Abstract
In mammalian cells, fragile X mental retardation protein (FMRP) has been reported to be part of a microRNA (miRNA)-containing effector ribonucleoprotien (RNP) complex believed to mediate translational control of specific mRNAs. Here, using recombinant proteins, we demonstrate that human FMRP can act as a miRNA acceptor protein for the ribonuclease Dicer and facilitate the assembly of miRNAs on specific target RNA sequences. The miRNA assembler property of FMRP was abrogated upon deletion of its single-stranded (ss) RNA binding K-homology domains. The requirement of FMRP for efficient RNA interference (RNAi) in vivo was unveiled by reporter gene silencing assays using various small RNA inducers, which also supports its involvement in an ss small interfering RNA (siRNA)-containing RNP (siRNP) effector complex in mammalian cells. Our results define a possible role for FMRP in RNA silencing and may provide further insight into the molecular defects in patients with the fragile X syndrome.
INTRODUCTION
MiRNAs form a class of small regulatory RNAs (∼ 21−23 nt) involved in guiding translational repression or cleavage of mRNAs [1]. Biosynthesis of miRNAs is initiated upon transcription of miRNA-encoding genes into primary miRNAs (pri-miRNAs) by RNA polymerase II [2]. Pri-miRNAs are then processed by the nuclear ribonclease (RNase) III Drosha into miRNA precursors (pre-miRNAs) [3]. Following export to the cytoplasm via Exportin 5 [4], the imperfectly paired stem-loop pre-miRNAs are cleaved into miRNA:miRNA* duplexes by the RNase III Dicer [5–8]. Based upon the stability of the base pairs at the 5′ ends of the two strands [9], the mature miRNA will be incorporated into the RNA-induced silencing complex (RISC) [10], whereas the opposite miRNA* strand is encountered much less frequently and is presumably degraded [1]. The mRNA specifically recognized by the RISC will either be cleaved or translationally repressed, depending on whether the miRNA:mRNA pairing is perfect or not [1].
Hundreds of miRNAs have been identified in Drosophila, C elegans [11], A thaliana [12], zebrafish, mice, human cells [13–15], and viruses [16]. The ability of some of these miRNAs to regulate translation of specific mRNAs has been demonstrated experimentally. For example, cel-let-7 silences lin-41 mRNA through binding to its 3′ untranslated region (UTR) in C elegans [17], whereas miR-196 mediates cleavage of the HOXB8 mRNA in mouse embryos [18]. A mechanistic insight into miRNA-mediated translational repression was recently provided, as endogenous let-7 miRNPs were shown to inhibit translation initiation [19]. mRNA functional regulation by miRNAs has been involved in various cellular processes, such as leaf morphogenesis in plants, developmental timing and left/right asymmetry in nematodes, cell proliferation and apoptosis in flies, and hematopoietic cell differentiation in mice [1].
In humans, loss of expression of the FMR1 (fragile mental retardation 1) gene product, the fragile X mental retardation protein (FMRP), is the etiologic factor of the fragile X syndrome, the most frequent cause of inherited mental retardation [20, 21]. FMRP is an RNA-binding protein that contains two K-homology (KH) domains and an RGG box and is involved in RNA regulation of translation, RNA transfer, and local modulation of synaptic mRNA translation. However, its exact roles are still unclear and the mechanisms by which it controls translation are poorly understood. FMRP has been reported to behave as a negative regulator of translation both in vitro and in vivo [22–26], and it is deduced that the miRNA-guided RNA silencing pathway could be a cellular process through which FMRP could regulate translation of target mRNAs.
Indeed, a relation between FMRP and components of the RNAi machinery was uncovered. The ortholog of FMRP (dFMR1) was found to be associated with the effector RISC as well as miRNAs in Drosophila S2 cells [27, 28]. In mammalian cells, FMRP has been reported to be part of a ribonucleoprotein (RNP) complex with miRNAs and Argonaute 2 (Ago2) [29]. However, how FMRP functions in miRNA-mediated translational control remains unknown.
In this study, we have used recombinant proteins to show that human FMRP can accept miRNAs derived from Dicer cleavage and facilitate the formation of specific miRNA: target transition complexes in vitro. Reporter gene silencing assays, using various small regulatory RNAs, revealed the requirement of FMRP for efficient RNAi in vivo. The results obtained with single-stranded (ss) antisense siRNA also support its involvement in an ss siRNP effector complex in mammalian cells.
MATERIALS AND METHODS
Protein expression, purification, and analysis
Recombinant Dicer [6], FMRP deleted variant ΔKHT, FMRP mutant I304N, and FXR1P [30] proteins were expressed and purified as previously reported. Immunoblot analysis was performed with previously described antibodies recognizing FMRP [30] and FXR1P [31] proteins, and the immunoreactive proteins visualized with peroxydase-labeled affinity-purified goat anti-rabbit or mouse IgG secondary antibody using Western Lightning Chemiluminescent Reagent (PerkinElmer).
Preparation of RNAs
For in vitro experiments, all RNA substrates contained 5′-p and 3′-OH ends (for complete sequence and structure information, see Supplementary text available online at DOI 10.1155/JBB/2006/64347). Synthetic RNAs (Dharmacon) devoid of 5′-p were labeled at their 5′ end using T4 polynucleotide kinase (New England Biolabs) and [γ-32 P] ATP (PerkinElmer), and purified by denaturing PAGE. The following siRNA duplexes were used: CLP siRNA (5′-pAGGAGGUCGUACAGAAUUUdtdt) and Dicer siRNA (5′-pUGCUUGAAGCAGCUCUGGAdtdt).
Target RNAs (∼ 60 or ∼ 100 nt), encompassing the miRNA/siRNA binding sites and flanking regions, were synthesized from DNA templates by in vitro transcription using the T7 RNA polymerase (Ambion) and purified by denaturing PAGE. Templates were obtained either by annealing DNA oligonucleotides or by introducing a T7 RNA polymerase promoter by PCR followed by gel purification. The following target RNAs were used: CLP target RNA (nt 414−514, Acc no L54057), Dicer target RNA (nt 353−453, Acc no AJ132261), lin-41 target RNA (nt 4100−4162, Acc no AF195610), and HOXB8 target RNA (nt 2237−2297, Acc no X13721).
Prior to use, miRNAs and target RNAs were denatured at 90°C for 2 min and chilled on ice. Pre-miRNAs and miRNA:miRNA* were renaturated, and complementary strands of siRNA duplexes annealed, by incubation at 95°C for 5 min, and then slowly cooled down to room temperature.
For cell transfection, the siRNA duplexes siRluc (5′-GGCCUUUCACUACUCCUACdtdt) and siCtl (5′-GUAUAAUACACCGCGCUACdtdt) were used, whereas the ss antisense siRNAs asRluc (5′-GUAGGAGUAGUGAAAGGCCdtdt) and asCtl (5′-GUAGCGCGGUGUAUUAUACdtdt) were phosphorylated in vitro using T4 polynucleotide kinase, followed by ethanol precipitation, prior to use.
Dicer RNase assays
32P-labeled pre-miR-31 RNA (40000 cpm) was incubated in the absence or presence of recombinant Dicer (50−400 ng) and/or FMRP (0.18−2.8 μg) with MgCl2 (5 mM) at 37°C for 1 h, as described [6]. The samples were analyzed by denaturing PAGE and autoradiography. For the experiments with FMRP, the samples were treated with 40 μg proteinase K (Ambion) at 55°C for 45 min prior to analysis. RNA derived from Dicer cleavage was gel-purified and incubated with increasing amounts of recombinant FMRP (0.35−1.4 μg), without or with BSA (20 μg), for 30 min on ice prior to EMSA analysis.
EMSA
32P-labeled human GFP siRNA (0.25 pmol), hsa-miR-196a-1 miRNA:miRNA* duplex (0.50 pmol), or miRNA (0.50 pmol) (40000 cpm) was incubated in the absence or presence of recombinant FMRP (0.35−1.4 μg) for 30 min on ice prior to electrophoretic mobility shift assay (EMSA) analysis, as described [18]. FMRP-RNA complex formation was analyzed by nondenaturing PAGE (6%) and autoradiography, and analyzed quantitatively by AlphaImaging.
RNA strand annealing assays
These assays were adapted from Gabus et al [32]. Briefly, 0.015 pmol of 32P-labeled RNA was incubated, with or without target RNA, in the presence of recombinant FMRP, FMRP I304N, FMRP ΔKHT, or FXR1P [30] protein at 1 : 1, 2 : 1, or 4 : 1 (protein:RNA), or BSA (B) at 4 : 1 molar ratio. Assays were performed in 10 μL containing 20 mM Tris-HCl (pH 7.0), 0.1 mM MgCl2, 30 mM NaCl, 10 μM ZnCl2, 5 mM DTT, 5% Superase-In at 37°C for 10 min. The reaction was stopped by chilling on ice. After adding 0.5 volume of stop mix (20% glycerol, 20 mM EDTA pH 8.0, 0.2% SDS, 0.4 mg/mL yeast tRNA, and 0.25% bromophenol blue) to chase FMRP, the samples were analyzed by nondenaturing 10% PAGE in TBE buffer. The gels were dried and the radioactive RNA complexes were visualized by autoradiography, and analyzed quantitatively by PhosphorImaging.
DNA constructs, cell culture, and luciferase assays
The psiSTRIKE and psiCHECK vectors were obtained from Promega. Mouse embryonic Fmr1 KO (STEK TSV-40, 3T3A, 3T6A, or TpBSVE) [24, 30, 33], Fxr1 KO (KETS) (to be described elsewhere) and wild-type (Naïves or NIH/3T3) fibroblasts were grown in DMEM supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin in a humidified incubator under 5% CO2 at 37°C. Expression of FMRP, FXR1P, and actin proteins was verified by Western blot using anti-mAb1C3 [34], mAb2FX [31], and anti-actin AC-40 (Sigma) antibodies, respectively. Cells grown in 12-well plates to 70% confluency were transfected with 0.8 μg of the psiCHECK reporter vector and either 0.0005−0.5 μg of the psiSTRIKE construct using Lipofectamine 2000 (Invitrogen) or 100−400 nM of ss antisense Rluc siRNA (asRluc) or 100 nM of Rluc siRNA duplex (siRluc) using Oligofectamine (Invitrogen). Twenty-four hours later, cells were harvested, lysates were prepared, and Rluc and Fluc activities were measured successively using the Dual-Luciferase Reporter Assay System (Promega).
RESULTS
FMRP acts as an acceptor of miRNAs derived from Dicer cleavage
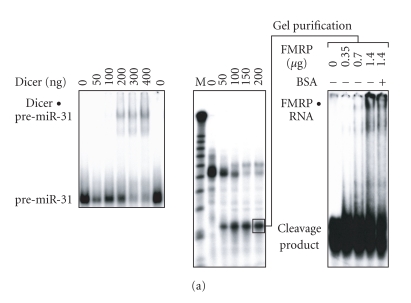
Dicer activity has been reported to be associated with FMRP in EBV-transformed human B cells [29], suggesting that Dicer and FMRP are part of a complex in vivo. Since FMRP is likely acting downstream of Dicer in the RNA silencing pathway, we asked whether FMRP can act as a miRNA acceptor protein for the ribonuclease Dicer. To test this hypothesis, the 32P-labeled miRNA precursor pre-miR-31 was incubated in the presence of recombinant Dicer, which bound (Figure 1(a), left panel) and cleaved pre-miR-31 into a ∼ 21-nt RNA product (Figure 1(a), center panel). When this 32P-labeled RNA derived from Dicer cleavage was gel-purified and incubated with increasing amounts of recombinant FMRP, a gel mobility shift was observed, compatible with the formation of an FMRP-RNA complex (Figure 1(a), right panel).
Figure 1.
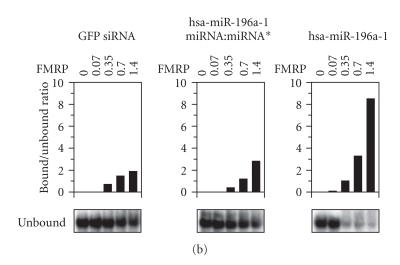
FMRP can act as an acceptor of miRNA derived from Dicer cleavage of pre-miRNA. (a) Recombinant FMRP interacts with Dicer RNA cleavage products. 32P-labeled pre-miR-31 was incubated in the absence or presence of recombinant Dicer, without (left) or with (center) MgCl2. The samples were analyzed by nondenaturing (left) or denaturing (center) PAGE and autoradiography. RNA derived from Dicer cleavage was gel-purified and incubated with increasing amounts of recombinant FMRP (0.35–1.4 μg), without or with BSA (20 μg), prior to EMSA analysis (right). M indicates a 10-nt RNA size marker. (b) FMRP preferentially interacts with miRNAs. 32P-labeled green fluorescent protein (GFP) siRNA (left), hsa-miR-196a-1 miRNA:miRNA* duplex (center), or miRNA (right) was incubated in the absence or presence of increasing amounts of recombinant FMRP. The samples were analyzed by EMSA and autoradiography, and analyzed quantitatively by PhosphorImaging. Bound RNA was expressed as a bound to unbound ratio.
The relative ability of FMRP to interact with RNA species involved in RNA silencing was further quantified by EMSA. Initial analyses showed that recombinant FMRP associates with the human immunodeficiency virus transactivating response DNA [32] and RNA (I Plante and P Provost, unpublished data) structure, the latter of which closely resembles that of a pre-miRNA. We observed that FMRP preferentially recognized mature miRNA (Figure 1(b), right), as monitored by the concomitant decrease in unbound RNA levels. Under these conditions, FMRP only weakly bound a perfect siRNA (Figure 1(b), left), or an imperfect miRNA:miRNA* (Figure 1(b), center) duplex. These results confirm the ability of FMRP to bind to the effector mature miRNA.
FMRP assembles miRNAs and ss siRNAs on target RNAs in vitro
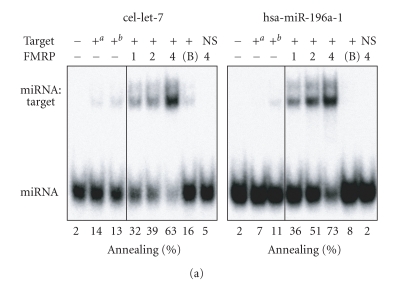
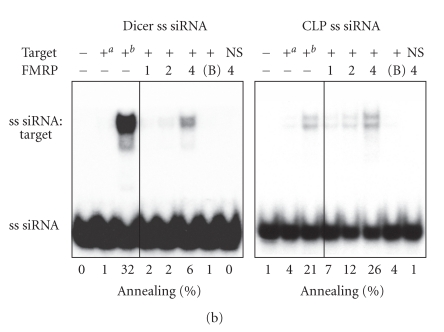
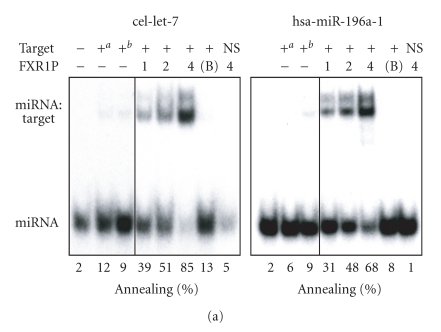
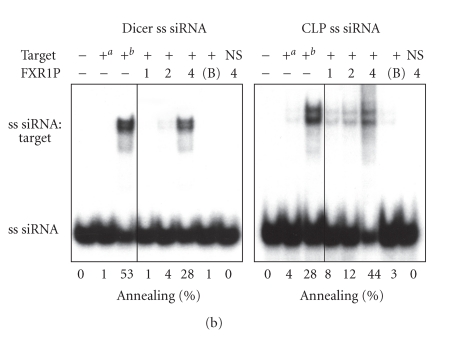
Given the ability of FMRP to interact with several hundreds of mRNAs [35, 36] and its recently described nucleic acid chaperone properties [32], we asked if FMRP could facilitate the assembly of miRNAs on target RNAs. To this end, single miRNA strands were incubated with a specific target RNA in the presence or absence of recombinant FMRP. Formation of imperfectly paired miRNA:target RNA complexes was visualized by nondenaturing PAGE after FMRP removal. We observed FMRP-mediated assembly of cel-let-7 (Figure 2(a), left panel) and hsa-miR-196a-1 (Figure 2(a), right panel) miRNAs on their specific 60-nt RNA targets. No annealing was observed in the presence of bovine serum albumin (BSA) (Figure 2(a), 7th lane of each panel) or when the cel-let-7 target lin-41 was swapped with that of hsa-miR-196a-1 (HOXB8) (Figure 2(a), last lane of each panel). Whether this activity could be extended to siRNAs was tested using siRNAs directed against Dicer and coactosin-like protein (CLP) mRNAs. In both cases, assembly of single siRNA strands on their 100-nt RNA targets was observed (Figure 2(b)), but not when the targets were interchanged (Figure 2(b), last lane of each panel). Used as hybridization control reactions, temperature-mediated annealing of ss siRNAs to their targets was superior to that of miRNAs, as expected considering the increased stability of perfectly complementary ss siRNA:target RNA versus imperfect miRNA:target RNA duplexes. In contrast, FMRP favored annealing of miRNAs to their targets, more than that of ss siRNAs (compare Figure 2(a) versus Figure 2(b)), thereby illustrating its preference for imperfect, but natural miRNA:target RNA complex formation.
Figure 2.
FMRP preferentially facilitates natural miRNA:target RNA complex formation. FMRP mediates miRNA (a) and siRNA (b) annealing to specific RNA targets. (a) 32P-labeled cel-let-7 (left) or hsa-miR-196a-1 (right) was incubated with a specific lin-41 or HOXB8 RNA target, respectively, in the absence or presence of increasing amounts of FMRP, or BSA at 4 : 1 (protein:RNA) molar ratio. (b) The RNA strand annealing assays using a single 32P-labeled strand of Dicer (left) or CLP (right) siRNA duplex incubated with a specific or nonspecific RNA target were performed as in (a). Control hybridization was conducted at 37°C for 10 mina or 65°C for 30 minb. RNA complexes were analyzed by nondenaturing 10% PAGE and autoradiography. B, BSA; NS, nonspecific target.
FMRP mediates miRNA annealing to specific RNA targets through its KH domains
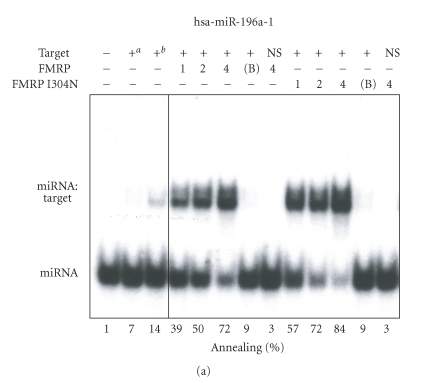
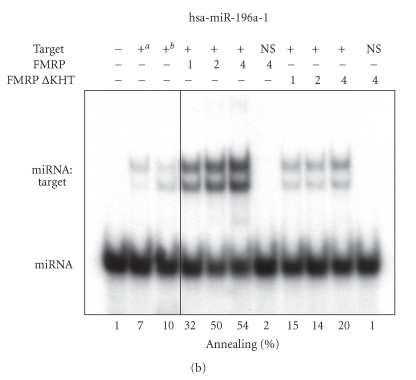
To get further insights into the miRNA annealing properties of FMRP, we assessed the activity of the biologically relevant FMRP I304N mutant and the deleted ΔKHT FMRP version. A single substitution (I304N) in the Fmr1 gene product FMRP was found in a fragile X patient suffering of a severe form of fragile X syndrome [37]. When the recombinant FMRP I304N mutant protein was tested in RNA strand annealing assays, it exhibited properties comparable to the wild-type protein (Figure 3(a)). On the other hand, deletion of the KH domains markedly compromised its miRNA annealing properties (Figure 3(b)). Since the KH domains present in RNA-binding proteins are known to mediate binding to ss nucleic acids, as reviewed recently [38], these results support the requirement of the ssRNA-binding KH domains for FMRP-mediated miRNA annealing to specific RNA targets.
Figure 3.
FMRP mediates miRNA annealing to specific RNA targets through its KH domains. RNA strand annealing properties of the FMRP I304N (a) and ΔKHT (b) mutants. (a) 32P-labeled hsa-miR-196a-1 miRNA was incubated with a specific HOXB8 RNA target in the absence or presence of FMRP or FMRP I304N or with BSA at 4 : 1 (protein:RNA) molar ratio. (b) The RNA strand annealing assays using the FMRP ΔKHT mutant were performed as in (a). Control hybridization was conducted at 37°C for 10 mina or 65°C for 30 minb. RNA complexes were analyzed by nondenaturing 10% PAGE and autoradiography. B, BSA; NS, nonspecific lin-41 RNA target.
FMRP is required for efficient RNA silencing in mammalian cells
In order to assess whether the activity displayed by FMRP in vitro is important for RNA silencing in vivo, we set up a reporter-based cellular assay using cultured mouse embryonic fibroblasts established from Fmr1 KO [24] or isogenic wild-type mice. In these assays, a dual reporter gene construct encoding Renilla (Rluc) and Firefly (Fluc) luciferase was cotransfected with a construct encoding an shRNA directed against Rluc. We used a second Fluc reporter that, in addition to correct for cell-to-cell variability, allowed us to examine the function of FMRP pertaining to small RNA-mediated RNA silencing. This was essential considering that FMRP can act as a negative regulator of translation [22–24].
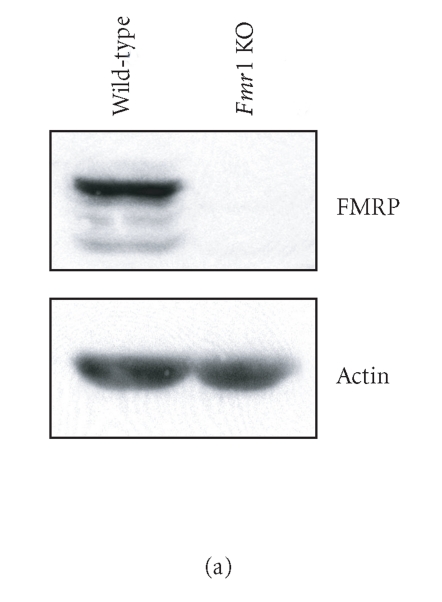
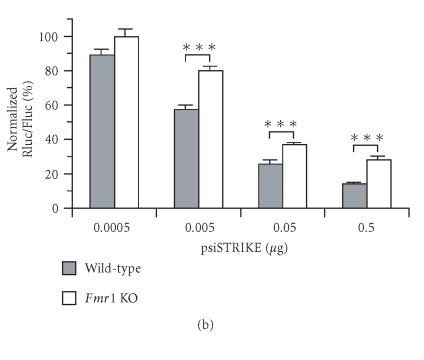
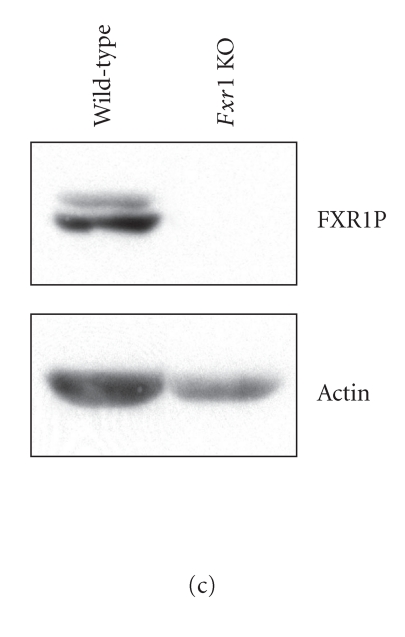
Immunoblot analyses of Fmr1 KO cell protein extracts confirmed the absence of FMRP (Figure 4(a), right lane), whereas FMRP was detected in wild-type cells (Figure 4(a), left lane). Expression of Rluc shRNA, which had no effect on interferon regulatory factor-1 (IRF-1) protein levels (see Supplementary Data, Figure S1), induced a dose-dependent decrease in Rluc activity in wild-type cells (Figure 4(b)). The efficiency of Rluc shRNA to induce RNA silencing was significantly impaired in Fmr1 KO cells, as compared to wild-type cells.
Figure 4.
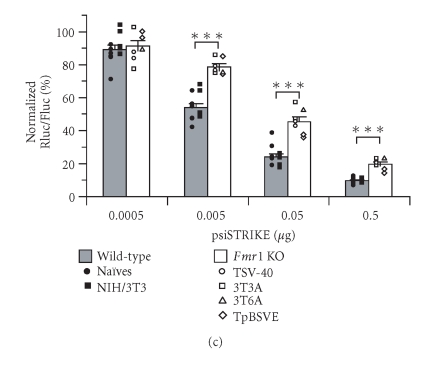
FMRP is required for efficient RNA silencing in mammalian cells. (a) Expression of FMRP in wild-type and Fmr1 KO (TSV-40) cells was verified by immunoblot analysis, in parallel with actin. (b) Fmr1 KO (TSV-40) and wild-type (Naïves) cells were cotransfected with psiSTRIKE encoding Rluc shRNA and psiCHECK reporter construct (n = 6). (c) Fmr1 KO cell lines STEK TSV-40 (n = 2), STEK 3T3A (n = 2), STEK 3T6A (n = 1), and STEK TpBSVE (n = 2), and wild-type Naïves (n = 6) and NIH/3T3 (n = 5) cell lines were cotransfected as in (b). Results of Rluc activity were normalized with Fluc activity and expressed as a percentage of Rluc activity obtained with an shRNA directed against a sequence deleted in the Rluc reporter mRNA. Results were expressed as mean ± SEM and analyzed by analysis of variance followed by unpaired Student's t test. ***P < .001.
The observed variations in RNA silencing responses might be a consequence of looking at different cell line clones which, apart from being deficient in FMRP, may have different properties with respect to transfection, nucleic acid release, or amount of available Exportin 5 [39] or RISC [40]. A recent study reported that the efficiency of the RNAi pathway may differ between cell types [41]. In order to confirm our data and to avoid possible misinterpretation due to cell line-specific RNA silencing responses, we have tested three additional Fmr1 KO cell lines (STEK 3T3A, STEK 3T6A, and STEK TpBSVE) and one additional control wild-type cell line (NIH/3T3). The absence or presence of FMRP in these cell lines was confirmed by immunoblot analysis. As shown in Figure 4(c), impairment of shRNA-induced Rluc silencing was observed in all Fmr1 KO cell lines tested, as compared to FMRP-expressing control cell lines. Thus, the observed defect of the RNA silencing pathway in Fmr1 KO cell lines is not specific to a particular cell line clone, thereby confirming the importance of FMRP for efficient RNA silencing in mammalian cells.
FMRP is essential for ss siRNA-induced RNA silencing
In order to get further insights into the role of FMRP in RNA silencing, we compared in Fmr1 KO and wild-type cells the efficiency of two other RNA silencers (siRNA duplex and ss antisense siRNA) that are known to enter the endogenous machinery at levels downstream to that of shRNAs or pre-miRNAs. Duplex siRNA-induced silencing was efficient in wild-type cells, with a > 90% inhibition of Rluc activity at 100 nM, but was defective in Fmr1 KO cells (P < .001) (Figure 5). These results suggest that the defect associated with the absence of FMRP is situated at or downstream of the siRNA duplex entry level into the RNAi pathway.
Figure 5.
FMRP is required for ss siRNA-induced RNA silencing. Fmr1 KO (TSV-40) and wild-type (Naïves) cells were cotransfected with ss antisense Rluc siRNA (asRluc) or Rluc siRNA duplex (siRluc) and psiCHECK reporter construct (n = 6). Results were expressed as mean ± SEM and analyzed as described in the legend of Figure 4. *P < .05; **P < .01; ***P < .001.
To characterize this perturbation further, we induced silencing of the Rluc reporter gene by using ss antisense siRNA, which may enter the RNA silencing machinery downstream of the strand selection/separation step(s). In wild-type cells, the extent of Rluc silencing induced by ss antisense Rluc siRNA reached ∼ 35% at the highest concentration (Figure 5), which was less pronounced than that achieved with the corresponding duplex, as previously reported [42, 43]. However, cells lacking FMRP were incompetent at mediating Rluc silencing induced by ss antisense Rluc siRNA (P < .05). These data imply the involvement of FMRP in a distal ss siRNP effector complex in mammalian cells.
The properties of FMRP are shared by its paralog FXR1P
In contrast to the single dFMR1 gene in Drosophila, three paralogs, namely, FMRP and the fragile X-related proteins FXR1P and FXR2P, are expressed in mammalian cells [31]. In order to examine if the properties of FMRP are unique or representative of this gene family, we studied the function of FXR1P. As illustrated in Figures 6(a) and 6(b), the miRNA and ss siRNA annealing properties of FXR1P are similar to those exhibited by FMRP in vitro (compare with Figures 2(a) and 2(b)). We then studied the importance of FXR1P in RNA silencing in vivo using Fxr1 KO cells. Immunoblot analyses confirmed the absence of FXR1P in these cells (Figure 6(c), compare right lane versus left lane). As Fmr1 KO cells, Fxr1 KO cells also exhibited, although to a lesser extent, a compromised RNA silencing response relative to wild-type cells (Figure 6(d)). These results suggest a common, possibly complementary, role for members of the fragile X protein family in RNA silencing in mammals.
Figure 6.
The paralog FXR1P shares the RNA strand annealing properties of FMRP. (a)-(b) RNA strand annealing assays with FXR1P were performed and analyzed as described in the legend of Figure 2. (c) Expression of FXR1P in wild-type and Fxr1 KO cells was verified by immunoblot analysis, in parallel with actin. (D) Fxr1 KO and wild-type (Naïves) cells were cotransfected with psiSTRIKE encoding Rluc shRNA and psiCHECK reporter construct (n = 6). Results were expressed as mean ± SEM and analyzed as described in the legend of Figure 4. *P < .05; **P < .01.
DISCUSSION
A cooperation between Dicer and FMRP, in which FMRP would act as an acceptor of miRNAs derived from Dicer processing of pre-miRNAs, is consistent with a concerted action in regulating mRNA expression. Independent groups have shown that target cleavage directed by ss siRNA is supported in extracts of HeLa cells [42, 43], from which a human RISC containing ss siRNA was characterized [42]. The presence of a single siRNA strand in mRNA-cleaving RNP complexes, previously found to contain dFMR1 [27, 28], has also been demonstrated in Drosophila S2 cells [44, 45]. Although the human and fly RNA silencing machineries differ mechanistically and in terms of mediating translational repression or mRNA cleavage, they may share two features in common: an ssRNA-containing effector RNP complex and a member of the fragile X protein family.
Recent findings indicate that siRNA-bound Dicer-2 assembles into RISC in Drosophila [44–46]. Although the assembly of miRNA-bound Dicer into human RISC remains to be demonstrated, the observations that (i) the product of human Dicer remains associated with the enzyme [7], that (ii) Dicer interacts directly with the human RISC component Ago2 [47], and that (iii) FMRP forms an RNP complex with Ago2 and miRNAs in mammalian cells [29] are compatible with this scenario. This assembly scheme may determine the nature of the RNA species to be loaded into RNPs and confer specificity to the general RNA binding and chaperone properties of FMRP operating in miRNA-mediated RNA silencing. Although a recent study demonstrating the presence of Dicer preassembled in a human RISC that can cleave target RNA by utilization of a pre-miRNA Dicer substrate offers a slightly different perspective [48].
When assessing cellular responses to silencing RNAs, we found that FMRP was required for optimal RNA silencing in mammalian cells, as previously reported in Drosophila [27]. Rluc silencing mediated by Rluc shRNA is likely executed by an ss siRNP through a reaction in which the strand annealing properties of FMRP are solicited. This is in agreement with the fact that FMRP is unable to exchange strands of perfectly paired siRNA duplexes to target RNAs in vitro (see Supplementary Data, Figure S2) and recent evidences suggesting the involvement of an ss siRNP in target RNA cleavage mediated by human RISC [48]. Considering the transposition of our in vitro annealing assays, using synthetic RNAs and recombinant proteins, to RNA silencing in vivo, the caveat has to be taken into account that the presence of other protein or nucleic acid components as well as postranslational modifications may influence the role and function of FMRP.
The relative importance of fragile X proteins in RNA silencing observed in our study may have been underestimated given that expression of FXR1P and FMRP may have contributed to alleviate and compensate for their respective functional loss in Fmr1 and Fxr1 KO cells. A relatively higher expression levels of FMRP and Dicer in Fxr1 KO fibroblasts was observed (I Plante and P Provost, unpublished data). This may be related either to a compensatory mechanism, in the case of the former, or a dysfunctional miRNA-based regulation of their expression. The fact that FMRP and FXR1P exhibit similar activities, conjugated with the differential tissue expression patterns of the fragile X proteins observed in mammals, suggests that RNP complexes of various composition may be functionally redundant.
A fragile X patient was found to carry a single substitution (I304N) in the Fmr1 gene product FMRP. We observed that recombinant FMRP I304N mutant protein exhibited miRNA strand annealing properties comparable to the wild-type protein. In contrast, the ss nucleic acid binding KH domains appear to be required for the miRNA annealing activity of FMRP. However, neither of these two mutants, that is, FMRP ΔKHT [30] and FMRP I304N [49], cosedimented with polyribosomes, thereby dissociating the polyribosomal association and miRNA annealing activity of FMRP. These observations suggest that the phenotype observed in fragile X patients may be related either to the loss of its chaperone activity [32] or the mislocalization of a still active mutant protein, as demonstrated for the FMRP I304N mutant [49].
The hypothesis that the subset of mRNAs bound by FMRP may be subjected to miRNA regulation, as proposed by Jin et al [50], is attractive. FMRP may recognize its target mRNAs through recognition of the G quartet structures [51, 52] and/or via the kissing complex motifs [53]. The silencing state of the bound mRNA might then be determined by a specific regulatory miRNA. The specificity of miRNA-based translational regulation may thus reside in both the sequence of the miRNA as well as the recognition of selected mRNA targets by FMRP. In turn, the presence and importance of FMRP in effector miRNP complexes may explain why FMRP is found associated with so many mRNAs. It will be interesting to see what proportion of the several hundreds of mRNAs bound by FMRP [54, 55] are experimentally validated and physiologically relevant miRNA targets.
The biological significance of pre-miRNA binding by FMRP (I Plante and P Provost, unpublished data), an event that would occur upstream of Dicer, is unclear. However, it is relevant to note that FMRP contains a nuclear localization signal (NLS) as well as a nuclear export signal (NES) [56]. These sequences appear to be functional, as FMRP was subsequently characterized as a nucleocytoplasmic shuttling protein [57]. Together, these observations raise the possibility that FMRP may play a role in nuclear export of pre-miRNA. The issues as to whether FMRP can be found in processing bodies mediating translation repression, as suggested by trapping of mRNAs by FMRP into cytoplasmic granules [24], or accompanies miRNP complexes involved in other cellular processes requiring mRNA repression, such as transfer of mRNAs from the cell body to dendrites of neurons [58], will also need to be addressed.
Several findings suggest the possible presence of Dicer in FMRP-containing complexes present in the translation machinery: (i) fractionation of competent RISC with ribosomes [10]; (ii) the demonstration that Dicer is part of the human RISC [48]; (iii) the documented existence of polyribosomal miRNAs [59, 60]; and (iv) transcripts engaged in translation regulation are sensitive to RNAi [61]. Our findings provide yet another functional link between the initiation and effector steps of RNA silencing.
FMRP may function in RNA silencing as a downstream effector of Dicer and miRNA assembler on target mRNAs. FMRP is particularly abundant in the brain due to its high expression in neurons [34, 62], and the extent of neuronal miRNA-mediated mRNA regulation, as deduced by the number and diversity of miRNAs cloned from mammalian neurons [60], appears to be important. A suboptimal miRNA assembly on their natural mRNA targets, resulting from the lack of FMRP expression, may contribute to the molecular basis for the fragile X syndrome, thereby suggesting a causal link between dysfunction of the RNA silencing machinery and a human disease.
Supplementary Material
The Supplementary text shows sequence and structure of RNAs used in this study. Supplementary Figure S1 shows that expression of shRNA has no effect on interferon regulatory factor-1 (IRF-1) protein expression and Supplementary Figure S2 shows that FMRP is unable to exchange strands of perfectly paired siRNA duplexes to target RNAs in vitro.
ACKNOWLEDGMENTS
We thank Stéphane Richard for critical reading of a previous version of the manuscript and Guy Rousseau for helpful discussions. E. W. Khandjian is indebted to Gideon Dreyfuss for the gift of Fxr1 KO mouse embryos. L. Davidovic was supported by a postdoctoral fellowship from the FRAXA Research Foundation and L.-A. Gobiel by a master studentship from the Natural Sciences and Engineering Research Council of Canada (NSERC). P Provost is a New Investigator of the Canadian Institutes of Health Research (CIHR). This work was supported by Grant NDG-70190 from CIHR-Institute of Genetics and CIHR-Institute of Neuroscience, Mental Health and Addiction (to P. Provost and E. W. Khandjian), Grant IGI-44012 from CIHR (to E W Khandjian), and Grant EOP-64706 from Health Canada/CIHR (to P. Provost). P. Provost is the author of correspondence.
Abbreviations
- EMSA
electrophoretic mobility shift assay
- FMRP
fragile X mental retardation protein
- KH
K-homology
- KO
knockout
- miRNA
microRNA
- pre-miRNA
miRNA precursor
- pri-miRNA
primary miRNA
- RISC
RNA-induced silencing complex
- RNAi
RNA interference
- RNase
ribonuclease
- RNP
ribonucleoprotein
- sh
short hairpin
- siRNA
small interfering RNA
- ss
single-stranded
- FXR1P
fragile X-related protein 1
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO Journal. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 4.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes and Development. 2003;17(24):3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 6.Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO Journal. 2002;21(21):5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO Journal. 2002;21(21):5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118(1):57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 9.Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 10.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404(6775):293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 11.Lim LP, Lau NC, Weinstein EG, et al. The microRNAs of Caenorhabditis elegans. Genes and Development. 2003;17(8):991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes and Development. 2002;16(13):1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 14.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294(5543):858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 15.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294(5543):862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 16.Pfeffer S, Zavolan M, Grässer FA, et al. Identification of virus-encoded microRNAs. Science. 2004;304(5671):734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 17.Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 18.Yekta S, Shih I-H, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304(5670):594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 19.Pillai RS, Bhattacharyya SN, Artus CG, et al. Inhibition of translational initiation by let-7 microRNA in human cells. Science. 2005;309(5740):1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 20.O'Donnell WT, Warren ST. A decade of molecular studies of fragile X syndrome. Annual Review of Neuroscience. 2002;25:315–338. doi: 10.1146/annurev.neuro.25.112701.142909. [DOI] [PubMed] [Google Scholar]
- 21.Bardoni B, Mandel J-L. Advances in understanding of fragile X pathogenesis and FMRP function, and in identification of X linked mental retardation genes. Current Opinion in Genetics and Development. 2002;12(3):284–293. doi: 10.1016/s0959-437x(02)00300-3. [DOI] [PubMed] [Google Scholar]
- 22.Laggerbauer B, Ostareck D, Keidel E-M, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Human Molecular Genetics. 2001;10(4):329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- 23.Li Z, Zhang Y, Ku L, Wilkinson KD, Warren ST, Feng Y. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Research. 2001;29(11):2276–2283. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazroui R, Hout ME, Tremblay S, Fillion C, Labelle Y, Khandjian EW. Trapping of messenger RNA by fragile X mental retardation protein into cytoplasmic granules induces translation repression. Human Molecular Genetics. 2002;11(24):3007–3017. doi: 10.1093/hmg/11.24.3007. [DOI] [PubMed] [Google Scholar]
- 25.Costa A, Wang Y, Dockendorff TC, et al. The Drosophila fragile X protein functions as a negative regulator in the orb autoregulatory pathway. Developmental Cell. 2005;8(3):331–342. doi: 10.1016/j.devcel.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 26.Zhang YQ, Bailey AM, Matthies HJG, et al. Drosophila fragile x-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107(5):591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]
- 27.Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes and Development. 2002;16(19):2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes and Development. 2002;16(19):2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin P, Zarnescu DC, Ceman S, et al. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nature Neuroscience. 2004;7(2):113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 30.Mazroui R, Huot M-E, Tremblay S, Boilard N, Labelle Y, Khandjian EW. Fragile X mental retardation protein determinants required for its association with polyribosomal mRNPs. Human Molecular Genetics. 2003;12(23):3087–3096. doi: 10.1093/hmg/ddg335. [DOI] [PubMed] [Google Scholar]
- 31.Khandjian EW. Biology of the fragile X mental retardation protein, an RNA-binding protein. Biochemistry and Cell Biology. 1999;77(4):331–342. [PubMed] [Google Scholar]
- 32.Gabus C, Mazroui R, Tremblay S, Khandjian EW, Darlix J-L. The fragile X mental retardation protein has nucleic acid chaperone properties. Nucleic Acids Research. 2004;32(7):2129–2137. doi: 10.1093/nar/gkh535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castets M, Schaeffer C, Bechara E, et al. FMRP interferes with the Rac1 pathway and controls actin cytoskeleton dynamics in murine fibroblasts. Human Molecular Genetics. 2005;14(6):835–844. doi: 10.1093/hmg/ddi077. [DOI] [PubMed] [Google Scholar]
- 34.Devys D, Lutz Y, Rouyer N, Bellocq J-P, Mandel J-L. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nature Genetics. 1993;4(4):335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- 35.Darnell JC, Mostovetsky O, Darnell RB. FMRP RNA targets: identification and validation. Genes, Brain and Behavior. 2005;4(6):341–349. doi: 10.1111/j.1601-183X.2005.00144.x. [DOI] [PubMed] [Google Scholar]
- 36.Khandjian EW, Bechara E, Davidovic L, Bardoni B. Fragile X mental retardation protein: many partners and multiple targets for a promiscuous function. Current Genomics. 2005;6(7):515–522. [Google Scholar]
- 37.De Boulle K, Verkerk AJMH, Reyniers E, et al. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nature Genetics. 1993;3(1):31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- 38.Singh R, Valcárcel J. Building specificity with nonspecific RNA-binding proteins. Nature Structural and Molecular Biology. 2005;12(8):645–653. doi: 10.1038/nsmb961. [DOI] [PubMed] [Google Scholar]
- 39.Yi R, Doehle BP, Qin Y, Macara IG, Cullen BR. Overexpression of Exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA. 2005;11(2):220–226. doi: 10.1261/rna.7233305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harborth J, Elbashir SM, Vandenburgh K, et al. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense and Nucleic Acid Drug Development. 2003;13(2):83–105. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]
- 41.Oberdoerffer P, Kanellopoulou C, Heissmeyer V, et al. Efficiency of RNA interference in the mouse hematopoietic system varies between cell types and developmental stages. Molecular and Cellular Biology. 2005;25(10):3896–3905. doi: 10.1128/MCB.25.10.3896-3905.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez J, Patkaniowska A, Urlaub H, Lührmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110(5):563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 43.Schwarz DS, Hutvágner G, Haley B, Zamore PD. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Molecular Cell. 2002;10(3):537–548. doi: 10.1016/s1097-2765(02)00651-2. [DOI] [PubMed] [Google Scholar]
- 44.Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004;117(1):83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- 45.Tomari Y, Du T, Haley B, et al. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116(6):831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- 46.Lee YS, Nakahara K, Pham JW, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117(1):69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 47.Tahbaz N, Kolb FA, Zhang H, Jaronczyk K, Filipowicz W, Hobman TC. Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer. EMBO Reports. 2004;5(2):189–194. doi: 10.1038/sj.embor.7400070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123(4):631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 49.Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, Warren ST. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Molecular Cell. 1997;1(1):109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- 50.Jin P, Alisch RS, Warren ST. RNA and microRNAs in fragile X mental retardation. Nature Cell Biology. 2004;6(11):1048–1053. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]
- 51.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107(4):489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 52.Schaeffer C, Bardoni B, Mandel J-L, Ehresmann B, Ehresmann C, Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO Journal. 2001;20(17):4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Darnell JC, Fraser CE, Mostovetsky O, et al. Kissing complex RNAs mediate interaction between the fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes and Development. 2005;19(8):903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darnell JC, Warren ST, Darnell RB. The fragile X mental retardation protein, FMRP, recognizes G-quartets. Mental Retardation and Developmental Disabilities Research Reviews. 2004;10(1):49–52. doi: 10.1002/mrdd.20008. [DOI] [PubMed] [Google Scholar]
- 55.Brown V, Jin P, Ceman S, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107(4):477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 56.Eberhart DE, Malter HE, Feng Y, Warren ST. The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Human Molecular Genetics. 1996;5(8):1083–1091. doi: 10.1093/hmg/5.8.1083. [DOI] [PubMed] [Google Scholar]
- 57.Feng Y, Gutekunst C-A, Eberhart DE, Yi H, Warren ST, Hersch SM. Fragile X mental retardation protein: nucleocytoplasmic shuttling and association with somatodendritic ribosomes. Journal of Neuroscience. 1997;17(5):1539–1547. doi: 10.1523/JNEUROSCI.17-05-01539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Antar LN, Afroz R, Dictenberg JB, Carroll RC, Bassell GJ. Metabotropic glutamate receptor activation regulates fragile X mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. Journal of Neuroscience. 2004;24(11):2648–2655. doi: 10.1523/JNEUROSCI.0099-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mourelatos Z, Dostie J, Paushkin S, et al. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes and Development. 2002;16(6):720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim J, Krichevsky A, Grad Y, et al. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(1):360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kennerdell JR, Yamaguchi S, Carthew RW. RNAi is activated during Drosophila oocyte maturation in a manner dependent on aubergine and spindle-E. Genes and Development. 2002;16(15):1884–1889. doi: 10.1101/gad.990802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khandjian EW, Fortin A, Thibodeau A, et al. A heterogeneous set of FMR1 proteins is widely distributed in mouse tissues and is modulated in cell culture. Human Molecular Genetics. 1995;4(5):783–789. doi: 10.1093/hmg/4.5.783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Supplementary text shows sequence and structure of RNAs used in this study. Supplementary Figure S1 shows that expression of shRNA has no effect on interferon regulatory factor-1 (IRF-1) protein expression and Supplementary Figure S2 shows that FMRP is unable to exchange strands of perfectly paired siRNA duplexes to target RNAs in vitro.