosolic or periplasmic side. These discrepancies can often be resolved by studying fusions of reporters to the C termini of full-length or truncated membrane proteins. Typically, a reporter that is active only on one side of the membrane is used in parallel with another reporter that is active only on the opposite side (38). For our work, we chose to use bacterial alkaline phosphatase (PhoA) (22) as a reporter of periplasmic localization and green fluorescent protein (GFP) (30) as a reporter of cytoplasmic localization. All of the work with PhoA was done using Escherichia coli since it requires an alkaline phosphatase-deficient strain, and no V. cholerae strain of this kind was available. For the work with GFP, both E. coli and V. cholerae were used. The use of GFP in E. coli is well established, but to our knowledge, this is the first time that GFP fusions have been used in V. cholerae. In several cases, this was successful, but some of the GFP fusion products were apparently toxic to the cells.
By using reporter group gene fusion methods to resolve the discrepancies among topology modeling predictions, we were able to arrive at a single consensus model for each subunit of Na+-NQR (12, 18). This topological model will be useful for future mechanistic studies because each of the redox cofactors for which the amino acid binding site is known can now be localized with respect to membrane sidedness. Interestingly, all of the localized cofactors are found on the cytoplasmic (negative) side of the membrane, in contrast to what is seen in many other ion pumps.
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers.
Strains and plasmids used in this study are listed in Table 1. Primers were synthesized by either IDT, Inc., or Operon and were high-pressure liquid chromatography purified. Primers included appropriate restriction cleavage sites to allow the insertion of the PCR products into the pBAD vectors (KpnI for pBAD-PhoA and NheI or SpeI [NqrD] for pBAD-GFP).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| E. coli LMG194 | F− ΔlacX74 galE thi rpsL ΔphoA (PvuII) Δara714 leu::Tn10 | Invitrogen |
| V. cholerae O395N1 | Wild type, Smr | 25 |
| V. cholerae O395N1 Δnqr | Smr Δnqr | Unpublished results |
| Plasmids | ||
| pBAD-PhoA | pBAD22 derivative containing signal sequenceless phoA with a 5′ KpnI site | 26 |
| pBAD-GFP | pBAD plasmid with cycle 3 gfp | 20 |
| AANC | pBAD-PhoA nqrA; full-length C-terminal fusion | This study |
| ABN240 | pBAD-PhoA nqrB; C-terminal fusion at residue 240 | This study |
| ABN255 | pBAD-PhoA nqrB; C-terminal fusion at residue 255 | This study |
| ABN377 | pBAD-PhoA nqrB; C-terminal fusion at residue 377 | This study |
| ABNC | pBAD-PhoA nqrB; full-length C-terminal fusion | This study |
| ACN90 | pBAD-PhoA nqrC; C-terminal fusion at residue 90 | This study |
| ACN180 | pBAD-PhoA nqrC; C-terminal fusion at residue 180 | This study |
| ACNC | pBAD-PhoA nqrC; full-length C-terminal fusion | This study |
| ADN110 | pBAD-PhoA nqrD; C-terminal fusion at residue 110 | This study |
| ADN125 | pBAD-PhoA nqrD; C-terminal fusion at residue 125 | This study |
| ADNC | pBAD-PhoA nqrD; full-length C-terminal fusion | This study |
| AENC | pBAD-PhoA nqrE; full-length C-terminal fusion | This study |
| AFN80 | pBAD-PhoA nqrF; C-terminal fusion at residue 80 | This study |
| AFN160 | pBAD-PhoA nqrF; C-terminal fusion at residue 160 | This study |
| AFNC | pBAD-PhoA nqrF; full-length C-terminal fusion | This study |
| GANC | pBAD-GFP nqrA; full-length C-terminal fusion | This study |
| GCN90 | pBAD-GFP nqrC; C-terminal fusion at residue 90 | This study |
| GCN180 | pBAD-GFP nqrC; C-terminal fusion at residue 180 | This study |
| GCNC | pBAD-GFP nqrC; full-length C-terminal fusion | This study |
| GDN125 | pBAD-GFP nqrD; C-terminal fusion at residue 125 | This study |
| GDN125+L | PBAD-GFP nqrD; C-terminal fusion at residue 125 with 18-amino-acid linker | This study |
| GDNC | pBAD-GFP nqrD; full-length C-terminal fusion | This study |
| GDNC+L | pBAD-GFP nqrD; full-length C-terminal fusion with 18-amino-acid linker | This study |
| GENC | pBAD-GFP nqrE; full-length C-terminal fusion | This study |
Membrane topology prediction methods.
Seven current Web version topology prediction algorithms were utilized: TMHMMfix (27), MEMSAT3 (24), TOPPRED II (10), TMpred (http://www.ch.embnet.org/software/TMPRED_form.html), HMMTOP 2.0 (37), DAS (11), and SOUSI (17). Primary amino acid sequences of each subunit were individually submitted, and default options were used. A summary of the predictions for each of the six subunits of Na+-NQR is shown in Table 2. C-terminal fusions were then made to discriminate between the various predictions, focusing on the point of discrepancy. By using the results of the fusion studies together with the topological models, a consensus model was constructed according to a method described previously by Ikeda et al. (18).
TABLE 2.
Membrane topology predictions among seven Web-based algorithms for each of the six subunits of Na+-NQR
| Subunit | Predictiona
|
|||||||
|---|---|---|---|---|---|---|---|---|
| TMHMMfix | MEMSAT3 | TopPred | TMPred | HMMTOP | DASb | SOUSIb | Consensusc | |
| NqrA | 0P | 1C | 0P | 0P | 0P | 0 | 0 | 0 |
| NqrB | 9C | 10C | 10C | 8P | 11C | 9 | 9 | 9C |
| NqrC | 1P | 1P | 2C | 3C | 2P | 1 | 1 | 1P |
| NqrD | 5C | 6C | 5P | 5P | 6C | 6 | 5 | 5C |
| NqrE | 6C | 5C | 6P | 6P | 6P | 6 | 6 | 6P |
| NrqF | 1P | 1C | 3C | 2P | 2P | 2 | 2 | 2P |
Values indicate predicted numbers of transmembrane helices, and the P or C denotes the predicted periplasmic or cytoplasmic C-terminal location, respectively.
DAS and SOUSI do not provide predictions of N-terminal locations; therefore, the C-terminal locations cannot be predicted.
Consensus model based on rules described previously by Ikeda et al. (18).
Molecular genetic techniques.
V. cholerae O395N1 genomic DNA was used as a source of the nqr operon. Genomic DNA was extracted from a culture of V. cholerae grown overnight by using the Wizard purification kit (Promega) according to the manufacturer's instructions. Full-length or partial fragments of individual subunits of Na+-NQR were amplified by PCR using high-fidelity polymerase (Platinum SuperMix High Fidelity; Invitrogen) and subjected to agarose gel electrophoresis, and single bands of the correct size were purified from the agarose (MinElute or Qiaquick gel extraction kit; QIAGEN). Fragments were first cloned into pCR2.1-TOPO TA vectors (Invitrogen) and transformed into E. coli Top10 cells (Invitrogen). Cloned PCR fragments were excised from the pCR2.1 TOPO TA vector with the appropriate restriction enzymes (New England Biolabs [NEB]). The resulting restriction enzyme digestion products were purified from agarose gels. Fusion expression vectors were digested with the corresponding restriction enzymes and dephosphorylated with Antarctic phosphatase (NEB), which was subsequently heat inactivated (65°C for 15 min). Vectors and fragments were ligated using T4 ligase (NEB or Invitrogen) overnight at 16°C. The resulting plasmids were transformed into E. coli Top10 cells, and the orientation of the insert was checked by restriction enzyme digestion. Plasmids with the correct orientation relative to the reporter gene were transformed by electroporation (Bio-Rad) into E. coli LMG194 for PhoA fusions and V. cholerae O395N1 or V. cholerae Δnqr for GFP fusions.
For GDNC and GDN125, an 18-amino-acid linker was inserted in frame between the nqr gene fragment and the GFP gene (40) using a mutagenesis kit (Quikchange; Stratagene). Fusions for GDNC and GDN125 without the linker were also constructed.
Reporter gene assays.
Cultures of strains containing pBAD-PhoA/GFP fusions grown overnight were reinoculated into 50-ml cultures of Luria broth (Miller) with antibiotics (100 μg/ml ampicillin and, in the case of V. cholerae, 50 μg/ml streptomycin) and allowed to grow to early log phase before the addition of l-(+)-arabinose (Sigma) to a final concentration of 0.2% to induce expression. Growth was allowed to continue, and at late log phase, cells were harvested for analysis of PhoA or GFP activity (see below). At the same time, a 1-ml aliquot of culture was removed for sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and Western blot analysis. These cells were harvested by centrifugation, and the cell pellets were stored at −20°C until use.
A quantitative assay of alkaline phosphatase activity was conducted using a method described previously (21). After growth of cultures as described above, a 5-ml aliquot of culture was removed, 50 μl of 100 mM iodoacetamide (Sigma) was added, and the cells were incubated for 10 min at 37°C. Cells were then harvested by centrifugation, washed once with 5 ml cold PhoA buffer (10 mM Tris, pH 8.0, 10 mM MgSO4, 1 mM iodoacetamide), and resuspended in 1 ml 1 M Tris (pH 8.0)-1 mM iodoacetamide. The cell density was measured by the absorbance at 600 nm using an Agilent spectrophotometer (model 8453). PhoA activity was measured in cells permeabilized with SDS and chloroform, and incubated for 45 min at 37°C with p-nitrophenyl phosphate (pNPP) as a substrate. Absorbances at 420 nm and 550 nm were measured and used to calculate activity using the equation described previously by Manoil (21).
To observe GFP fluorescence, 10-ml aliquots of late-log-phase induced cells were harvested and resuspended in 0.5 ml of 10 mM Tris buffer, pH 8. Concentrated cells were mounted on 3% agarose pads, covered with a coverslip, and observed using a Zeiss Axiovert 200 M epifluorescent microscope equipped with a Plan-Neofluor 100×/1.3-numerical-aperture oil immersion objective, 1.6× Optavar, and an Endow GFP bandpass filter set. Images were obtained using a Zeiss AxioCam MRm charge-coupled-device digital camera and Axiovision software. Different samples were photographed with the same exposure time.
Expression and analysis of fusions.
For SDS-PAGE, thawed cell pellets were added to 100 μl of 2× sample loading buffer and incubated in a boiling water bath for 10 min. Denatured samples (10 μl) were loaded into a 4 to 12% Tris-Bis acrylamide gel (Invitrogen). Following electrophoresis, the gel was first photographed under UV illumination to detect the presence of fluorescent cofactors, and proteins were then transferred onto a polyvinylidene difluoride membrane. Western immunoblotting was performed on the polyvinylidene difluoride membranes according to the protocol for the Western Breeze kit (Invitrogen). The primary antibody was either mouse anti-alkaline phosphatase or mouse anti-GFP (Chemicon).
Membrane isolation.
The PhoA fusions ABNC and ACNC in V. cholerae Δnqr were grown in LB medium (with the appropriate antibiotics) to the beginning of early log phase, at which point arabinose was added to a final concentration of 0.2%. The cultures were allowed to grow to the beginning of stationary phase and then harvested. Cells were washed with a solution containing 10 mM Tris buffer, pH 8, 200 mM NaCl, and 5 mM MgSO4. Cells were broken using a microfluidizer (model 110S; Microfluidics) in the presence of DNase I (Sigma) and the protease inhibitor AEBSF [4-(2-aminoethyl)benzenesulfonyl fluoride]. Cell debris and unbroken cells were removed by centrifugation at 3,500 × g for 30 min. The supernatant containing the membrane fraction was then centrifuged at 100,000 × g for 12 h. The resulting pellet was resuspended in the Tris buffer using a glass-Teflon homogenizer, and membrane aliquots were frozen at −80°C until needed.
Protein concentration of the membrane fractions was determined by using the BCA method (Pierce).
RESULTS
Cloning and expression.
For all six subunits, full-length C-terminal fusions with PhoA were constructed. In addition, truncated C-terminal fusions with PhoA were constructed for NqrB, NqrC, NqrD, and NqrF. No truncated C-terminal fusions were constructed for NqrA and NqrE because topology prediction algorithms were in near agreement and could be discriminated based on the full-length fusion. A select set of fusions with GFP were constructed to provide confirmation of PhoA results. C-terminal fusions of full-length NqrA, NqrC, NqrD, and NqrE and truncated C-terminal fusions of NqrC and NqrD to GFP were constructed.
All PhoA fusions were expressed upon the addition of arabinose. Several GFP fusions, particularly the NqrD fusions, showed inhibition of growth upon the addition of arabinose while still showing limited levels of expression (not shown).
PhoA fusion analysis.
The activities of PhoA fusions are reported in Table 3. Based on the PhoA activity of E. coli LMG194 cells (typically observed to be between 0 and 20 units), a threshold of 40 units (a unit is the amount of substrate, pNPP, hydrolyzed per minute per optical density at 600 nm) was set for positive activity; high PhoA activity was defined as >100 units. The full-length NqrA fusion showed no PhoA activity, indicating a cytoplasmic location of the C terminus. Full-length fusions of NqrC and NqrE to PhoA yielded high PhoA activity. Only two of nine truncated fusions showed positive PhoA activity: ABN377 and AFN80.
TABLE 3.
Alkaline phosphatase (PhoA) activities of constructed fusions
| Fusiona | Alkaline phosphatase activityb | Locationc |
|---|---|---|
| AANC | −0.14 | C |
| ABN240 | 33 | C |
| ABN255 | 24 | C |
| ABN377 | 390 | P |
| ABNC | −7.4 | C |
| ACN90 | 20 | C |
| ACN180 | 0.2 | C |
| ACNC | 630 | P |
| ADN110 | 20 | C |
| ADN125 | 1.4 | C |
| ADNC | 9.5 | C |
| AENC | 400 | P |
| AFN80 | 230 | P |
| AFN160 | 6.2 | C |
| AFNC | 6.0 | C |
Descriptions are given in Table 1.
Activity is measured as the pNPP hydrolyzed per minute per optical density at 600 nm of cells.
C, cytoplasm; P, periplasm.
GFP fusion analysis.
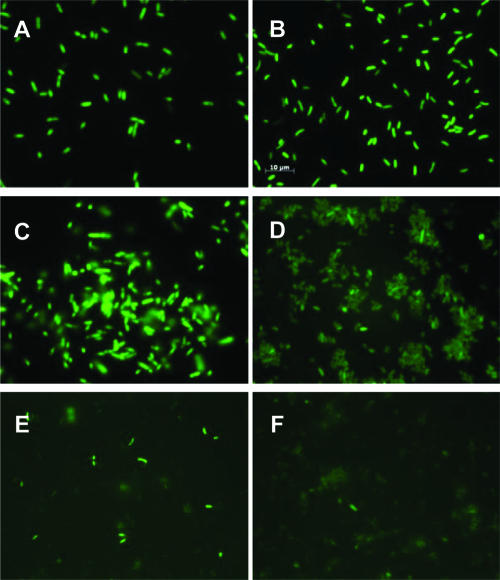
Observation of GFP activity in V. cholerae wild-type and Δnqr strains confirmed the ability of GFP to serve as a reporter of cytoplasmic localization (Fig. 1). Full-length C-terminal GFP fusions for NqrC and NqrE did not fluoresce under UV illumination, strengthening the conclusions from the PhoA analysis, which show that the C termini of NqrC and NqrE both lie on the periplasmic side of the membrane (Fig. 2 and 3). The full-length C-terminal GFP fusion for NqrA fluoresced under UV illumination, again confirming the results with the PhoA fusions and showing that the C terminus of this subunit lies on the cytoplasmic side of the membrane (Fig. 1C and D). Two GFP fusions with NqrD (GDN125 and GDNC) where the reporter group is expected to be in the cytoplasm, and therefore fluorescent, showed only low levels of fluorescence under the microscope (Fig. 1E and F). This is consistent with the fact that the addition of an inducer (arabinose) to these cells led to the inhibition of growth and low levels of expression as judged by Western blot analyses (not shown).
FIG. 1.
Epifluorescent micrograph of GFP fusions. (A) E. coli pBAD-GFP. (B) Wild-type V. cholerae pBAD-GFP. (C) GFP fusion to the complete nqrA gene (GANC) in E. coli Top10 cells. (D) GANC in wild-type V. cholerae. (E) GFP fusion to nqrD from the N terminus to amino acid 125 (GDN125) in V. cholerae Δnqr. (F) GFP fusion to the complete nqrD gene plus a linker (GDNC+L) in V. cholerae Δnqr. The scale bar in panel B applies to all panels.
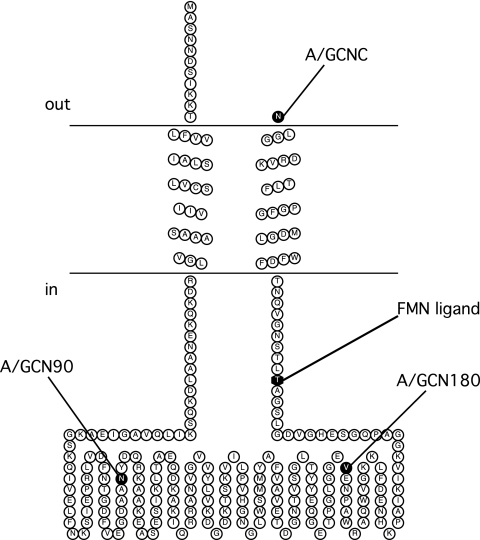
FIG. 2.
Membrane topology model of NqrC. The FMN binding site at residue T225 is shown. C-terminal fusion sites are indicated in the figure using the codes shown in Table 1.
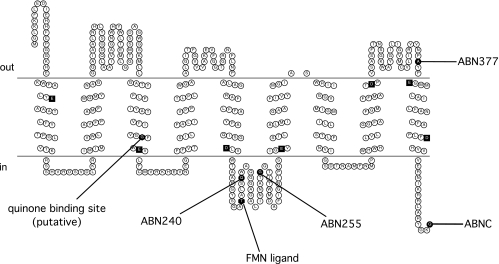
FIG. 3.
Membrane topology model of NqrB. Conserved negatively charged residues located in the membrane spans are represented as black squares. The FMN binding site at residue T236 and a predicted quinone binding site at residue G141 are shown. C-terminal fusion sites are indicated in the figure using the codes shown in Table 1.
Topology of Na+-NQR.
The topologies of the six subunits of Na+-NQR from V. cholerae were determined through a combination of topology prediction algorithms and gene fusion analysis. Topology prediction algorithms and rules for generating a consensus model were useful in reducing the total number of fusions to construct. C-terminal full-length and truncated fusions to PhoA were constructed and analyzed, yielding data that were used to model the topologies of the six subunits. Fusions with GFP were used to confirm a subset of the PhoA fusions.
The results of both the PhoA and GFP fusions with full-length NqrA are in agreement with a cytoplasmic location of this subunit. No transmembrane helices were predicted by the majority of the algorithms, which can be taken to indicate that NqrA is a soluble protein. We can therefore conclude that the entire subunit A resides on the cytoplasmic side of the membrane. The fact that NqrA consistently copurifies with the other five subunits indicates that it must be tightly attached to the complex by noncovalent interactions.
The PhoA activity results for NqrB are consistent with the topology model shown in Fig. 3. NqrB has nine transmembrane helices containing seven negatively charged residues that could be involved in sodium translocation. All of these residues are conserved across the Na+-NQR sequences of several organisms (Fig. 3). Analysis of the NqrB subunit also showed that the putative quinone binding site, G141 (G140 in Vibrio alginolyticus) (16), is localized to the cytoplasmic half of transmembrane helix III. The FMN binding site at residue T236 was localized to the cytoplasmic loop between transmembrane helices V and VI.
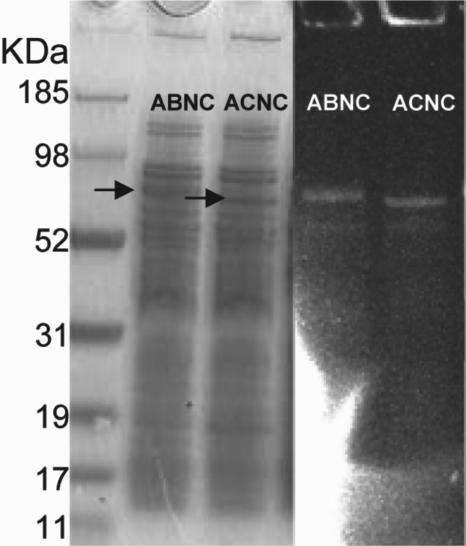
The PhoA fusion constructs also provided evidence that the FMN cofactors of NqrB and NqrC are incorporated, even when the remaining subunits of Na+-NQR are not present. When the membrane fraction from V. cholerae (Δnqr) containing the complete NqrB subunit fused to alkaline phosphatase (ABNC fusion) was run on an SDS-PAGE gel (Fig. 4), a fluorescent band was observed at the predicted molecular mass for the fusion protein (∼80 kDa), indicating the presence of the FMN. Similarly, the PhoA fusion for NqrC (ACNC fusion) also gave a fluorescent band at the predicted molecular mass for the fusion protein (∼70 kDa), indicating that the FMN in the subunit is also bound (Fig. 4). Since the SDS-PAGE gels denature the protein, these results also indicate that the flavins in these two subunits are already covalently bound.
FIG. 4.
Coomassie blue-stained and UV-illuminated SDS-PAGE gel of membrane fractions of the alkaline phosphatase fusion to the complete nqrB gene (ABNC) and the alkaline phosphatase fusion to the complete nqrC gene (ACNC) in V. cholerae Δnqr. The fluorescent bands indicate the presence of the covalently bound FMNs. Thirty micrograms of membrane protein was used in each lane.
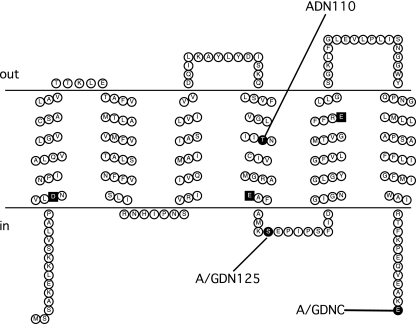
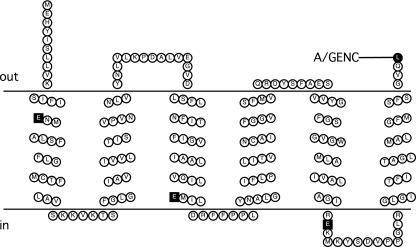
The topologies of homologues of NqrD and NqrE (YdgQ and YdgL, respectively) in E. coli have been previously determined and consist of six transmembrane helices each but with opposite overall orientations (YdgQ has its N and C termini in the cytoplasm, while YdgL has its N and C termini in the periplasm) (33). Our results are in agreement with the topologies of these homologues (Fig. 5 and 6). While the consensus model for NqrD had five transmembrane helices, three of the seven predictions included a sixth transmembrane helix at the N terminus. Our method of constructing fusions (via PCR and subcloning into the fusion vector) was unable to generate a fusion construct to test for the presence of this initial helix. However, analysis of the primary sequence in conjunction with the “positive-inside” rule (39) suggest that the N terminus is located in the cytoplasm, which would indicate that the initial helix exists. Both subunits contain negatively charged amino acids within their six transmembrane helices, and several of these residues are conserved.
FIG. 5.
Membrane topology model of NqrD. Conserved negatively charged residues located in the membrane spans are represented as black squares. C-terminal fusion sites are indicated in the figure using the codes shown in Table 1.
FIG. 6.
Membrane topology model of NqrE. Conserved negatively charged residues located in the membrane spans are represented as squares. C-terminal fusion sites are indicated in the figure using the codes shown in Table 1.
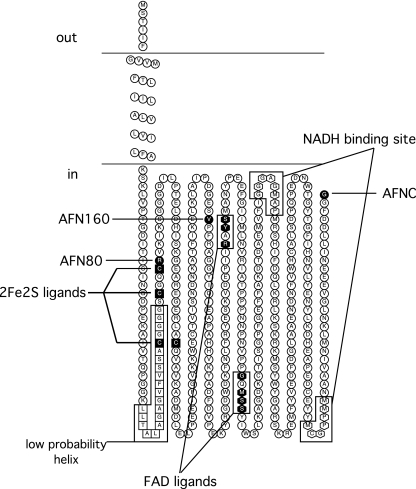
The fusion activity results indicated that the topology of NqrC consists of two transmembrane helices with the FMN binding site, residue T225, residing in the cytoplasm (Fig. 2). Fusion activity results for NqrF indicate a cytoplasmic localization of the cofactor binding regions (Fig. 7). A single transmembrane helix is most likely for NqrF, despite the two helices predicted in the consensus. All five topology algorithms that predicted a second transmembrane helix scored this helix as having a low probability of formation. Assuming that a single-helix topology is correct, an explanation of the positive PhoA activity for AFN80 is that the weakly hydrophobic region (the second predicted helix, indicated by white squares in Fig. 7) and the lack of a strong positively charged region preceding the fusion site lead to the translocation of the PhoA moiety to the periplasm, resulting in the observed activity. However, the existence of this second predicted helix is unlikely, since this two-helix model would locate one of the four cysteines that ligate the 2Fe-2S in the middle of membrane span. In addition, Turk et al. (36) were previously able to produce a soluble variant of NqrF by removing residues threonine 3 to alanine 25, consistent with this stretch of the protein being the membrane anchor.
FIG. 7.
Membrane topology model of NqrF. The binding sites for NADH, the 2Fe-2S center, and FAD are indicated. The residues of the weak hydrophobic region predicted to be a possible second transmembrane helix are shown as white squares. C-terminal fusion sites are indicated in the figure using the codes shown in Table 1.
DISCUSSION
Protein reporter group fusions are a well-established and proven method of studying the topology of membrane proteins (12, 22). Using this method, a marker is attached at a specific point in the amino acid sequence of a membrane protein so as to reveal the location of that point with respect to the membrane. This is accomplished by making a fusion on the gene level. The gene for the membrane protein is truncated at a point of interest, and the gene for a “reporter group” protein is spliced in. The objective is that when the modified gene is expressed, the remaining part of the membrane protein will be inserted into the membrane in the normal fashion, guiding the reporter group to one side of the membrane or the other, depending on where the membrane protein was truncated. A reporter group must be a protein that exhibits different properties depending on whether it resides in the cytosol or the periplasmic space. To be useful, the properties of the reporter group must manifest themselves in whole cells. We have used two reporter proteins in this study: GFP as the cytoplasmic reporter (12, 13) in both E. coli and V. cholerae (34) and PhoA (26) as the periplasmic reporter in a PhoA-deficient strain of E. coli.
Using a combination of computer-based topology predictions and gene fusion studies, we have obtained a consensus model for each of the six subunits of the Na+-NQR complex from V. cholerae. For each subunit, at least one fusion was made, providing an experimental point of reference to establish sidedness for the computer predictions. In several cases, additional fusions were made at intermediate points in the nqr genes to resolve discrepancies among the predictions from different algorithms (Table 2). For example, in the case of NqrC, the seven topology algorithms gave four different predictions (Table 2), and the consensus algorithm produced a prediction of one transmembrane helix with the C terminus located in the periplasm. We constructed reporter fusions at three different points in the sequence of this subunit. The results indicate that NqrC has two transmembrane helices and that the C terminus lies in the periplasm, consistent with the predictions from the HMMTOP algorithm. This result allows us to definitively localize the FMN cofactor to the cytoplasmic side of the membrane, a result which would have been impossible to obtain from theoretical models alone.
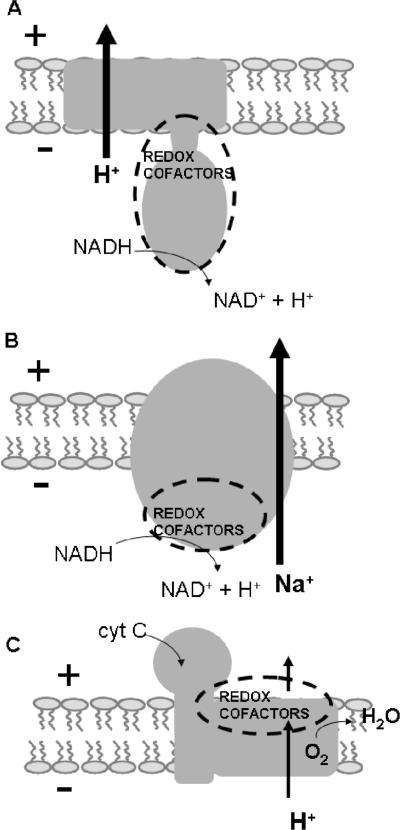
The results presented in this paper indicate that all the redox-active cofactors of Na+-NQR are located on the cytoplasmic (negative) side of the membrane, with the possible exception of riboflavin, whose location in the protein has not been established. This topological placement is analogous to that of the H+-pumping NADH:quinone oxidoreductase (complex I) but contrasts with those of some other redox-driven ion pumps. These differences almost certainly reflect important aspects of the ion-pumping mechanism (9, 23, 32). In the case of cytochrome c oxidase, a proton pump, the redox cofactors all lie to the positive side of the membrane, that is, toward the side where positively charged ions are ejected. The binuclear oxygen-reducing site, which is believed to play an important role in proton pumping, is located near the electrical center of the membrane (Fig. 8). For protons to reach the binuclear center, they must cross a fraction of the membrane, and therefore, a significant part of the work of ion translocation is done during proton uptake (41, 42). In Na+-NQR, with the redox cofactors near the negative side of the membrane, if we assume that there is a discrete binding site for sodium pumping associated with a redox cofactor, most of the electric work of ion translocation would need to be done during sodium release. We hope to study this question further by biophysical methods.
FIG. 8.
Scheme illustrating the relationship between redox cofactor sidedness and the directionality of ion translocation in primary pumps. (A) H+-pumping NADH:quinone oxidoreductase (complex I). (B) Na+-pumping NADH:quinone oxidoreductase (Na+-NQR). (C) Cytochrome c (cyt C) oxidase.
The results reported in this paper will help to guide future structure-function studies of Na+-NQR. They will be of particular importance for investigations of the chemiosmotic properties of this redox-driven Na+ pump, where it is essential to know the locations of cofactors and amino acid residues with respect to the membrane dielectric.
Acknowledgments
This study was funded by NIH grant R01 GM069936 to B.B.
We greatly appreciate the help, support, and critical reading of the manuscript of Joel Morgan. We thank Fern Finger and Christine Morton for use of and assistance with the epifluorescence microscope. We are grateful to Chengyen Wu for his skillful assistance in the preparation of the nqr deletion strain. We also thank Nadia Dolganov and Gary Schoolnik at Stanford University for their help in the preparation of strains.
Footnotes
Published ahead of print on 13 October 2006.
REFERENCES
- 1.Barquera, B., C. C. Hase, and R. B. Gennis. 2001. Expression and mutagenesis of the NqrC subunit of the NQR respiratory Na(+) pump from Vibrio cholerae with covalently attached FMN. FEBS Lett. 492:45-49. [DOI] [PubMed] [Google Scholar]
- 2.Barquera, B., P. Hellwig, W. Zhou, J. E. Morgan, C. C. Hase, K. K. Gosink, M. Nilges, P. J. Bruesehoff, A. Roth, C. R. Lancaster, and R. B. Gennis. 2002. Purification and characterization of the recombinant Na(+)-translocating NADH:quinone oxidoreductase from Vibrio cholerae. Biochemistry 41:3781-3789. [DOI] [PubMed] [Google Scholar]
- 3.Barquera, B., M. J. Nilges, J. E. Morgan, L. Ramirez-Silva, W. Zhou, and R. B. Gennis. 2004. Mutagenesis study of the 2Fe-2S center and the FAD binding site of the Na(+)-translocating NADH:ubiquinone oxidoreductase from Vibrio cholerae. Biochemistry 43:12322-12330. [DOI] [PubMed] [Google Scholar]
- 4.Barquera, B., W. Zhou, J. E. Morgan, and R. B. Gennis. 2002. Riboflavin is a component of the Na+-pumping NADH-quinone oxidoreductase from Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:10322-10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertsova, Y., and A. V. Bogachev. 2004. The origin of the sodium-dependent NADH oxidation by the respiratory chain of Klebsiella pneumoniae. FEBS Lett. 563:207-212. [DOI] [PubMed] [Google Scholar]
- 6.Bogachev, A., Y. V. Bertsova, B. Barquera, and M. I. Verkhovsky. 2001. Sodium-dependent steps in the redox reactions of the Na+-motive NADH:quinone oxidoreductase from Vibrio harveyi. Biochemistry 40:7318-7323. [DOI] [PubMed] [Google Scholar]
- 7.Bogachev, A., Y. V. Bertsova, E. K. Ruuge, M. Wikstrom, and M. I. Verkhovsky. 2002. Kinetics of the spectral changes during reduction of the Na+-motive NADH:quinone oxidoreductase from Vibrio harveyi. Biochim. Biophys. Acta 1556:113-120. [DOI] [PubMed] [Google Scholar]
- 8.Bogachev, A., and M. I. Verkhovsky. 2005. Na+-translocating NADH:quinone oxidoreductase: progress achieved and prospects of investigations. Biochemistry (Moscow) 70:143-149. [DOI] [PubMed] [Google Scholar]
- 9.Brandt, U. 2006. Energy converting NADH:quinone oxidoreductase (complex I). Annu. Rev. Biochem. 75:69-92. [DOI] [PubMed] [Google Scholar]
- 10.Claros, M. G., and G. von Heijne. 1994. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10:685-686.7704669 [Google Scholar]
- 11.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 12.Drew, D., D. Sjostrand, J. Nilsson, T. Urbig, C. N. Chin, J. W. de Gier, and G. von Heijne. 2002. Rapid topology mapping of Escherichia coli inner-membrane proteins by prediction and PhoA/GFP fusion analysis. Proc. Natl. Acad. Sci. USA 99:2690-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feilmeier, B. J., G. Iseminger, D. Schroeder, H. Webber, and G. J. Phillips. 2000. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J. Bacteriol. 182:4068-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi, M., Y. Nakayama, and T. Unemoto. 2001. Recent progress in the Na(+)-translocating NADH-quinone reductase from the marine Vibrio alginolyticus. Biochim. Biophys. Acta 1505:37-44. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi, M., Y. Nakayama, M. Yasui, M. Maeda, K. Furuishi, and T. Unemoto. 2001. FMN is covalently attached to a threonine residue in the NqrB and NqrC subunits of Na(+)-translocating NADH-quinone reductase from Vibrio alginolyticus. FEBS Lett. 488:5-8. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi, M., N. Shibata, Y. Nakayama, K. Yoshikawa, and T. Unemoto. 2002. Korormicin insensitivity in Vibrio alginolyticus is correlated with a single point mutation of Gly-140 in the NqrB subunit of the Na(+)-translocating NADH-quinone reductase. Arch. Biochem. Biophys. 401:173-177. [DOI] [PubMed] [Google Scholar]
- 17.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 18.Ikeda, M., M. Arai, D. M. Lao, and T. Shimizu. 2002. Transmembrane topology prediction methods: a re-assessment and improvement by a consensus method using a dataset of experimentally-characterized transmembrane topologies. In Silico Biol. 2:19-33. [PubMed] [Google Scholar]
- 19.Kojima, S., K. Yamamoto, I. Kawagishi, and M. Homma. 1999. The polar flagellar motor of Vibrio cholerae is driven by an Na+ motive force. J. Bacteriol. 181:1927-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu, C., W. E. Bentley, and G. Rao. 2004. A high-throughput approach to promoter study using green fluorescent protein. Biotechnol. Prog. 20:1634-1640. [DOI] [PubMed] [Google Scholar]
- 21.Manoil, C. 1991. Analysis of membrane protein topology using alkaline phosphatase and beta-galactosidase gene fusions. Methods Cell Biol. 34: 61-75. [DOI] [PubMed] [Google Scholar]
- 22.Manoil, C., J. J. Mekalanos, and J. Beckwith. 1990. Alkaline phosphatase fusions: sensors of subcellular location. J. Bacteriol. 172:515-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathiesen, C., and C. Hagerhall. 2002. Transmembrane topology of the NuoL, M and N subunits of NADH:quinone oxidoreductase and their homologues among membrane-bound hydrogenases and bona fide antiporters. Biochim. Biophys. Acta 1556:121-132. [DOI] [PubMed] [Google Scholar]
- 24.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 25.Mekalanos, J. J., D. J. Swartz, G. D. Pearson, N. Harford, F. Groyne, and M. de Wilde. 1983. Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature 306:551-557. [DOI] [PubMed] [Google Scholar]
- 26.Melchers, K., A. Schuhmacher, A. Buhmann, T. Weitzenegger, D. Belin, S. Grau, and M. Ehrmann. 1999. Membrane topology of CadA homologous P-type ATPase of Helicobacter pylori as determined by expression of phoA fusions in Escherichia coli and the positive inside rule. Res. Microbiol. 150:507-520. [DOI] [PubMed] [Google Scholar]
- 27.Melen, K., A. Krogh, and G. von Heijne. 2003. Reliability measures for membrane protein topology prediction algorithms. J. Mol. Biol. 327:735-744. [DOI] [PubMed] [Google Scholar]
- 28.Nakayama, Y., M. Yasui, K. Sugahara, M. Hayashi, and T. Unemoto. 2000. Covalently bound flavin in the NqrB and NqrC subunits of Na(+)-translocating NADH-quinone reductase from Vibrio alginolyticus. FEBS Lett. 474:165-168. [DOI] [PubMed] [Google Scholar]
- 29.Pfenninger-Li, X., S. P. Albracht, R. van Belzen, and P. Dimroth. 1996. NADH:ubiquinone oxidoreductase of Vibrio alginolyticus: purification, properties, and reconstitution of the Na+ pump. Biochemistry 35:6233-6242. [DOI] [PubMed] [Google Scholar]
- 30.Phillips, G. J. 2001. Green fluorescent protein—a bright idea for the study of bacterial protein localization. FEMS Microbiol. Lett. 204:9-18. [DOI] [PubMed] [Google Scholar]
- 31.Rich, P. R., B. Meunier, and F. B. Ward. 1995. Predicted structure and possible ion motive mechanism of the sodium-linked NADH-ubiquinone oxidoreductase of Vibrio alginolyticus. FEBS Lett. 375:5-10. [DOI] [PubMed] [Google Scholar]
- 32.Roth, R., and C. Hargerhall. 2001. Transmembrane orientation and topology of the NADH:quinone oxidoreductase putative quinone binding subunit NuoH. Biochim. Biophys. Acta 1504:352-362. [DOI] [PubMed] [Google Scholar]
- 33.Saaf, A., M. Johansson, E. Wallin, and G. von Heijne. 1999. Divergent evolution of membrane protein topology: the Escherichia coli RnfA and RnfE homologues. Proc. Natl. Acad. Sci. USA 96:8540-8544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott, M. E., Z. Y. Dossani, and M. Sandkvist. 2001. Direct polar secretion of protease from single cells of Vibrio cholerae via the type II secretion pathway. Proc. Natl. Acad. Sci. USA 98:13978-13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steuber, J. 2001. Na(+) translocation by bacterial NADH:quinone oxidoreductases: an extension to the complex-I family of primary redox pumps. Biochim. Biophys. Acta 1505:45-56. [DOI] [PubMed] [Google Scholar]
- 36.Turk, K., A. Puhar, F. Neese, E. Bill, G. Fritz, and J. Steuber. 2004. NADH oxidation by the Na+-translocating NADH:quinone oxidoreductase from Vibrio cholerae: functional role of the NqrF subunit. J. Biol. Chem. 279:21349-21355. [DOI] [PubMed] [Google Scholar]
- 37.Tusnady, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849-850. [DOI] [PubMed] [Google Scholar]
- 38.van Geest, M., and J. S. Lolkema. 2000. Membrane topology and insertion of membrane proteins: search for topogenic signals. Microbiol. Mol. Biol. Rev. 64:13-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Heijne, G., and Y. Gavel. 1988. Topogenic signals in integral membrane proteins. Eur. J. Biochem. 174:671-678. [DOI] [PubMed] [Google Scholar]
- 40.Waldo, G. S., B. M. Standish, J. Berendzen, and T. C. Terwilliger. 1999. Rapid protein-folding assay using green fluorescent protein. Nat. Biotechnol. 17:691-695. [DOI] [PubMed] [Google Scholar]
- 41.Wikstrom, M. 2004. Cytochrome c oxidase: 25 years of the elusive proton pump. Biochim. Biophys. Acta 1655:241-247. [DOI] [PubMed] [Google Scholar]
- 42.Wikstrom, M. 1998. Proton translocation by bacteriorhodopsin and heme-copper oxidases. Curr. Opin. Struct. Biol. 8:480-484. [DOI] [PubMed] [Google Scholar]
- 43.Zhou, W., Y. V. Bertsova, B. Feng, P. Tsatsos, M. L. Verkhovskaya, R. B. Gennis, A. V. Bogachev, and B. Barquera. 1999. Sequencing and preliminary characterization of the Na+-translocating NADH:ubiquinone oxidoreductase from Vibrio harveyi. Biochemistry 38:16246-16252. [DOI] [PubMed] [Google Scholar]