Abstract
Two yeast strains, strains XH 1026 and XH 1164, isolated from the sputum of an intensive care unit patient with acute pneumonia, were originally identified as Candida albicans and C. tropicalis, respectively. Sequence analysis of the 26S rRNA gene D1/D2 domain and the internal transcribed spacer (ITS) region indicated that the two strains represent a novel yeast species closely related to C. rogusa. The name Candida pseudorugosa sp. nov. is therefore proposed (type strain, AS 2.3107 [CBS 10433]). The new species is able to grow at 42°C and is resistant or insusceptible to amphotericin B (MIC, 2 μg/ml), caspofungin (MIC, 64 μg/ml), itraconazole (MIC, 1 μg/ml), and nystatin (MIC, 16 μg/ml); dose-dependent susceptible to fluconazole (MIC, 16 μg/ml); and susceptible to flucytosine (MIC, 0.125 μg/ml) and voriconazole (MIC, 0.125 to 0.25 μg/ml). The code for C. pseudorugosa sp. nov. provided by the API 20C AUX system is identical to that for C. rugosa. The colonies of the new species on CHROMagar Candida appear blue-green, similar to those of C. albicans. In addition to the molecular method based on D1/D2 domain or ITS region sequencing, use of the combination of the API system and CHROMagar Candida is helpful for the correct identification of C. pseudorugosa sp. nov.
Developments in medical and surgical therapy over the past two decades have changed the type of inpatient populations. Newer technologies and therapies, such as bone marrow or solid-organ transplantations, chemotherapy, monitoring with invasive devices, parenteral nutrition, broad-spectrum antimicrobial treatment, and assisted ventilation, have helped to treat patients with previously devastating or fatal diseases. However, these successes have resulted in the admission of increasing numbers of critically ill or immunocompromised patients to hospitals (4). Furthermore, the AIDS epidemic has added to the growing population of immunocompromised individuals (16). These patients are highly susceptible to opportunistic fungal pathogens (4, 7, 13). Candidiasis is most frequent in immunocompromised hosts, and Candida albicans is responsible for the majority of the infections (20, 21). However, data obtained over the past two decades clearly indicate that invasive candidiasis in critically ill, nonimmunocompromised patients is of increasing importance (1, 8, 18) and that serious life-threatening infections due to new and emerging fungal pathogens, including non-C. albicans Candida species, have increased significantly (3, 13, 17, 29, 30, 40, 41, 43).
Triazole-based antifungal treatment and prophylaxis applied since the 1980s have reduced the proportion of cases of invasive candidiasis caused by C. albicans. In parallel, a marked increase in the proportion of non-C. albicans Candida bloodstream isolates has been reported in several countries or regions (30). This trend is evident in patients with cancer. Neutropenia in tumor patients and acute leukemia and antifungal prophylaxis in hematology patients were significantly associated with non-C. albicans candidemia (10, 39). Species other than C. albicans have accounted for nearly 50% of all systemic Candida infections in many cancer centers since the late 1980s (2, 19, 34, 39, 42).
During a survey of the species and the genetic diversity of Candida strains from clinical sources in Beijing, China, two strains isolated from the sputum of an intensive care unit (ICU) patient who died of acute pulmonary infection after an esophageal tumor resection was revealed to represent a novel species closely related to C. rugosa by rRNA gene sequence comparison. The two strains were originally misidentified as C. albicans and C. tropicalis, respectively. The new Candida species is of special clinical importance because it is insusceptible to several commonly used antifungal agents and cannot be correctly identified by the methods commonly used in clinical laboratories.
CASE REPORT
The patient was a 66-year-old woman who used to live in Tianjin, China. On 19 February 2003, the patient was treated by an esophageal tumor resection in a local hospital. One day after the operation, she presented with an episode of hyperpyrexia associated with mass viscous expectoration. Chest X rays indicated double-lung pneumonia. Laboratory test results did not reveal tuberculosis. Empirical treatment with a combination of antibiotics failed to control the infection. Beginning on 24 February, the patient was placed on mechanical ventilation, with a few short intervals of voluntary respiration. On 18 March 2003 the patient was transferred to the ICU of a hospital in Beijing. Repeated aerobic and anaerobic microbiological cultures of blood were negative. Examination and cultivation of a lung tissue sample, performed on 21 March, were negative. The following bacteria were detected from sputum cultures: Acinetobacter baumannii (8 and 18 March), Escherichia coli (21 March), and Staphylococcus aureus and Stenotrophomonas maltophilia (24 March). Yeasts were detected from the sputum cultures on 14, 24, and 25 March and were identified as C. albicans, a Candida sp., and C. tropicalis, respectively, by using CHROMagar Candida (28). The antifungal agent fluconazole (200 mg/day) had been administered since 18 March; and antibiotic treatments were adjusted beginning on 20 March so that they included impenem-cilastatin sodium (1,500 mg/day), vancomycin (1,500 mg/day), and metronidazole (1,000 mg/day), according to the results of susceptibility testing of the bacteria detected. No clinical improvements were achieved. Chest X rays and a computed tomography scan showed a progressive infection with tiny nodes and multiple patchy infiltrates in both lungs. On 25 March 2003, the patient died of respiratory failure, septic shock, and deterioration.
MATERIALS AND METHODS
Yeast strains and phenotypic characterization.
Strains XH 1026 and XH 1164 were isolated from the sputum of the patient on 14 and 25 March 2003, respectively. The morphological, physiological, and biochemical characteristics were examined by standard methods commonly used in yeast taxonomy (45). Chromogenic testing of the colony was performed on CHROMagar Candida (CHROMagar Company, Paris, France), according to the manufacturer's instructions. The API 20C AUX kit (bioMérieux, Lyon, France) was used for identification of the yeast strains, according to the manufacturer's instruction.
Sequencing and molecular phylogenetic analysis.
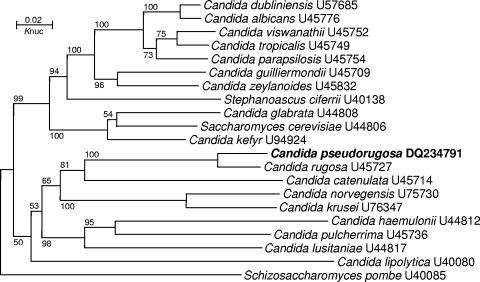
Nuclear DNA was extracted by the method of Makimura et al. (25). The internal transcribed spacer (ITS) region (including the 5.8S rRNA gene) and the 26S rRNA gene D1/D2 domain were amplified with primer pair ITS1 (5′-GTC GTA ACA AGG TTT CCG TAG GTG-3′) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′) and primer pair NL1 (5′-GCA TAT CAA TAA GCG GAG GAA AAG-3′) and NL4 (5′-GGT CCG TGT TTC AAG ACG G-3′), respectively. The PCR was performed for 36 cycles, with denaturation at 95°C for 1 min, annealing at 52°C for 1 min, and extension at 72°C for 1 min. The PCR products were purified by using SUPREC-02 centrifugal filter tubes (TaKaRa Bio, Shiga, Japan), according to the instructions of the manufacturer. After purification, the PCR products were directly sequenced with forward primer ITS1 or NL1 and reverse primer ITS4 or NL4 by using the ABI BigDye Terminator cycle sequencing kit (Applied Biosystems, Foster City, CA). The sequences of the D1/D2 domains or ITS regions of the strains determined in this study and those of the reference strains obtained from GenBank were aligned by use of the Clustal X program (38). Phylogenetic analysis was preformed by the neighbor-joining method, as described previously (5). The accession numbers of the reference sequences were indicated in the tree shown in Fig. 2.
FIG. 2.
Phylogenetic tree drawn from neighbor-joining analysis of 26S rRNA gene D1/D2 domain sequences depicting the relationships of Candida pseudorugosa sp. nov. with closely related yeast taxa. Bootstrap percentages over 50% from 1,000 bootstrap replicates are shown. Reference sequences were from the type strains of the species and were retrieved from GenBank under the accession numbers indicated.
Antifungal susceptibility.
Antifungal susceptibility testing was performed by the broth dilution method, according to the guidelines outlined in Clinical and Laboratory Standards Institute (formerly National Committee for Clinical Laboratory Standards) document M27-A2 (26).
Nucleotide sequence accession numbers.
The D1/D2 domain and ITS region sequences of strain XH 1164 have been released in GenBank with accession numbers DQ234791 and DQ234792, respectively.
RESULTS
Phenotypic characteristics.
Strains XH 1026 and XH 1164 were morphologically and physiologically very similar. They grew well at 42°C. In yeast extract-malt extract broth (45), after 3 days at 30°C, the cells were ovoid, ellipsoid, or cylindrical. Budding was multilateral. On YM agar (45), after 10 days at 30°C, the steak culture was butyrous, cream colored, raised, semiglossy, and wrinkled; the margin was slightly undulating. Abundant pseudohyphae were formed in Dalmau plate culture on cornmeal agar. The carbon and nitrogen assimilation patterns of the two strains were similar to those of C. rugosa. Sexual structures were not observed.
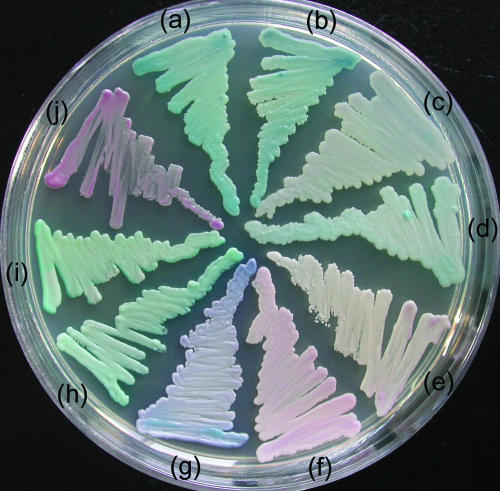
On CHROMagar Candida, the colonies of strains XH 1026 and XH 1164 appeared dark blue-green, similar to those of C. albicans and C. dubliniensis (Fig. 1). An API system code (code 6442104) typical of that for C. rugosa was obtained for both strains by using the API 20C AUX system.
FIG. 1.
Colonies grown on CHROMagar Candida for 48 h at 30°C: (a) C. pseudorugosa XH 1026, (b) C. pseudorugosa XH 1164, (c) C. rugosa CBS 613, (d) C. rugosa AS 2.1498, (e) C. parapsilosis ATCC 22019, (f) C. krusei AS 2.3194, (g) C. tropicalis AS 2.3195, (h) C. dublinensis CBS 7988, (i) C. albicans ATCC 90028, and (j) C. glabrata ATCC 90030.
Sequence comparison.
Strains XH 1026 and XH 1164 had identical D1/D2 domain and ITS region sequences, confirming their conspecificity. The results of a BLAST search of the sequences in GenBank by use of the D1/D2 domain and ITS region sequences of the two strains as the queries showed that the closest matches were the corresponding sequences of the species C. rugosa. In the phylogenetic tree drawn from the D1/D2 domain sequence alignment, the two strains clustered together with C. rugosa (Fig. 2). They differed from the type strain of C. rugosa by 23 (4.7%) mismatches (21 substitutions and 2 indels) in the D1/D2 domain and by 64 (20.4%) mismatches (61 substitutions and 3 indels) in the ITS region (including the 5.8S rRNA gene).
Antifungal susceptibilities.
The in vitro susceptibilities of strains XH 1026 and XH 1164 to the following nine antifungal agents were the same: amphotericin B (MIC, 2 μg/ml), caspofungin (MIC, 64 μg/ml), clotrimoxazole (MIC, 1 μg/ml), fluconazole (MIC, 16 μg/ml), flucytosine (MIC, 0.125 μg/ml), itraconazole (MIC, 1 μg/ml), miconazole (MIC, 2 μg/ml), nystatin (MIC, 16 μg/ml), and terbinafine (MIC, ≥16 μg/ml). The MICs of voriconazole for strains XH 1026 and XH 1164 were 0.25 and 0.125 μg/ml, respectively.
DISCUSSION
Previous studies have shown that yeast strains with a greater than 1% substitution in the D1/D2 domain or ITS region usually represent separate species (15, 23, 24, 35, 36). The sequence comparison performed in the present study clearly indicated that strains XH 1026 and XH 1164 represent a distinct novel species with a close phylogenetic relationship to C. rugosa. The name Candida pseudorugosa sp. nov. is therefore proposed for the new species.
The original clinical records, which contained no histopathologic evidence, are not sufficient to definitely ascribe the lung infection of the patient to the new Candida species. There is no more evidence at present to indicate that this organism is a human pathogen. However, the origin and special properties of the species suggest that it is highly possibly a new opportunistic fungus worthy of note. It is common to isolate C. albicans strains from the sputum of patients or even healthy people, for this species is a commensal organism found frequently in healthy humans (27). However, it is uncommon to isolate strains of a new Candida species repeatedly from the sputum of an ICU patient with acute pneumonia. Although a few pathogenic bacterial species were also isolated from the sputum of the patient, the susceptibilities of the bacteria to antibiotics were tested in vitro and the antibacterial treatments were adjusted accordingly, but the lung infection failed to be controlled.
The resistance or insusceptibility of C. pseudorugosa sp. nov. to multiple antifungal agents is noticeable. If the breakpoints for Candida species tentatively adopted in NCCLS document M27-A2 (26) and other literature (4, 6, 9, 13, 22, 29, 32, 33, 44) are used as references, the susceptibility of the new species to the antifungal agents tested can be interpreted as resistance to amphotericin B, caspofungin, itraconazole, and nystatin; dose-dependent susceptibility to fluconazole; and susceptibility to flucytosine and voriconazole.
The antifungal-insusceptible property of the new species is most probably intrinsic, since strain XH 1026 was isolated before the use of fluconazole and other antifungals were not used during the whole course of treatment. Among the opportunistic non-C. albicans Candida species, C. glabrata, C. krusei, C. lusitaniae, and C. rugosa are well-known species that may exhibit innate or acquired resistance to one or more established antifungal agents (32). An amphotericin B- and azole-resistant species, C. pseudohaemulonii, isolated from the blood of a patient was reported recently (37). C. rugosa, the closest relative of the new species, is an emerging recognized opportunistic pathogen for humans. A cluster of six episodes of candidemia caused by C. rugosa was recently reported from Brazil (11). Fifteen episodes of candidemia due to C. rugosa in burn patients receiving topical nystatin treatment were reported in 1994 (12). The C. rugosa isolates were all shown to be resistant to nystatin (12). C. rugosa also has decreased susceptibilities to amphotericin B and fluconazole (12, 14). As a close relative of C. rugosa with a broader antifungal resistance spectrum, C. pseudorugosa sp. nov. is worthy of note because of its possible ability to infect humans.
Because of the resistance or insusceptibility of C. pseudorugosa sp. nov. to commonly used antifungal agents, the correct and rapid identification of the new species is clinically important. However, C. pseudorugosa sp. nov. tends to be easily misidentified by the methods commonly used in clinical laboratories. In agreement with the close phylogenetic relationship to C. rugosa, the new species is similar to C. rugosa physiologically. By using the present kit and database of the API 20C AUX system, it can be unambiguously identified as C. rugosa. However, on CHROMagar Candida, the color of the colonies of the new species is more similar to that of C. albicans than to that of C. rugosa (Fig. 1). This was the reason that strain XH 1026 was originally misidentified as C. albicans. The original identification of strain XH 1164 as C. tropicalis was probably misguided by its blue tinge, but C. pseudorugosa sp. nov. is greener than C. tropicalis and does not produce a dark halo, as C. tropicalis usually does on CHROMagar Candida (28). Although a side-by-side comparison showed slight differences in colors between C. pseudorugosa sp. nov. and C. albicans or C. dubliniensis, without the awareness of this new species, isolates of the new species may easily be misidentified as the most common species C. albicans on CHROMagar Candida. If the maximum growth temperature is tested, the isolates of the new species may be misidentified as C. dubliniensis (31), since C. pseudorugosa sp. nov. grew well at 42°C but not at 45°C.
The special antifungal resistance and physiological and morphological characteristics of C. pseudorugosa sp. nov. suggest that the clinical yeast strains identified as C. albicans, C. dubliniensis, or C. tropicalis by chromogenic testing or as C. rugosa by the methods based on physiological reactions, such as those in the API 20C AUX system or a similar system, but that were refractory to amphotericin B, fluconazole, or other antifungal therapy should be reidentified. The combination of the testing results obtained from the API system and CHROMagar Candida is helpful for the correct recognition of C. pseudorugosa sp. nov. The new species can be distinguished from C. rugosa on CHROMagar Candida by its deeper color (Fig. 1). The lengths of the D1/D2 domain (484 bp) and the ITS1-5.8S rRNA-ITS2 region (310 bp) of C. pseudorugosa sp. nov. are remarkably shorter than those of C. albicans, C. dubliniensis, and C. tropicalis (570 to 571 bp for the D1/D2 domain, 440 to 450 bp for the ITS region). Direct comparison of the lengths of the PCR products of the D1/D2 domain or the ITS region by agarose gel electrophoresis may clearly separate the new species from the last three ones. The identification of the new species should be confirmed by sequencing of the D1/D2 domain or the ITS region.
Latin description of Candida pseudorugosa Bai & Li sp. nov.
In medio liquido YM post dies 3 ad 30°C, cellulae elipsoideae, 2.5-4.5 × 3.0-6.0 μm, singulae, binae et adhaerents. Per gemmationem multipolarem reproducents. Post 1 mensem sedimentum et annulus formatur. In agaro YM post 1 mensem ad 30°C, butyrose, cremea, infimo-convexa, seminitida, et margo undulato. Fermentatio nulla. Glucosum, galactosum, l-sorbosum (lente), maltosum, melibiosum (infirme), d-xylosum, ethanolum, glycerolum, d-mannitouml, d-glucitolum, methyl-α-d-glucosidum, salicinum, acidum dl-lacticum, acidum succinium et hexdecanum assimilanturat non sucrosum, cellobiosum, trehalosum, lactosum, raffinsum, melezitosum, inulinum, amylum soluible, l-arabinosum, d-arabinosum, d-ribosum, l-rhamnosum, d-glucosaminum, methanolum, erythritolum, acidum citricum nec inositolum. Ethylaminum, l-lysinum, cadaverinum, ammonium sulfatum assimilantur at non natrum nitrosun nec kalium nitricum. Vitaminae externae ad crescentiam necessaria sunt. Maxima temperature crescentiae: 42°C. Materiae ammyloidea iodophila non formantur. Diazonium caeruleum B non respondens. Ureum non hydrolysatur. Typus: isolatus ex saliva, XH 1164, depositus in collectione China General Microbiological Culture Collection Centre, Academia Sinica (AS 2.3107).
Description of Candida pseudorugosa Bai & Li sp. nov.
In YM broth, after 3 days at 30°C, the cells are ovoid, ellipsoid, or cylindrical and 2.5 to 4.5 by 3.0 to 6.0 μm and occur singly, in pairs, or in chains. Budding is multilateral. After 1 month at 30°C, sediment and a thin ring are present. On YM agar, after 10 days at 30°C, the streak culture is butyrous, cream colored, raised, semiglossy, and wrinkled; the margin is slightly undulating. In Dalmau plate culture on cornmeal agar, abundant pseudohyphae are formed. Sporulation was not observed. Fermentation is negative. Glucose, galactose, l-sorbose (delayed), maltose, melibiose (weak), d-xylose, ethanol, glycerol, d-mannitol, d-glucitol, methyl α-d-glucoside, salicin, dl-lactic acid, succinic acid, and hexadecane are assimilated. Sucrose, cellobiose, trehalose, lactose, raffinsoe, melezitose, inulin, soluble starch, l-arabinose, d-arabinose, d-ribose, l-rhamnose, d-glucosamine, methanol, erythritol hydrochloride, citric acid, and inositol are not assimilated. Ethylamine hydrochloride, l-lysine, cadaverine dihydrochloride, and ammonium sulfate are assimilated. Sodium nitrate and potassium nitrate are not assimilated. Growth in vitamin-free medium is negative. The maximum growth temperature is 42°C. Starch-like free compounds are not produced. The diazonium blue B reaction is negative. Urease activity is negative. The type strain, XH 1164, was isolated from the sputum of an ICU patient with acute pneumonia in Beijing, China, on 25 March 2003. This strain has been deposited in the China General Microbiological Culture Collection Center (CGMCC), Academia Sinica, Beijing, China, as AS 2.3107 and in the Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands, as CBS 10433. The specific epithet Candida pseudorugosa refers to the close relationship of the species to Candida rugosa.
Acknowledgments
We thank Ruoyu Li, Department of Dermatology/Research Center of Medical Mycology, Peking University First Hospital, Beijing, for help with antifungal susceptibility testing.
This study was supported by grant 30570098 from the National Natural Science Foundation of China.
Footnotes
Published ahead of print on 4 October 2006.
REFERENCES
- 1.Abi-Said, D., E. Anaissie, O. Uzun, I. Raad, H. Pinzcowski, and S. Vartivarian. 1997. The epidemiology of hematogenous candidiasis by different Candida species. Clin. Infect. Dis. 24:1122-1128. [DOI] [PubMed] [Google Scholar]
- 2.Anaissie, E. J., J. H. Rex, O. Uzun, and S. Vartivarian. 1998. Predictors of adverse outcome in cancer patients with candidemia. Am. J. Med. 104:238-245. [DOI] [PubMed] [Google Scholar]
- 3.Anaissie, E. J., G. P. Bodey, and M. G. Rinaldi. 1989. Emerging fungal pathogens. Eur. J. Clin. Microbiol. Infect. Dis. 8:323-330. [DOI] [PubMed] [Google Scholar]
- 4.Baddley, J. W., and S. A. Moser. 2004. Emerging fungal resistance. Clin. Lab. Med. 24:721-735. [DOI] [PubMed] [Google Scholar]
- 5.Bai, F. Y., J. H. Zhao, M. Takashima, J. H. Jia, T. Boekhout, and T. Nakase. 2002. Reclassification of the Sporobolomyces roseus and Sporidiobolus pararoseus complexes, with the description of Sporobolomyces phaffii sp. nov. Int. J. Syst. Evol. Microbiol. 52:2309-2314. [DOI] [PubMed] [Google Scholar]
- 6.Blignaut, E., J. Molepo, C. Pujol, D. R. Soll, and M. A. Pfaller. 2005. Clade-related amphotericin B resistance among South African Candida albicans isolates. Diagn. Microbiol. Infect. Dis. 53:29-31. [DOI] [PubMed] [Google Scholar]
- 7.Bodey, G. P. 1988. The emergence of fungi as major hospital pathogens. J. Hosp. Infect. 11(Suppl. A):411-426. [DOI] [PubMed] [Google Scholar]
- 8.Bodey, G. P., M. Mardani, H. A. Hanna, M. Boktour, J. Abbas, E. Girgawy, R. Y. Hachem, D. P. Kontoyiannis, and I. I. Raad. 2002. The epidemiology of Candida glabrata and Candida albicans fungemia in immunocompromised patients with cancer. Am. J. Med. 112:380-385. [DOI] [PubMed] [Google Scholar]
- 9.Chaturvedi, V., R. Ramani, and J. H. Rex. 2004. Collaborative study of antibiotic medium 3 and flow cytometry for identification of amphotericin B-resistant Candida isolates. J. Clin. Microbiol. 42:2252-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng, M. F., Y. L. Yang, T. J. Yao, C. Y. Lin, J. S. Liu, R. B. Tang, K. W. Yu, Y. H. Fan, K. S. Hsieh, M. Ho, and H. J. Lo. 2005. Risk factors for fatal candidemia caused by Candida albicans and non-albicans Candida species. BMC Infect. Dis. 5:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colombo, A. L., A. S. A. Melo, R. F. C. Rosas, R. Salomao, M. Briones, R. J.Hollis, S. A. Messer, and M. A. Pfaller. 2003. Outbreak of Candida rugosa candidemia: an emerging pathogen that may be refractory to amphotericin B therapy. Diagn. Microbiol. Infect. Dis. 46:253-257. [DOI] [PubMed] [Google Scholar]
- 12.Dubé, M. P., P. N. R. Heseltine, M. G. Rinaldi, S. Evans, and B. Zawachi. 1994. Fungemia and colonization with nystatin-resistant Candida rugosa in a burn unit. Clin. Infect. Dis. 18:77-82. [DOI] [PubMed] [Google Scholar]
- 13.Eggimann, P., J. Garbino, and D. Pittet. 2003. Epidemiology of Candida species infections in critically ill nonimmunosuppressed patients. Lancet Infect. Dis. 3:685-702. [DOI] [PubMed] [Google Scholar]
- 14.Espinel-Ingroff, A. 1998. Comparison of in vitro activities of the new triazole SCH56592 and the echinocandins MK-0991 (L-743,872) and LY303366 against opportunistic filamentous and dimorphic fungi. J. Clin. Microbiol. 36:2950-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fell, J. W., T. Boekhout, A. Fonseca, G. Scorzetti, and A. Statzell-Tallman. 2000. Biodiversity and systematics of basidiomycetous yeasts as determined by larger subunit rDNA D1/D2/domain sequence analysis. Int. J. Bacteriol. 50:1351-1371. [DOI] [PubMed] [Google Scholar]
- 16.Fisher-Hoch, S. P., and L. Hutwagner. 1995. Opportunistic candidiasis: an epidemic of the 1980s. Clin. Infect. Dis. 21:897-904. [DOI] [PubMed] [Google Scholar]
- 17.Fridkin, S., and W. Jarvis. 1996. Epidemiology of nosocomial fungal infections. Clin. Microbiol. Rev. 9:499-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girmenia, C., and P. Martino. 1998. Fluconazole and the changing epidemiology of candidemia. Clin. Infect. Dis. 27:232-234. [DOI] [PubMed] [Google Scholar]
- 19.Horn, R., B. Wong, T. E. Kiehn, and D. Armstrong. 1985. Fungemia in a cancer hospital: changing frequency, earlier onset, and results of therapy. Rev. Infect. Dis. 7:646-655. [DOI] [PubMed] [Google Scholar]
- 20.Hughes, W. T., D. Armstrong, G. P. Bodey, E. J. Bow, A. E. Brown, T. Calandra, R. Feld, P. A. Pizzo, K. V. Rolston, J. L. Shenep, and L. S. Young. 2002. Guidelines for the use of antimicrobial agents in neutropenic patients with cancer. Clin. Infect. Dis. 34:730-751. [DOI] [PubMed] [Google Scholar]
- 21.Jarvis, W. R. 1995. Epidemiology of nosocomial fungal infections, with emphasis on Candida species. Clin. Infect. Dis. 20:1526-1530. [DOI] [PubMed] [Google Scholar]
- 22.Kuriyama, T., D. W. Williams, J. Bagg, W. A. Coulter, D. Ready, and M. A. O. Lewis. 2005. In vitro susceptibility of oral Candida to seven antifungal agents. Oral Microbiol. Immunol. 20:349-353. [DOI] [PubMed] [Google Scholar]
- 23.Kurtzman, C. P., and C. J. Robnett. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 35:1216-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurtzman, C. P., and C. J. Robnett. 1998. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Leeuwenhoek 73:331-371. [DOI] [PubMed] [Google Scholar]
- 25.Makimura, K., Y. S. Murayama, and H. Yamaguchi. 1994. Detection of a wide range of medically important fungi by the polymerase chain reaction. J. Med. Microbiol. 40:358-364. [DOI] [PubMed] [Google Scholar]
- 26.National Committee for Clinical Laboratory Standards. 2002. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard, 2nd ed. M27-A2. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 27.Odds, F. C. 1988. Candida and candidiasis: a review and bibliography. Bailliere Tindall, London, United Kingdom.
- 28.Odds, F. C., and R. Bernaerts. 1994. Chromagar Candida, a new differential isolation medium for presumptive identification of clinically important Candida species. J. Clin. Microbiol. 32:1923-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfaller, M. A., and D. J. Diekema. 2004. Rare and emerging opportunistic fungal pathogens: concern for resistance beyond Candida albicans and Aspergillus fumigatus. J. Clin. Microbiol. 42:4419-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfaller, M. A., D. J. Diekema, R. N. Jones, H. S. Sader, A. C. Fluit, R. J. Hollis, S. A. Messer, and the SENTRY Participant Group. 2001. International surveillance of bloodstream infections due to Candida species: frequency of occurrence and in vitro susceptibility to fluconazole, ravuconazole, and voriconazole of isolates collected from 1997 through 1999 in the SENTRY Antimicrobial Surveillance Program. J. Clin. Microbiol. 39:3254-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinjon, E., D. Sullivan, I. Salkin, D. Shanley, and D. Coleman. 1998. Simple, inexpensive, reliable method for differentiation of Candida dubliniensis from Candida albicans. J. Clin. Microbiol. 36:2093-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rex, J. H., M. A. Pfaller, J. N. Galgiani, M. S. Bartlett, A. Espinel-Ingroff, M. A. Ghannoum, M. Lancaster, F. C. Odds, M. G. Rinaldi, T. J. Walsh, and A. L. Barry. 1997. Development of interpretive breakpoints for antifungal susceptibility testing: conceptual framework and analysis of in vitro-in vivo correlation data for fluconazole, itraconazole, and Candida infections. Clin. Infect. Dis. 24:235-247. [DOI] [PubMed] [Google Scholar]
- 33.Rex, J. H., T. J. Walsh, J. D. Sobel, S. G. Filler, P. G. Pappas, W. E. Dismukes, and J. E. Edwards. 2000. Practice guidelines for the treatment of candidiasis. Clin. Infect. Dis. 30:662-678. [DOI] [PubMed] [Google Scholar]
- 34.Safdar, A., V. Chaturvedi, E. W. Cross, S. Park, E. M. Bernard, D. Armstrong, and D. S. Perlin. 2001. Prospective study of Candida species in patients at a comprehensive cancer center. Antimicrob. Agents Chemother. 45:2129-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scorzetti, G., J. W. Fell, A. Fonseca, and A. Statzell-Tallman. 2002. Systematics of basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res. 2:495-517. [DOI] [PubMed] [Google Scholar]
- 36.Sugita, T., A. Nishikawa, R. Ikeda, and T. Shinoda. 1999. Identification of medically relevant Trichosporon species based on sequences of internal transcribed spacer regions and construction of a database for Trichosporon identification. J. Clin. Microbiol. 37:1985-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugita, T., M. Takashima, N. Poonwan, and N. Mekha. 2006. Candida pseudohaemulonii sp. nov., an amphotericin B- and azole-resistant yeast species, isolated from the blood of a patient from Thailand. Microbiol. Immunol. 50:469-473. [DOI] [PubMed] [Google Scholar]
- 38.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL-X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Viscoli, C., C. Girmenia, A. Marinus, L. Collette, P. Martino, B. Vandercam, C. Doyen, B. Lebeau, D. Spence, V. Krcmery, B. De Pauw, and F. Meunier. 1999. Candidemia in cancer patients: a prospective, multicenter surveillance study by the Invasive Fungal Infection Group (IFIG) of the European Organization for Research and Treatment of Cancer (EORTC). Clin. Infect. Dis. 28:1071-1079. [DOI] [PubMed] [Google Scholar]
- 40.Weems, J. J. J. 1992. Candida parapsilosis: epidemiology, pathogenicity, clinical manifestations, and antimicrobial susceptibility. Clin. Infect. Dis. 14:756-766. [DOI] [PubMed] [Google Scholar]
- 41.Wenzel, R. P., and M. A. Pfaller. 1991. Candida species: emerging hospital bloodstream pathogens. Infect. Control Hosp. Epidemiol. 12:523-524. [DOI] [PubMed] [Google Scholar]
- 42.Wingard, J. R. 1995. Importance of Candida species other than C. albicans as pathogens in oncology patients. Clin. Infect. Dis. 20:115-125. [DOI] [PubMed] [Google Scholar]
- 43.Wingard, J. R., W. G. Merz, M. G. Rindali, T. R. Johnson, J. E. Karp, and R. Saral. 1991. Increase in Candida krusei infection among patients with bone marrow transplantation and neutropenia treated prophylactically with fluconazole. N. Engl. J. Med. 325:1274-1277. [DOI] [PubMed] [Google Scholar]
- 44.Yang, Y. L., S. Y. Li, H. H. Cheng, and H. J. Lo. 2005. Susceptibilities to amphotericin B and fluconazole of Candida species in TSARY 2002. Diagn. Microbiol. Infect. Dis. 51:179-183. [DOI] [PubMed] [Google Scholar]
- 45.Yarrow, D. 1998. Methods for the isolation, maintenance and identification of yeasts, p. 77-100. In C. P. Kurtzman and J. W. Fell. (ed.), The yeasts, a taxonomic study, 4th ed. Elsevier, Amsterdam, The Netherlands.