Abstract
The archaeal DNA replication machinery bears striking similarity to that of eukaryotes and is clearly distinct from the bacterial apparatus. In recent years, considerable advances have been made in understanding the biochemistry of the archaeal replication proteins. Furthermore, a number of structures have now been obtained for individual components and higher-order assemblies of archaeal replication factors, yielding important insights into the mechanisms of DNA replication in both archaea and eukaryotes.
INTRODUCTION
Since the pioneering work of Carl Woese in the late 1970s, it has been well established that the archaea constitute a defined domain of life (118). With the availability of archaeal genome sequences in the mid- to late 1990s, it became apparent that the archaeal DNA replication machinery has striking similarity to that in eukaryotes and is evolutionarily distinct from that in bacteria. How this curious dichotomy arose in a process central to the very propagation of life has been the subject of much debate. A wide range of theories have been put forward to account for this observation, ranging from the proposal that DNA replication arose twice in cellular organisms, suggesting that the last common ancestor of all living organisms may not have had a DNA genome, to the possibility that the last common ancestor had a defined replication system but that it was displaced by nonorthologous gene transfer from, for example, a viral source (26, 65, 82). Regardless of the derivation of the archaeon-eukaryote DNA replication system, it is apparent that the archaeal machinery is a simplified, and presumably ancestral, form of that in eukaryotes. The organizational simplicity of the archaeal machinery (Fig. 1), coupled with the biochemical advantages conferred by the study of hyperthermophilic archaea, has led to considerable interest in the archaeal machinery as a model of that in eukaryotes.
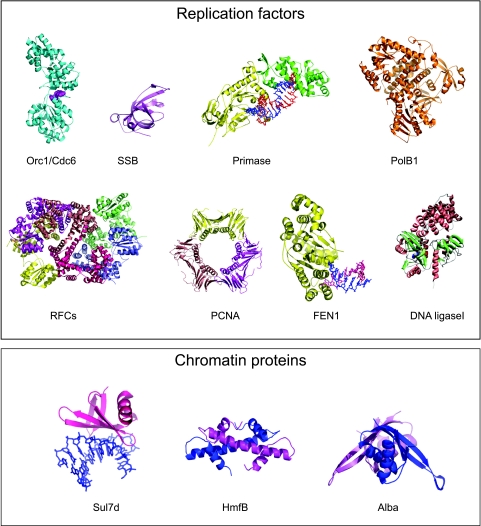
FIG. 1.
Components of the archaeal DNA replication machinery and chromatin proteins. Structure figures were prepared using Pymol (www.pymol.org), using the following PDB coordinates: Pyrobaculum Cdc6, 1FNN; Sulfolobus SSB, 1O71; Pyrococcus RFC small subunit, 1IQP; Sulfolobus Pol B1, 1S5J; Pyrococcus PCNA, 1ISQ; Archaeoglobus Fen1, 1RXW; Sulfolobus Alba, 1H0Y; Sulfolobus Sul7d, 1WTP; and Methanothermus histone HmfB, 1BFM. The image of the ligase structure was supplied by Y. Ishino (Fukuoka, Japan), and we obtained the image of the primase complex in collaboration with L. Pellegrini (Cambridge, United Kingdom).
REPLICATION ORIGINS
The replicon hypothesis, proposed by Jacob et al., predicted that a trans-acting initiator protein binds to a cis-acting replicator DNA sequence to initiate DNA replication in bacteria (47). This model has proved extremely accurate for bacteria. Bacterial chromosomes contain a single replication origin, oriC, which consists of an A-T-rich region of DNA containing multiple copies of the DnaA box, which is bound by the initiator protein DnaA. In many species, the gene for DnaA is carried adjacent to the origin, so the two may be coregulated (75).
In contrast to those of bacteria, eukaryotic chromosomes contain multiple replication origins. So far, only Saccharomyces cerevisiae (budding yeast) has been shown to have clearly defined replication origins, known as autonomously replicating sequences. These contain conserved sequence elements, similar to the situation in bacteria, and are bound by the origin recognition complex (ORC). The ORC contains six separate polypeptides, Orc1-6, several of which contain AAA+ (ATPases associated with various cellular activities) ATPase domains. Interestingly, Orc1 is also closely related to another replication factor, Cdc6 (Cdc18 in Schizosaccharomyces pombe), which presumably is indicative of the derivation of Orc1 and Cdc6 from a common ancestor. Although ORC acts as a sequence-specific DNA binding complex in budding yeast, in S. pombe and higher eukaryotes there is no clear consensus sequence for origins, although in many cases they do tend to be A-T-rich regions. Indeed, in Xenopus laevis eggs, any sequence seems capable of initiating DNA replication (16). Instead of relying on sequence-specific DNA recognition by ORC, a growing body of evidence suggests that in higher eukaryotes, origins are defined by facilitated recruitment of ORC by a variety of other DNA binding proteins. The extent to which this is a direct effect or mediated by secondary chromatin alterations is not fully understood (98).
It was initially thought that because the chromosome structure in archaea is similar to that in bacteria, archaeal chromosomes were also likely to contain one origin of replication. The first origin to be identified was the single origin of Pyrococcus abyssi (80, 89). The origin binding proteins in archaea are homologues of the related eukaryotic Orc1 and Cdc6 proteins (discussed below). The origin in P. abyssi is located adjacent to the gene for Orc1/Cdc6, in a situation similar to that for DnaA in many bacteria. Although bioinformatic analysis using the Z curve method showed that there were likely to be two origins in Halobacterium (121), a genetic screen found that only one of them had autonomously replicating sequence activity (3). However, two origins of replication were subsequently found and mapped for Sulfolobus solfataricus by two-dimensional gel analysis (99). S. solfataricus has three Orc1/Cdc6 genes, encoding Cdc6-1, Cdc6-2, and Cdc6-3. The two identified origins, oriC1 and oriC2, are located upstream of the genes for Cdc6-1 and Cdc6-3, respectively (99). No origin was found adjacent to the gene for Cdc6-2. A third origin, oriC3, was subsequently identified in both S. solfataricus and Sulfolobus acidocaldarius by marker frequency analysis but was located at least 50 kb away from Cdc6-2 (69). Thus, at least some archaea contain multiple origins of replication.
Fine mapping of the three replication origins in S. solfataricus led to the identification of origin recognition boxes (ORBs), which are inverted repeat sequence elements bound by Cdc6-1, at oriC1. These sequence elements are well conserved across many archaeal species, although most archaeal origins have not been proven experimentally. oriC2 in S. solfataricus contains sequences homologous to the central elements of ORBs. These sequences, termed mini-ORBs, are also found in the predicted origin of Methanobacterium thermautotrophicum. It therefore seems that, like the case for bacteria and S. cerevisiae, archaeal origins are defined by specific sequence elements (99).
In eukaryotes, not all origins are used in each S phase, and those that are used are fired asynchronously. Whether an origin is used and whether it fires early or late in S phase vary depending on chromatin structure, the transcriptional status of the surrounding regions, and the developmental stage and cell type for higher eukaryotes (98). Marker frequency analysis combined with computational modeling suggested that all three origins in S. acidocaldarius and S. solfataricus fire synchronously in all cells and that all three are used in each cell cycle (69). However, some differential origin usage cannot be ruled out, especially as the three Cdc6 proteins in S. solfataricus bind with different affinities to the different origins in vitro (99; discussed below).
ORIGIN BINDING BY Orc1/Cdc6
In the replicon hypothesis, Jacob et al. propose that a trans-acting factor recognizes and binds the replicator sequence and recruits other replication factors (47). As mentioned above, in bacteria this protein is DnaA, multiple monomers of which bind the DnaA boxes at the origin and melt the DNA. In eukaryotes, the ORC, consisting of proteins Orc1-6, binds at replication origins. In many eukaryotes, ORC remains bound throughout the cell cycle, whereas in bacteria DnaA is released as replication starts and then rebinds before the next round (70, 75). ORC recruits many proteins to the replication origins, including Cdc6 (Cdc18 in S. pombe). With the exception of three methanogenic archaeal species, all archaeal genomes sequenced to date contain at least one gene with homology to both Orc1 and Cdc6 (Table 1). The identities of the initiator proteins in the three methanogen exceptions remain unknown. Although all archaeal Orc/Cdc6 genes contain regions of homology to both ORC and Cdc6 genes, in different archaeal genome sequences they are generally annotated as either Orc-x or Cdc6-x. Like components of the eukaryotic ORC and the Cdc6 protein, archaeal Orc1/Cdc6 proteins are members of the AAA+ protein family. The three Orc1/Cdc6 proteins from S. solfataricus, i.e., Cdc6-1, Cdc6-2, and Cdc6-3, have been shown to bind to origins, with Cdc6-1 binding specifically to ORB elements (99). The single Orc1/Cdc6 protein from Pyrococcus was shown by chromatin immunoprecipitation to bind the region containing the single known P. abyssi origin (80). Therefore, the Orc1/Cdc6 proteins are thought to act as the origin recognition and binding proteins in archaea. In addition, Cdc6-1 from S. solfataricus can bind to ORB elements present in the Halobacterium NRC1 and P. abyssi origins in vitro (99), supporting the idea that archaeal Orc1/Cdc6 proteins recognize specific sequence motifs and that these motifs are conserved across archaea. It is not known whether multiple Orc1/Cdc6 proteins bind to each origin or whether binding is cooperative, but this has been suggested based on the structure of Orc1/Cdc6 and the symmetry of ORB elements (110).
TABLE 1.
Genes for Orc1/Cdc5, MCM, and PCNA in sequenced archaeal genomesa
| Organism | No. of genes
|
||
|---|---|---|---|
| Orc1/Cdc6 | MCM | PCNA | |
| Aeropyrum pernix | 2 | 1 | 3 |
| Archaeoglobus fulgidus | 2 | 1 | 1 |
| Cenarchaeum symbiosum | 1 | 1 | 1 |
| Haloarcula marismortui | 17 | 3 | 1 |
| Halobacterium sp. strain NRC-1 | 9 | 1 | 1 |
| Ignicoccussp. strain Kin4-1 | 2 | 1 | 3 |
| Methanobacterium thermautotrophicum | 2 | 1 | 1 |
| Methanococcoides burtonii | 2 | 1 | 1 |
| Methanococcus jannaschii | 0 | 4 | 1 |
| Methanococcus maripaludis | 0 | 4 | 1 |
| Methanopyrus kandleri | 0 | 2 | 1 |
| Methanosarcina acetivorans | 2 | 2 | 1 |
| Methanosarcina barkeri | 3 | 1 | 1 |
| Methanosarcina mazei | 2 | 1 | 1 |
| Methanospirillum hungatei | 2 | 1 | 1 |
| Methanosphaera stadtmanae | 2 | 1 | 1 |
| Nanoarchaeum equitans | 1 | 1 | 1 |
| Natronomonas pharaonis | 5 | 2 | 1 |
| Picrophilus torridus | 1 | 1 | 1 |
| Pyrobaculum aerophilum | 1 | 1 | 2 |
| Pyrococcus abyssi | 1 | 1 | 1 |
| Pyrococcus furiosus | 1 | 1 | 1 |
| Pyrococcus horikoshii | 1 | 1 | 1 |
| Sulfolobus solfataricus | 3 | 1 | 3 |
| Sulfolobus acidocaldarius | 3 | 1 | 3 |
| Sulfolobus tokodaii | 3 | 1 | 3 |
| Thermococcus kodakarensis | 1 | 3 | 2 |
| Thermoplasma acidophilum | 2 | 1 | 1 |
| Thermoplasma volcanium | 2 | 1 | 1 |
Crenarchaeal species and their values are indicated in bold. Genes were identified by Blast searching using the server at http://www-archbac.u-psud.fr/projects/sulfolobus/Blast_Search.html.
Most archaeal genomes carry from one (Pyrococcus species) to nine (Halobacterium) Orc/Cdc6 genes. Sequence analysis has shown that these can be classified into three major groups (3), and all species that have more than one Orc1/Cdc6 protein have at least one from the SsoCdc6-1 and SsoCdc6-2 subgroups (99). The three Orc1/Cdc6 proteins in S. solfataricus show different DNA binding footprints for the origins, which suggests that they could function differently or play different roles in replication. Work with S. acidocaldarius, which also contains three Orc1/Cdc6 proteins, showed different patterns of protein levels following perturbation of the cell cycle by treatment with acetic acid. Treatment of Sulfolobus cells with low concentrations of acetic acid leads to an accumulation of cells in the G2 period of the cell cycle. Following washing of the cells and transfer to fresh medium, cells reenter the cell cycle. However, this entry appears to be very asynchronous. Nevertheless, it was demonstrated that Cdc6-1 and Cdc6-3 levels were elevated in G1- and S-phase cells, whereas the Cdc6-2 level was highest in G2-arrested cells. This observation, along with the fact that Cdc6-1 and Cdc6-3 binding sites overlap with Cdc6-2 binding sites in S. solfataricus origins, suggests that Cdc6-2 may act as a repressor of replication. The expression of all three proteins was also reduced in stationary-phase cells compared to that in exponentially growing cells (99).
The structures of an Orc1/Cdc6 protein from Pyrobaculum aerophilium and of Aeropyrum pernix ORC2 (it should be emphasized that this protein is related to S. solfataricus Cdc6-2, not eukaryotic Orc2) have been solved. These structures show that the C-terminal region of Orc1/Cdc6 contains a winged-helix (WH) domain, and sequence alignments show that this is conserved throughout archaeal and eukaryotic Cdc6 proteins (67, 110). This domain is found in several DNA binding proteins and thus has been postulated to be the region of Cdc6 responsible for contacting DNA. In support of this hypothesis, mutation of WH domain residues in S. solfataricus Cdc6-1 reduced its ability to bind origin DNA (99), and the WH domain from A. pernix ORC2 was shown to be necessary and sufficient for DNA binding (110).
The crystal structure of A. pernix ORC2 was determined in the presence of both ADP and the nonhydrolyzable ATP analogue ADPNP. The structures showed substantial conformational flexibility in the ADP-bound form, but all ADPNP-bound proteins adopted the same conformation. This suggests that ATP binding may stabilize a single conformation of ORC2. The in vivo relevance of this is unclear as yet, but ATP binding and hydrolysis are likely to play an important part in Cdc6 function in the cell (110).
The two Orc1/Cdc6 homologues from M. thermautotrophicum and P. aerophilium Cdc6 have been shown to autophosphorylate at serine residues. This autophosphorylation activity is inhibited by DNA, but this inhibition is severely reduced in the absence of the WH domain. This further supports the idea that the WH domain interacts with DNA. It is interesting that the S. pombe Cdc6 homolog (called Cdc18) can also autophosphorylate, but it is not clear what functional significance this autophosphorylation activity might have (40).
The ORC2 structure also reveals remarkable similarities to DnaA. Both DnaA and ORC2 are AAA+ proteins, and both contain a C-terminal DNA binding domain, although in DnaA this is a helix-turn-helix, not WH, domain. It is therefore likely that despite the lack of homology between the two proteins beyond their AAA+ domains, they function in similar ways.
REPLICATIVE HELICASE
In bacteria, the replicative helicase is DnaB, an AAA+ protein which functions as a homohexamer. In eukaryotes, the best candidate for the replicative helicase is MCM (minichromosome maintenance complex). The MCM proteins were originally identified in yeast, in a screen for genes whose mutation abrogated the ability of the cells to maintain a plasmid containing a centromere and a replication origin (72). MCM in eukaryotes is a heterohexamer of MCM2-7. MCM8, another member of the MCM family, was recently identified and seems to be required for replication elongation and meiotic recombination (5, 73). The function of MCM9, which is present only in higher eukaryotes, is currently unknown (74).
There is considerable debate concerning whether MCM is the replicative helicase in eukaryotes. Mcm2-7 are essential for the initiation and elongation phases of DNA replication in yeast and Xenopus (58, 91, 106). They are recruited to the replication origin by ORC, Cdc6, and Cdt1 (discussed below), and blocking this recruitment completely inhibits replication. However, the complex has no ATPase or helicase activity in vitro. A subcomplex of MCM4, -6, and -7 has weak 3′-5′ helicase activity in vitro, and thus has been suggested to form the active complex, whereas MCM2, -3, and -5 are regulatory (45, 63, 64). All six proteins are required for replication elongation, however. In this light, a weak helicase activity was recently found to be associated with an endogenous purified complex from Drosophila melanogaster containing MCM, Cdc45, and the GINS proteins (see below) (87). Many MCM molecules (10 to 100 in some organisms) are also loaded at each origin (7, 29), in contrast to the case in Escherichia coli, where only two molecules of DnaB are loaded, with one for each replication fork. In addition, many immunofluorescence studies of higher eukaryotes have found that the majority of MCM does not colocalize with replication forks but, instead, associates with unreplicated DNA (24, 56, 71). However, a model has been proposed for MCM function at a distance from forks (discussed below), so this does not necessarily preclude a function for MCM as the replicative helicase.
All archaeal genomes sequenced to date have at least one MCM homologue (Table 1). In contrast to eukaryotes, however, many archaea contain only one MCM gene, and the protein forms homohexamers in vitro. Like the eukaryotic MCM proteins and bacterial DnaB, archaeal MCM is an AAA+ protein. In vivo, MCM interacts functionally with Cdc6 (22, 23, 51, 108) and, via GINS, with the primase (76). It localizes to replication origins in P. abyssi (80); however, since genetic systems for archaea are still in their infancy, it is not known whether MCM is essential for replication. Despite this, the very conservation of MCM from archaea to eukaryotes argues for an essential role, such as that of a replicative helicase.
Loading of the Replicative Helicase
In eukaryotes, MCM is loaded onto DNA in a process that requires ORC, Cdc6, and Cdt1 (70). Cdc6 binding to DNA is an ATP-independent process; however, MCM loading requires ATP hydrolysis by Cdc6 but not by MCM (67, 116).
In bacteria, both DnaA and DnaC are required to load DnaB onto DNA at origins. Like Cdc6, DnaC is an AAA+ protein. DnaC binds hexameric DnaB and, presumably, alters the conformation of the ring, thereby facilitating its loading onto DNA. It is thought that DnaC binds DnaB while bound to ATP. This increases its affinity for single-stranded DNA (ssDNA), but DnaC-ATP inhibits DnaB helicase activity. Once DnaB is loaded at the origin, the presence of both DnaB and ssDNA stimulates the ATPase activity of DnaC, causing it to stimulate instead of inhibit DnaB (20, 21).
For archaea, little is known about MCM loading. Open forms of the hexameric MCM ring from M. thermautotrophicum have been detected by electron microscopy (EM). These may represent a loading intermediate, as the MCM ring may be broken and reformed around DNA in a way similar to that for DnaB (21, 39). There is apparently no homologue of DnaC or Cdt1 in archaea, suggesting that the Orc/Cdc6 proteins may perform the functions carried out by ORC, Cdc6, and Cdt1 in eukaryotes. However, the sequence similarity between yeast and human Cdt1 proteins is only around 10%, so it is conceivable that there is a protein with low or even no homology in archaea which performs the same functions as Cdt1. Like the case in eukaryotes, binding of archaeal Orc1/Cdc6 proteins to origins is apparently ATP independent (99), despite the fact that they have a functional AAA+ domain (40, 110). ATP hydrolysis by Cdc6 may be important for MCM loading, similar to the situation in eukaryotes. The Cdc6 proteins in M. thermautotrophicum have been shown to inhibit MCM helicase activity in an ATP-dependent manner, but the in vivo significance of this is not clear. Interestingly, MCM from M. thermautotrophicum also modulates autophosphorylation of Cdc6-1 and Cdc6-2 (51, 108).
Structure and Function of MCM Proteins
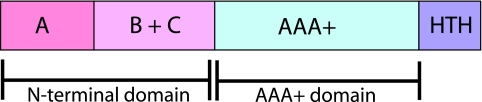
All MCM proteins have a conserved domain structure (Fig. 2). The C-terminal AAA+ catalytic domain is well conserved between MCM proteins. The N termini of the proteins are less well conserved and are thought to be responsible for multimerization and regulation (30, 33). In eukaryotic MCM proteins, the phosphorylation sites for cyclin-dependent kinases and other regulatory kinases are mostly located in this region (54, 86). There is also a helix-turn-helix domain at the extreme C terminus of the protein. This does not seem to be responsible for DNA binding in S. solfataricus MCM, and its in vivo function is unknown.
FIG. 2.
Domain organization of MCM proteins. The N-terminal region, consisting of three domains, A, B, and C, is poorly conserved between different MCM proteins and is thought to be involved in regulation. Eukaryotic MCM proteins often have an additional N-terminal extension. The catalytic AAA+ domain is shown in blue-green. The helix-turn-helix (HTH) domain at the C terminus is not involved in DNA binding but may play a role in regulation of the complex.
In solution, archaeal MCM proteins usually form double hexamers, although single hexameric, heptameric, and filamentous forms have also been reported (12, 15, 30, 31, 39, 92, 120). Unlike the heterohexameric eukaryotic complex, the double hexamer has DNA-stimulated ATPase and helicase activities in vitro (13, 52, 83, 107). The double- and single-hexamer forms of M. thermautotrophicum MCM have equivalent ATPase and DNA binding activities in vitro, but the double hexamer is a more active helicase than the single hexamer, implying that this may be the form responsible for active unwinding in the cell (31). In contrast to DnaB, a 5′-3′ helicase, but like eukaryotic MCM-4, -6, and -7, archaeal MCM is a 3′-5′ helicase, and thus it probably tracks along the leading strand during replication.
The crystal structure of the dodecameric N-terminal region of M. thermautotrophicum MCM has been solved, as has the EM structure of the full-length protein (30, 92). These structures revealed a large, positively charged channel in the center of the ring, with a diameter of between 23 Å and 47 Å, which is easily wide enough to accommodate single- or double-stranded DNA. The mechanisms by which ATP hydrolysis is coupled to helicase activity are unclear. However, the crystal structure of the N terminus revealed a conserved β-hairpin motif. Analysis of the C-terminal sequence showed that there was another conserved insertion likely to form a β-hairpin in the AAA+ domain. Mutation of conserved basic residues in either of these had only a modest effect on the ability of the protein to bind DNA. Mutation of both, however, caused a loss of DNA binding. In addition, mutation of the N-terminal β-hairpin caused a slight reduction in helicase activity, whereas mutation of the C-terminal β-hairpin completely abrogated helicase activity (83). For the superfamily 3 helicase simian virus 40 (SV40) large T antigen, a similar β-hairpin has been shown to move upon ATP hydrolysis and thus has been proposed to effect a power stroke, driving the protein along DNA (37, 66). It is therefore highly possible that MCM may translocate along DNA by a similar mechanism.
The mechanism by which MCM effects unwinding is still unknown. MCM from M. thermautotrophicum is able to translocate along double- and single-stranded DNA, as well as being able to unwind a forked substrate (109). It may act as a molecular bulldozer, separating strands as it translocates along DNA. Consistent with this possibility, the EM structure of MCM revealed the presence of holes in the side of the complex which may act as exit pores for DNA (92). Alternatively, a rotary pumping model for eukaryal MCM function at a distance from forks has been put forward. This proposes that after MCM proteins are loaded, they translocate along dsDNA away from the origin in both directions and are then immobilized by attachment to the nuclear matrix. After immobilization, further translocation causes the DNA to rotate and thus leads to unwinding at the fork (62). This would explain why many MCM proteins are loaded per origin and why they do not colocalize with replication forks. It remains to be seen which model is correct and, indeed, whether MCM functions in the same way in archaea and eukaryotes.
Hel308a
It is likely that other helicases also function in archaeal DNA replication. It was recently shown that Hel308a, a helicase from M. thermautotrophicum, interacts with stalled replication forks in vivo in E. coli and in assays performed in vitro. Strikingly similar results were observed with the Pyrococcus homolog, termed Hjm. When expressed in an E. coli strain lacking RecQ (a DNA helicase associated with recovery of stalled replication forks), Hel308a/Hjm complemented the recQ phenotype, strongly suggesting that Hel308a/Hjm may play a similar role in archaea (6, 35, 42). Interestingly, in some archaea the Hel308a homologue is encoded within an operon-like structure along with MCM and GINS, suggestive of a linked function of these proteins.
SSBs
Single-stranded binding proteins (SSBs) are present in all three domains of life. They protect single-stranded DNA from nuclease degradation and chemical modification during DNA replication, recombination, and other processes which require DNA to be unwound. All SSBs bind DNA via a common oligonucleotide/oligosaccharide binding (OB) fold (88). Bacterial SSB is a homotetramer. Each monomer contains an OB fold for contacting DNA and an acidic C-terminal domain (CTD) responsible for protein-protein interactions (94, 95). Eukaryotes contain a heterotrimer called replication protein A (RPA) that contains four DNA-binding OB folds. The largest subunit, RPA70, contains two of these in addition to a zinc binding motif, whereas the smaller two subunits, RPA30 and RPA14, both contain single OB folds.
Archaea contain a variety of SSB arrangements, although overall they show more similarity to eukaryotic RPA than to bacterial SSBs. The best-studied archaeal SSB is the 16-kDa single SSB of S. solfataricus. It contains a single OB fold and constitutes 2 to 5% of total soluble protein in the cell. The sequence shows that the domain structure is most similar to that of a bacterial SSB, and it contains an acidic CTD similar to that of E. coli SSB (114). Like the case in E. coli, this CTD is not required for DNA binding but mediates protein-protein interactions (96, 114). However, the crystal structure of S. solfataricus SSB revealed that the OB fold is actually more similar to that of human RPA than to those of bacterial SSBs (53).
Pyrococcus furiosis has three SSBs, namely, RPA41, RPA32, and RPA14, which form a heterotrimer with high affinity for ssDNA. RPA41 shows homology to eukaryotic RPA70, and like RPA70 it contains a zinc binding motif (55). Methanosarcina acetivorans also has three SSBs, namely, RPA1, RPA2, and RPA3. However, unlike the Pyrococcus proteins, they do not interact, and all form homodimers (97).
In addition to its role in stabilizing ssDNA, S. solfataricus SSB has also been implicated in DNA damage recognition (17). The M. acetivorans SSBs have also been shown to stimulate the primer extension activity of polymerase B1 (Pol B1) (97). There is some debate over the effect of SSB on MCM. Although one group reported that SSB stimulates MCM helicase activity (10), most data suggest that the presence of SSB at low concentrations has no effect on MCM, whereas at higher concentrations it is slightly inhibitory (52, 77).
PRIMASE
DNA polymerases are unable to initiate synthesis de novo and therefore require a DNA or RNA primer which they can elongate. This is synthesized by a primase, which in eukaryotes and bacteria is a DNA-dependent RNA polymerase. Bacterial primase is DnaG, a monomer, whereas eukaryotic primase is a dimer consisting of a small catalytic (PriS) and a large noncatalytic (PriL) subunit. This associates with Polα and the B subunit to form the Polα/primase complex (34). The primase synthesizes RNA primers of 8 to 12 nucleotides (nt) in length. These are then elongated to around 30 nt by Polα to produce a DNA-RNA hybrid before handoff to the replicative polymerase (34).
Archaea have homologues of eukaryotic PriS and PriL, but they lack Polα and the B subunit. The small subunits of primase from both S. solfataricus and Pyrococcus species can synthesize both RNA and DNA primers in vitro (59, 68, 79). Pyrococcus furiosis PriS preferentially synthesis long (up to 6 kb) DNA oligonucleotides. However, the addition of PriL increases the RNA polymerase activity, decreases the DNA polymerase activity, and decreases the average product length, suggesting that PriL plays a regulatory role (68). Despite the fact that S. solfataricus PriS can synthesize both DNA and RNA in vitro, it has a higher affinity for nucleoside triphosphates than for deoxynucleoside triphosphates, so it probably makes RNA primers in vivo (59, 81). In addition, Okazaki fragments have been isolated from archaea and found to be RNA at the 5′ end (80). Any in vivo relevance of the DNA polymerase activity of PriS is unclear, but given the lack of a Polα homologue in archaea, it is possible that the primase plays a role in primer elongation analogous to that of Polα in eukaryotes. However, there is currently no evidence for this.
The structures of the P. furiosis and Pyrococcus horikoshii primase small subunits and the S. solfataricus heterodimer have been solved (1, 46, 60) In all cases, PriS consists of a large α/β domain containing the catalytic prim domain and a smaller α-helical domain. It also contains a zinc binding motif which is conserved in eukaryotes and has been suggested to be involved in interaction of the enzyme with DNA in S. solfataricus. PriL is largely made up of an α-helical domain with a small α/β domain that mediates interaction with PriS. The interface between the two subunits is conserved between archaea and eukaryotes. The structure of the heterodimer shows that PriL does not directly contact the active site and can probably interact with the primer only once it reaches a length of 7 to 14 bp. This may trigger handoff to the polymerase and may explain why PriL inhibits the production of longer primers (60).
Archaeal PriS also contains a conserved catalytic triple-aspartate motif which is structurally similar to that found in the Pol X family of DNA polymerases. However, secondary structure elements surrounding this motif are very different, suggesting convergent evolution (60). There are five known Pol X family members in mammalian cells, and they function in processes involving DNA replication, repair, and recombination. With the exception of M. thermautotrophicum, archaea do not contain Pol X family proteins, so the similarity between these proteins and primase has led to suggestions that archaeal primase may play a role in DNA repair (61). This may explain why it possesses functions, such as DNA polymerase and 3′-nucleotidyl terminal transferase activities, not normally associated with primases.
GINS
The heterotetrameric eukaryotic GINS (go, ichi, nii, san [five, one, two, three in Japanese]) complex was first identified in yeast and Xenopus and consists of SLD5, PSF1, PSF2, and PSF3 (38, 57, 112). The complex is essential in yeast and interacts with MCM and CDC45 (38, 57). More recently, it was shown that GINS is necessary for the inclusion of MCM in replisome progression complexes, which include several replication and checkpoint proteins, during replication (38).
An archaeal homologue of GINS was originally identified in a yeast two-hybrid screen for interaction partners of MCM in S. solfataricus. The proteins interact both in vivo and in vitro. Further investigation showed that this protein, which is homologous to all the eukaryotic subunits but which has stronger homology to the proteins encoded by psf2 and psf3, interacts with another GINS homologue, with stronger homology to the sld5- and psf1-encoded proteins. The two genes are therefore known as gins23 and gins15. The proteins interact stably with RecJdbh, a protein homologous to the bacterial RecJ DNA binding domain, to form the archaeal GINS complex. The presence of a RecJ homologue in the complex has led to suggestions that the complex may be involved in stalled fork processing (76).
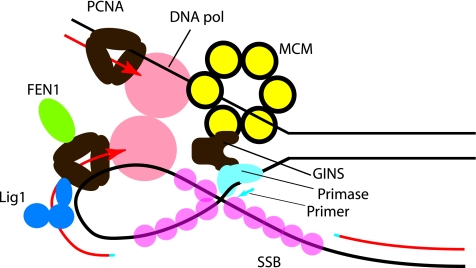
In addition to interacting with MCM, Gins23 also interacts with the primase. This solves a puzzle because in many replication systems, primase interacts with the replicative helicase. Indeed, a bacteriophage protein has been identified which contains homology to both MCM and primase (82). However, no interaction could be detected between primase and MCM in S. solfataricus. It therefore seems likely that the GINS complex forms a molecular bridge between MCM on the leading strand and primase on the lagging strand (Fig. 3) (76).
FIG. 3.
Model of the architecture of the archaeal DNA replication fork. Parental DNA is indicated by black lines, and newly synthesized DNA is shown in red. RNA primers, synthesized by primase, are shown in blue. MCM is shown as a yellow hexameric assembly surrounding the leading-strand template. We propose that the MCM helicase translocates along this strand, unwinding the parental duplex ahead of the replication fork. Single-stranded DNA is bound by SSB (Sulfolobus nomenclature), shown as pink circles. MCM interacts with the archaeal GINS complex (brown), and GINS, in turn, is additionally capable of binding primase (light blue). We propose that GINS acts to couple MCM translocation on the leading-strand template with deposition of primase on the lagging-strand template. DNA polymerase (in salmon pink) acts to extend the RNA primers, and we indicate that two polymerases are coupled, although there is currently no evidence for this in archaeal systems. Each DNA Pol interacts with a trimer of PCNA (brown). PCNA can act as a platform for additional assembly of the flap endonuclease FEN1 (green) and DNA ligase 1 (Lig1 [blue]), as cartooned on the lagging strand-associated PCNA only.
REPLICATIVE DNA POLYMERASES
As described above, the role of the primase is to synthesize a primer that is extended by a DNA polymerase. In common with bacteria and eukaryotes, archaea possess multiple DNA polymerases. Interestingly, there is a clear evolutionary divide in the distribution of polymerase families within the archaea. Organisms in the euryarchaeal phylum contain polymerases belonging to two distinct families, i.e., the ubiquitous family B polymerases and a family that thus far appears unique to the euryarchaea, called family D. The family D polymerases, first discovered by Ishino and coworkers, are two-subunit enzymes comprised of subunits DP1 and DP2 (9). It appears that the polymerization activity is contained within the larger subunit, DP2. The sequence of DP2, however, is very distinct from those of other DNA polymerases. Interestingly, the smaller subunit, DP1, possesses recognizable sequence homology to the noncatalytic B subunits of several eukaryotic family B DNA polymerases. Recent work has indicated that this subunit possesses intrinsic 3′-to-5′ exonuclease activity and that this activity is highest on substrates with mispaired nucleotides or single-stranded DNA, leading to speculation that this may be important for proofreading by the archaeal family D polymerases (50).
In addition to the family D polymerases, euryarchaea possess DNA polymerases belonging to family B, suggesting that the two classes of polymerases may play distinct roles in the cell. Indeed, an analogy may be found in the apparent discrimination between leading- and lagging-strand polymerases in Bacillus subtilis and eukaryotes. A recent study of the biochemical behavior of Pyrococcus D and B polymerases by Raffin and colleagues provided evidence for a model in which Pol B synthesizes the leading strand and Pol D replicates the lagging strand (43).
The crenarchaea do not carry a family D polymerase but generally have multiple family B polymerases. It is again possible, though as yet untested, that different crenarchaeal family B polymerases have distinct roles on the leading and lagging strands.
One intriguing feature that has been demonstrated for both euryarchaeal and crenarchaeal family B polymerases is the ability to sense uracil ahead of the polymerase in the DNA template and stall 4 nt before that residue, preventing its copying by the polymerase. Work by Connolly and colleagues revealed that this property is conferred upon the enzyme by a small, conserved pocket that lies in the N-terminal domain of the polymerase (14, 32). This pocket has the ability to bind uracil with high affinity, resulting in stalling of the polymerase as it moves along the template. Presumably, the polymerase then signals to the repair machinery to facilitate removal of the uracil base and lead to correction of the lesion. How this is achieved is currently unknown, although it is probable that some form of replication fork regression may be involved to facilitate template repair.
SLIDING CLAMPS
Although the leading-strand DNA polymerase in archaea may, in principle, have to synthesize over a million bases without disengaging from the template, the intrinsic processivity of purified DNA polymerases is actually quite low. The required processivity of polymerases is instead conferred upon them by association with an accessory factor, the sliding clamp. In bacteria, the sliding clamp is a homodimer, the β-clamp. In contrast, in archaea and eukaryotes, the sliding clamp, proliferating cell nuclear antigen (PCNA), is a trimer. However, despite the difference between the subunit compositions of bacterial and eukaryal/archaeal clamps, both classes of protein possess quasi-sixfold symmetry. As with the phylogenetic distribution of DNA polymerases, there appears to be a division within the archaea with regard to the identities of genes encoding PCNA (Table 1). In almost all euryarchaea, as in eukaryotes, there is a single PCNA homolog, and the protein forms a homotrimer. A single euryarchaeal species, Thermococcus kodakarensis, has two PCNA homologues, but it has been proposed that one of these (TK0582) may have been deposited in the genome relatively recently via a lateral gene transfer event (36). In contrast, the majority of crenarchaea for which genome sequences are available have multiple PCNA homologues. In Aeropyrum pernix, there are three PCNA homologs, and these have been shown to be capable of both homo- and heteromultimerization (19). Strikingly, S. solfataricus also possesses three PCNA homologs, but in this case they are only capable of heterotrimerization. In general, archaeal PCNAs have the capacity to interact with and stimulate the processivity of the replicative polymerases. In addition, as reviewed elsewhere, PCNA interacts with a number of other factors involved in replication and repair of DNA (113, 115).
The interaction between PCNA and a client protein is usually mediated via a short recognition motif, termed the PCNA-interacting protein (PIP) motif, most commonly found at either the N or C terminus of the protein (115). The structural basis of the interaction was revealed with the elucidation of the structure of human PCNA bound to the PIP peptide of p21. Importantly, this structure showed that one peptide could bind per PCNA monomer (41). It is therefore possible that PCNA is able to interact simultaneously with multiple partner proteins, forming a molecular “tool belt.” Interestingly, analysis of the heterotrimeric Sulfolobus PCNA led to the discovery that distinct PCNA subunits within this complex each have preferred client proteins. More specifically, Flap endonuclease 1 (FEN1), DNA polymerase B1, and DNA ligase I interact preferentially with distinct subunits. Furthermore, affinity chromatography indicated that the heterotrimeric Sulfolobus PCNA could bridge between FEN1 and ligase or polymerase (25). It appears, therefore, that an individual PCNA ring can organize and coordinate the activities of multiple factors simultaneously. The crystal structure of the Sulfolobus PCNA1-PCNA2 heterodimer in complex with Fen1 was recently elucidated (27). This has revealed that the basis of discrimination between PCNA subunits and client proteins lies in distinct geometries being adopted by the PIP motif-binding interdomain connector loop on the individual PCNA subunits.
RFC—the Clamp Loader
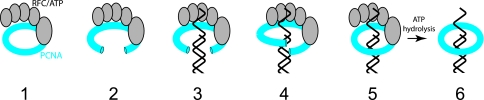
The PCNA trimer is a toroidal molecule that encircles DNA, thereby tethering its client proteins to the DNA substrate. However, PCNA does not normally spontaneously assemble around DNA, but rather requires a specific loading factor, replication factor C (RFC), to facilitate appropriate placing of the sliding clamp onto the DNA molecule. RFC catalyzes opening of PCNA and deposition of PCNA around double-stranded DNA at the site of a double- to single-strand transition, such as a primer-template junction (49). This process is dependent on ATP binding by RFC. Archaeal RFC is a pentamer containing four identical copies of a small subunit (RFCS) and a single copy of a large subunit (RFCL). Biochemical and structural analyses of the RFC of Archeoglobus fulgidus by Wigley and colleagues, along with structural studies of the Pyrococcus homolog by Ishino and colleagues, in conjunction with structural and biophysical studies of yeast RFC and PCNA, have shed considerable light on the molecular mechanisms of RFC action (84, 85, 90, 103-105). The RFC subunits are members of the AAA+ family of ATPases, and in common with other members of this family, ATP is bound at the junction of two subunits (8, 103-105). The intact RFC complex forms a rising right-handed spiral and therefore has four sites between subunits that can be occupied by ATP. ATP binding by all four sites is required for clamp loading. Fluorescence resonance energy transfer studies of yeast RFC and PCNA indicated that following binding of PCNA by RFC-ATP, PCNA is opened about 34 Å in the plane of the ring (122). This structure is likely the conformation that can bind DNA and mediate loading. The loading reaction then progresses via an intermediate where the PCNA ring is held open with a gap of about 5 Å, as seen in an EM structure of Pyrococcus PCNA (85). This appears to be a structure akin to a lock washer, with PCNA being open in and out of plane configuration (Fig. 4). Precisely how binding of ATP causes these modulations in the structure is not fully understood, but arginine fingers in the RFCS subunits that act to communicate with the ATP binding site of the neighboring subunit appear to play a pivotal role in the process (105). As stated above, ATP hydrolysis is not required for PCNA loading per se. Rather, hydrolysis drives the final stage of the process, i.e., release of the clamp loader from the loaded PCNA.
FIG. 4.
Cartoon of the clamp loading process. A pentameric RFC (gray) containing one large subunit and four identical small subunits binds ATP and interacts with a ring of PCNA (blue) (step 1). RFC opens PCNA 34 Å in the plane of the ring, and DNA enters the ring (steps 2 and 3). The PCNA ring then closes around the DNA but leaves an out-of-plane gap of approximately 5 Å (step 4) before sealing shut (step 5). ATP is then hydrolyzed by RFC, and RFC leaves PCNA bound at the primer-template junction.
PCNA-Interacting Proteins
Once loaded, PCNA can then bind DNA polymerase, and the primer can be extended. As alluded to above, PCNA also acts as an adaptor for a range of additional proteins. These include the flap endonuclease FEN1 and DNA ligase I (113, 115). During lagging-strand synthesis, downstream RNA primers must be removed in order for Okazaki fragments to be joined. These tasks are most likely performed by RNase H2, an RNase that cleaves RNA in RNA-DNA hybrids, and FEN1. FEN1 interacts with and is stimulated by PCNA. The crystal structures of a number of archaeal and eukaryal FEN1 enzymes have been solved, both alone and in complex with the DNA substrate and with PCNA (11, 44, 100).
The complex of human FEN1 with PCNA revealed that each subunit of the homotrimeric PCNA bound to a different monomer of FEN1. Interestingly, each individual PCNA subunit-FEN1 interaction showed a distinct geometry courtesy of a highly flexible hinge region adjacent to the PCNA interaction site (100). Two of the FEN1-PCNA monomer complexes did not interfere with the potential interaction between PCNA and DNA but held FEN1 in such a position that it was unlikely to be capable of catalyzing DNA strand cleavage. This has led to speculation that these geometries may correspond to translocating forms of the PCNA-FEN1 complex, with FEN1 in a “locked-down” and inactive conformation that would then undergo a structural transition in the presence of the appropriate DNA substrate, thereby activating FEN1. In the recent structure of Sulfolobus FEN1 in complex with PCNA1 and PCNA2, FEN1 was bound only to PCNA1, in agreement with previous biochemical studies, and was positioned in the complex in an orientation that was compatible with its accessing DNA.
The concept of carrier and active conformations of proteins on PCNA rings probably has relevance beyond the case of FEN1. For example, Sulfolobus PCNA was found to be able to simultaneously bind DNA ligase 1 (Lig1) and FEN1 in solution. However, the structure of human Lig1 in complex with DNA was recently solved, and it was seen that Lig1 effectively encircled the entire DNA molecule, such that even if it was bound to only a single subunit of PCNA, it would effectively sterically occlude access to PCNA by other factors (93). However, if Lig1 can adopt a carrier conformation on PCNA, like the case for FEN1, it may dock down on the substrate only transiently to catalyze the ligation step at the final stage in lagging-strand maturation. Indeed, it is tempting to speculate that the steric clash that would be induced by Lig1 engaging with the DNA template may displace other proteins from PCNA. This may permit access to PCNA by RFC after Lig1 has joined Okazaki fragments, whereupon RFC could unload the PCNA ring.
ROLE OF ARCHAEAL CHROMATIN
The vast majority of studies that have been performed on the archaeal DNA replication machinery have used naked DNA templates. Yet it is clear that within the cell, DNA is compacted by association with a range of small basic proteins and that these proteins have the potential to modulate access to the DNA template. As recently reviewed elsewhere, archaeal cells utilize an intriguing variety of different proteins to mediate genome compaction (102, 117). The majority of euryarchaea have homologues of eukaryotic histones, and extensive biochemical studies have revealed that these form structures analogous to the eukaryotic H3/H4 tetrasome. However, despite their discovery over 16 years ago, little is yet known about the role of archaeal histones in vivo. Differences in the expression of histone variants have been observed during culture growth, suggesting that alterations in levels of compaction and/or distribution of histone subtypes may modulate gene expression or even replication rates. Biochemical work using a highly defined in vitro transcription system derived from M. thermautotrophicum has revealed that an archaeal nucleosome positioned at an artificially selected high-affinity site has the capacity to slow transcription through the nucleosome (119). It is currently unclear whether transcription through the nucleosome displaces it from the template or if it remains bound.
Intriguingly, with the exception of some mesophilic marine organisms, histones are absent from the crenarchaea (18). The most highly studied crenarchaeal chromatin proteins come from the Sulfolobus genus. Species within this genus have a Sulfolobus-specific chromatin protein, Sul7d. In addition, there is a second, abundant, nonspecific nucleic acid binding protein, Alba. Alba is additionally found in a broad range of both crenarchaea and euryarchaea (117). Alba has both DNA and RNA binding activities but has been found to be associated with a range of genomic loci in Sulfolobus, suggesting that it has a significant role as a chromatin protein (78). Recent work has characterized a second homolog of Alba found in Sulfolobus cells (48). Interestingly, Alba2 forms obligate heterodimers with Alba1 and appears to alter higher-order DNA packing by Alba1. It is possible that differential levels of expression of Alba1 and Alba2 could modulate nucleoid structures in Sulfolobus.
In contrast to the situation with archaeal histones, both Sul7d and Alba show posttranslational modification in Sulfolobus cells. Sul7d shows monomethylation of lysine residues, but the consequence (if any) of this modification and the identity of the methyltransferase are currently unknown (28). Alba1 has been shown to be acetylated at an internal lysine residue, lysine 16 (2). The effect of acetylation of K16 is to lower Alba's affinity for DNA. Enzymes that acetylate and deacetylate Alba have been identified (78). Interestingly, the acetylase, Pat, and the deacetylase, Sir2, are conserved in many bacteria, where they act to regulate acetyl-coenzyme A synthetase by reversible acetylation (111). It therefore appears that Sulfolobus may have coopted this bacterial regulatory system to generate a primitive and simplified form of chromatin regulation. It is of particular interest that Sir2 is well conserved in eukaryotes, where it has expanded into a protein family whose members play roles in a variety of cellular processes, including regulation of chromatin structure, microtubule dynamics, and life span (4).
Recent work has revealed that both Alba and Sul7d can inhibit translocation by purified Sulfolobus MCM helicase (77). Acetylation of Alba by Pat alleviated this repression, leading to speculation that mechanisms may exist within Sulfolobus cells to couple Alba-modifying or -displacing activity to progression of the replication fork in vivo.
Considerable progress has been made in understanding the form and function of archaeal DNA replication and chromatin proteins. It is clear, however, that much remains to be discovered about how these proteins interact in the context of the macromolecular assemblies found at replication origins and during progression of the DNA replication fork. Furthermore, the manner in which these proteins are regulated during the course of the cell cycle in archaeal species remains largely unexplored. For Sulfolobus, in particular, where multiple DNA replication origins are used, it will be of great interest to determine whether a mechanism exists to allow coordinated regulation of origin activity, and if so, how this is mediated.
REFERENCES
- 1.Augustin, M. A., R. Huber, and J. T. Kaiser. 2001. Crystal structure of a DNA-dependent RNA polymerase (DNA primase). Nat. Struct. Biol. 8: 57-61. [DOI] [PubMed] [Google Scholar]
- 2.Bell, S. D., C. H. Botting, B. N. Wardleworth, S. P. Jackson, and M. F. White. 2002. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 296:148-151. [DOI] [PubMed] [Google Scholar]
- 3.Berquist, B. R., and S. DasSarma. 2003. An archaeal chromosomal autonomously replicating sequence element from an extreme halophile, Halobacterium sp. strain NRC-1. J. Bacteriol. 185:5959-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blander, G., and L. Guarente. 2004. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73:417-435. [DOI] [PubMed] [Google Scholar]
- 5.Blanton, H. L., S. J. Radford, S. McMahan, H. M. Kearney, J. G. Ibrahim, and J. Sekelsky. 2005. REC, Drosophila MCM8, drives formation of meiotic crossovers. PLoS Genet. 1:e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolt, E. L. 2005. Helicases that interact with replication forks: new candidates from archaea. Biochem. Soc. Trans. 33:1471-1473. [DOI] [PubMed] [Google Scholar]
- 7.Bowers, J. L., J. C. Randell, S. Chen, and S. P. Bell. 2004. ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol. Cell 16:967-978. [DOI] [PubMed] [Google Scholar]
- 8.Bowman, G. D., M. O'Donnell, and J. Kuriyan. 2004. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature 429:724-730. [DOI] [PubMed] [Google Scholar]
- 9.Cann, I. K., K. Komori, H. Toh, S. Kanai, and Y. Ishino. 1998. A heterodimeric DNA polymerase: evidence that members of Euryarchaeota possess a distinct DNA polymerase. Proc. Natl. Acad. Sci. USA 95:14250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carpentieri, F., M. De Felice, M. De Falco, M. Rossi, and F. M. Pisani. 2002. Physical and functional interaction between the mini-chromosome maintenance-like DNA helicase and the single-stranded DNA binding protein from the crenarchaeon Sulfolobus solfataricus. J. Biol. Chem. 277:12118-12127. [DOI] [PubMed] [Google Scholar]
- 11.Chapados, B. R., D. J. Hosfield, S. Han, J. Qiu, B. Yelent, B. Shen, and J. A. Tainer. 2004. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell 116:39-50. [DOI] [PubMed] [Google Scholar]
- 12.Chen, Y. J., X. Yu, R. Kasiviswanathan, J. H. Shin, Z. Kelman, and E. H. Egelman. 2005. Structural polymorphism of Methanothermobacter thermautotrophicus MCM. J. Mol. Biol. 346:389-394. [DOI] [PubMed] [Google Scholar]
- 13.Chong, J. P., M. K. Hayashi, M. N. Simon, R. M. Xu, and B. Stillman. 2000. A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc. Natl. Acad. Sci. USA 97:1530-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connolly, B. A., M. J. Fogg, G. Shuttleworth, and B. T. Wilson. 2003. Uracil recognition by archaeal family B DNA polymerases. Biochem. Soc. Trans. 31:699-702. [DOI] [PubMed] [Google Scholar]
- 15.Costa, A., T. Pape, M. van Heel, P. Brick, A. Patwardhan, and S. Onesti. 2006. Structural studies of the archaeal MCM complex in different functional states. J. Struct. Biol. 156:210-219. [DOI] [PubMed] [Google Scholar]
- 16.Coverley, D., and R. A. Laskey. 1994. Regulation of eukaryotic DNA replication. Annu. Rev. Biochem. 63:745-776. [DOI] [PubMed] [Google Scholar]
- 17.Cubeddu, L., and M. F. White. 2005. DNA damage detection by an archaeal single-stranded DNA-binding protein. J. Mol. Biol. 353:507-516. [DOI] [PubMed] [Google Scholar]
- 18.Cubonova, L., K. Sandman, S. J. Hallam, E. F. Delong, and J. N. Reeve. 2005. Histones in crenarchaea. J. Bacteriol. 187:5482-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daimon, K., Y. Kawarabayasi, H. Kikuchi, Y. Sako, and Y. Ishino. 2002. Three proliferating cell nuclear antigen-like proteins found in the hyperthermophilic archaeon Aeropyrum pernix: interactions with the two DNA polymerases. J. Bacteriol. 184:687-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davey, M. J., L. Fang, P. McInerney, R. E. Georgescu, and M. O'Donnell. 2002. The DnaC helicase loader is a dual ATP/ADP switch protein. EMBO J. 21:3148-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davey, M. J., and M. O'Donnell. 2003. Replicative helicase loaders: ring breakers and ring makers. Curr. Biol. 13:R594-R596. [DOI] [PubMed] [Google Scholar]
- 22.De Felice, M., L. Esposito, B. Pucci, F. Carpentieri, M. De Falco, M. Rossi, and F. M. Pisani. 2003. Biochemical characterization of a CDC6-like protein from the crenarchaeon Sulfolobus solfataricus. J. Biol. Chem. 278:46424-46431. [DOI] [PubMed] [Google Scholar]
- 23.De Felice, M., L. Esposito, B. Pucci, M. De Falco, M. Rossi, and F. M. Pisani. 2004. A CDC6-like factor from the archaea Sulfolobus solfataricus promotes binding of the mini-chromosome maintenance complex to DNA. J. Biol. Chem. 279:43008-43012. [DOI] [PubMed] [Google Scholar]
- 24.Dimitrova, D. S., I. T. Todorov, T. Melendy, and D. M. Gilbert. 1999. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J. Cell Biol. 146:709-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dionne, I., R. K. Nookala, S. P. Jackson, A. J. Doherty, and S. D. Bell. 2003. A heterotrimeric PCNA in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol. Cell 11:275-282. [DOI] [PubMed] [Google Scholar]
- 26.Dionne, I., N. P. Robinson, A. T. McGeoch, V. L. Marsh, A. Reddish, and S. D. Bell. 2003. DNA replication in the hyperthermophilic archaeon Sulfolobus solfataricus. Biochem. Soc. Trans. 31:674-676. [DOI] [PubMed] [Google Scholar]
- 27.Dore, A. S., M. L. Kilkenny, S. A. Jones, A. W. Oliver, S. M. Roe, S. D. Bell, and L. H. Pearl. 2006. Structure of an archaeal PCNA1-PCNA2-FEN1 complex: elucidating PCNA subunit and client enzyme specificity. Nucleic Acids Res. [Online.] 34:4515-4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edmondson, S. P., and J. W. Shriver. 2001. DNA binding proteins Sac7d and Sso7d from Sulfolobus. Methods Enzymol. 334:129-145. [DOI] [PubMed] [Google Scholar]
- 29.Edwards, M. C., A. V. Tutter, C. Cvetic, C. H. Gilbert, T. A. Prokhorova, and J. C. Walter. 2002. MCM2-7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J. Biol. Chem. 277:33049-33057. [DOI] [PubMed] [Google Scholar]
- 30.Fletcher, R. J., B. E. Bishop, R. P. Leon, R. A. Sclafani, C. M. Ogata, and X. S. Chen. 2003. The structure and function of MCM from archaeal M. thermoautotrophicum. Nat. Struct. Biol. 10:160-167. [DOI] [PubMed] [Google Scholar]
- 31.Fletcher, R. J., J. Shen, Y. Gomez-Llorente, C. S. Martin, J. M. Carazo, and X. S. Chen. 2005. Double hexamer disruption and biochemical activities of Methanobacterium thermoautotrophicum MCM. J. Biol. Chem. 280:42405-42410. [DOI] [PubMed] [Google Scholar]
- 32.Fogg, M. J., L. H. Pearl, and B. A. Connolly. 2002. Structural basis for uracil recognition by archaeal family B DNA polymerases. Nat. Struct. Biol. 9:922-927. [DOI] [PubMed] [Google Scholar]
- 33.Forsburg, S. L. 2004. Eukaryotic MCM proteins: beyond replication initiation. Microbiol. Mol. Biol. Rev. 68:109-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frick, D. N., and C. C. Richardson. 2001. DNA primases. Annu. Rev. Biochem. 70:39-80. [DOI] [PubMed] [Google Scholar]
- 35.Fujikane, R., K. Komori, H. Shinagawa, and Y. Ishino. 2005. Identification of a novel helicase activity unwinding branched DNAs from the hyperthermophilic archaeon, Pyrococcus furiosus. J. Biol. Chem. 280:12351-12358. [DOI] [PubMed] [Google Scholar]
- 36.Fukui, T., H. Atomi, T. Kanai, R. Matsumi, S. Fujiwara, and T. Imanaka. 2005. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 15:352-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gai, D., R. Zhao, D. Li, C. V. Finkielstein, and X. S. Chen. 2004. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell 119:47-60. [DOI] [PubMed] [Google Scholar]
- 38.Gambus, A., R. C. Jones, A. Sanchez-Diaz, M. Kanemaki, F. van Deursen, R. D. Edmondson, and K. Labib. 2006. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 8:358-366. [DOI] [PubMed] [Google Scholar]
- 39.Gomez-Llorente, Y., R. J. Fletcher, X. S. Chen, J. M. Carazo, and C. San Martin. 2005. Polymorphism and double hexamer structure in the archaeal minichromosome maintenance (MCM) helicase from Methanobacterium thermoautotrophicum. J. Biol. Chem. 280:40909-40915. [DOI] [PubMed] [Google Scholar]
- 40.Grabowski, B., and Z. Kelman. 2001. Autophosphorylation of archaeal Cdc6 homologues is regulated by DNA. J. Bacteriol. 183:5459-5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gulbis, J. M., Z. Kelman, J. Hurwitz, M. Odonnell, and J. Kuriyan. 1996. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell 87:297-306. [DOI] [PubMed] [Google Scholar]
- 42.Guy, C. P., and E. L. Bolt. 2005. Archaeal Hel308 helicase targets replication forks in vivo and in vitro and unwinds lagging strands. Nucleic Acids Res. 33:3678-3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henneke, G., D. Flament, U. Hubscher, J. Querellou, and J. P. Raffin. 2005. The hyperthermophilic euryarchaeota Pyrococcus abyssi likely requires the two DNA polymerases D and B for DNA replication. J. Mol. Biol. 350: 53-64. [DOI] [PubMed] [Google Scholar]
- 44.Hosfield, D. J., C. D. Mol, B. Shen, and J. A. Tainer. 1998. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell 95:135-146. [DOI] [PubMed] [Google Scholar]
- 45.Ishimi, Y. 1997. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem. 272:24508-24513. [DOI] [PubMed] [Google Scholar]
- 46.Ito, N., O. Nureki, M. Shirouzu, S. Yokoyama, and F. Hanaoka. 2003. Crystal structure of the Pyrococcus horikoshii DNA primase-UTP complex: implications for the mechanism of primer synthesis. Genes Cells 8:913-923. [DOI] [PubMed] [Google Scholar]
- 47.Jacob, F., S. Brenner, and F. Cuzin. 1963. On the regulation of DNA replication in bacteria. Cold Spring Harbor Symp. Quant. Biol. 28:329-348. [Google Scholar]
- 48.Jelinska, C., M. J. Conroy, C. J. Craven, A. M. Hounslow, P. A. Bullough, J. P. Waltho, G. L. Taylor, and M. F. White. 2005. Obligate heterodimerization of the archaeal Alba2 protein with Alba1 provides a mechanism for control of DNA packaging. Structure 13:963-971. [DOI] [PubMed] [Google Scholar]
- 49.Johnson, A., and M. O'Donnell. 2005. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 74:283-315. [DOI] [PubMed] [Google Scholar]
- 50.Jokela, M., A. Eskelinen, H. Pospiech, J. Rouvinen, and J. E. Syvaoja. 2004. Characterization of the 3′ exonuclease subunit DP1 of Methanococcus jannaschii replicative DNA polymerase D. Nucleic Acids Res. 32:2430-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kasiviswanathan, R., J. H. Shin, and Z. Kelman. 2005. Interactions between the archaeal Cdc6 and MCM proteins modulate their biochemical properties. Nucleic Acids Res. 33:4940-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kelman, Z., J. K. Lee, and J. Hurwitz. 1999. The single minichromosome maintenance protein of Methanobacterium thermoautotrophicum DeltaH contains DNA helicase activity. Proc. Natl. Acad. Sci. USA 96:14783-14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kerr, I. D., R. I. Wadsworth, L. Cubeddu, W. Blankenfeldt, J. H. Naismith, and M. F. White. 2003. Insights into ssDNA recognition by the OB fold from a structural and thermodynamic study of Sulfolobus SSB protein. EMBO J. 22:2561-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Komamura-Kohno, Y., K. Karasawa-Shimizu, T. Saitoh, M. Sato, F. Hanaoka, S. Tanaka, and Y. Ishimi. 2006. Site-specific phosphorylation of MCM4 during the cell cycle in mammalian cells. FEBS J. 273:1224-1239. [DOI] [PubMed] [Google Scholar]
- 55.Komori, K., and Y. Ishino. 2001. Replication protein A in Pyrococcus furiosus is involved in homologous DNA recombination. J. Biol. Chem. 276:25654-25660. [DOI] [PubMed] [Google Scholar]
- 56.Krude, T., C. Musahl, R. A. Laskey, and R. Knippers. 1996. Human replication proteins hCdc21, hCdc46 and P1Mcm3 bind chromatin uniformly before S-phase and are displaced locally during DNA replication. J. Cell Sci. 109:309-318. [DOI] [PubMed] [Google Scholar]
- 57.Kubota, Y., Y. Takase, Y. Komori, Y. Hashimoto, T. Arata, Y. Kamimura, H. Araki, and H. Takisawa. 2003. A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes Dev. 17:1141-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Labib, K., J. A. Tercero, and J. F. Diffley. 2000. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science 288:1643-1647. [DOI] [PubMed] [Google Scholar]
- 59.Lao-Sirieix, S. H., and S. D. Bell. 2004. The heterodimeric primase of the hyperthermophilic archaeon Sulfolobus solfataricus possesses DNA and RNA primase, polymerase and 3′-terminal nucleotidyl transferase activities. J. Mol. Biol. 344:1251-1263. [DOI] [PubMed] [Google Scholar]
- 60.Lao-Sirieix, S. H., R. K. Nookala, P. Roversi, S. D. Bell, and L. Pellegrini. 2005. Structure of the heterodimeric core primase. Nat. Struct. Mol. Biol. 12:1137-1144. [DOI] [PubMed] [Google Scholar]
- 61.Lao-Sirieix, S. H., L. Pellegrini, and S. D. Bell. 2005. The promiscuous primase. Trends Genet. 21:568-572. [DOI] [PubMed] [Google Scholar]
- 62.Laskey, R. A., and M. A. Madine. 2003. A rotary pumping model for helicase function of MCM proteins at a distance from replication forks. EMBO Rep. 4:26-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee, J. K., and J. Hurwitz. 2000. Isolation and characterization of various complexes of the minichromosome maintenance proteins of Schizosaccharomyces pombe. J. Biol. Chem. 275:18871-18878. [DOI] [PubMed] [Google Scholar]
- 64.Lee, J. K., and J. Hurwitz. 2001. Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6, and 7 complex requires forked DNA structures. Proc. Natl. Acad. Sci. USA 98:54-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leipe, D. D., L. Aravind, and E. V. Koonin. 1999. Did DNA replication evolve twice independently? Nucleic Acids Res. 27:3389-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li, D., R. Zhao, W. Lilyestrom, D. Gai, R. Zhang, J. A. DeCaprio, E. Fanning, A. Jochimiak, G. Szakonyi, and X. S. Chen. 2003. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature 423:512-518. [DOI] [PubMed] [Google Scholar]
- 67.Liu, J., C. L. Smith, D. DeRyckere, K. DeAngelis, G. S. Martin, and J. M. Berger. 2000. Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control. Mol. Cell 6:637-648. [DOI] [PubMed] [Google Scholar]
- 68.Liu, L., K. Komori, S. Ishino, A. A. Bocquier, I. K. Cann, D. Kohda, and Y. Ishino. 2001. The archaeal DNA primase: biochemical characterization of the p41-p46 complex from Pyrococcus furiosus. J. Biol. Chem. 276:45484-45490. [DOI] [PubMed] [Google Scholar]
- 69.Lundgren, M., A. Andersson, L. Chen, P. Nilsson, and R. Bernander. 2004. Three replication origins in Sulfolobus species: synchronous initiation of chromosome replication and asynchronous termination. Proc. Natl. Acad. Sci. USA 101:7046-7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Machida, Y. J., J. L. Hamlin, and A. Dutta. 2005. Right place, right time, and only once: replication initiation in metazoans. Cell 123:13-24. [DOI] [PubMed] [Google Scholar]
- 71.Madine, M. A., C. Y. Khoo, A. D. Mills, C. Musahl, and R. A. Laskey. 1995. The nuclear envelope prevents reinitiation of replication by regulating the binding of MCM3 to chromatin in Xenopus egg extracts. Curr. Biol. 5:1270-1279. [DOI] [PubMed] [Google Scholar]
- 72.Maine, G. T., P. Sinha, and B. K. Tye. 1984. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics 106:365-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maiorano, D., O. Cuvier, E. Danis, and M. Mechali. 2005. MCM8 is an MCM2-7-related protein that functions as a DNA helicase during replication elongation and not initiation. Cell 120:315-328. [DOI] [PubMed] [Google Scholar]
- 74.Maiorano, D., M. Lutzmann, and M. Mechali. 2006. MCM proteins and DNA replication. Curr. Opin. Cell Biol. 18:130-136. [DOI] [PubMed] [Google Scholar]
- 75.Marians, K. J. 1992. Prokaryotic DNA replication. Annu. Rev. Biochem. 61:673-719. [DOI] [PubMed] [Google Scholar]
- 76.Marinsek, N., E. R. Barry, K. S. Makarova, I. Dionne, E. V. Koonin, and S. D. Bell. 2006. GINS, a central nexus in the archaeal DNA replication fork. EMBO Rep. 7:539-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marsh, V. L., A. T. McGeoch, and S. D. Bell. 2006. Influence of chromatin and single strand binding proteins on the activity of an archaeal MCM. J. Mol. Biol. 357:1345-1350. [DOI] [PubMed] [Google Scholar]
- 78.Marsh, V. L., S. Y. Peak-Chew, and S. D. Bell. 2005. Sir2 and the acetyltransferase, Pat, regulate the archaeal chromatin protein, Alba. J. Biol. Chem. 280:21122-21128. [DOI] [PubMed] [Google Scholar]
- 79.Matsui, E., M. Nishio, H. Yokoyama, K. Harata, S. Darnis, and I. Matsui. 2003. Distinct domain functions regulating de novo DNA synthesis of thermostable DNA primase from hyperthermophile Pyrococcus horikoshii. Biochemistry 42:14968-14976. [DOI] [PubMed] [Google Scholar]
- 80.Matsunaga, F., P. Forterre, Y. Ishino, and H. Myllykallio. 2001. In vivo interactions of archaeal Cdc6/Orc1 and minichromosome maintenance proteins with the replication origin. Proc. Natl. Acad. Sci. USA 98:11152-11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Matsunaga, F., C. Norais, P. Forterre, and H. Myllykallio. 2003. Identification of short ‘eukaryotic’ Okazaki fragments synthesized from a prokaryotic replication origin. EMBO Rep. 4:154-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McGeoch, A. T., and S. D. Bell. 2005. Eukaryotic/archaeal primase and MCM proteins encoded in a bacteriophage genome. Cell 120:167-168. [DOI] [PubMed] [Google Scholar]
- 83.McGeoch, A. T., M. A. Trakselis, R. A. Laskey, and S. D. Bell. 2005. Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat. Struct. Mol. Biol. 12:756-762. [DOI] [PubMed] [Google Scholar]
- 84.Miyata, T., T. Oyama, K. Mayanagi, S. Ishino, Y. Ishino, and K. Morikawa. 2004. The clamp-loading complex for processive DNA replication. Nat. Struct. Mol. Biol. 11:632-636. [DOI] [PubMed] [Google Scholar]
- 85.Miyata, T., H. Suzuki, T. Oyama, K. Mayanagi, Y. Ishino, and K. Morikawa. 2005. Open clamp structure in the clamp-loading complex visualized by electron microscopic image analysis. Proc. Natl. Acad. Sci. USA 102:13795-13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Montagnoli, A., B. Valsasina, D. Brotherton, S. Troiani, S. Rainoldi, P. Tenca, A. Molinari, and C. Santocanale. 2006. Identification of Mcm2 phosphorylation sites by S-phase-regulating kinases. J. Biol. Chem. 281:10281-10290. [DOI] [PubMed] [Google Scholar]
- 87.Moyer, S. E., P. W. Lewis, and M. R. Botchan. 2006. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. USA 103:10236-10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Murzin, A. G. 1993. OB (oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 12:861-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Myllykallio, H., P. Lopez, P. Lopez-Garcia, R. Heilig, W. Saurin, Y. Zivanovic, H. Philippe, and P. Forterre. 2000. Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science 288:2212-2215. [DOI] [PubMed] [Google Scholar]
- 90.Oyama, T., Y. Ishino, I. K. Cann, S. Ishino, and K. Morikawa. 2001. Atomic structure of the clamp loader small subunit from Pyrococcus furiosus. Mol. Cell 8:455-463. [DOI] [PubMed] [Google Scholar]
- 91.Pacek, M., and J. C. Walter. 2004. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 23:3667-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pape, T., H. Meka, S. Chen, G. Vicentini, M. van Heel, and S. Onesti. 2003. Hexameric ring structure of the full-length archaeal MCM protein complex. EMBO Rep. 4:1079-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pascal, J. M., P. J. O'Brien, A. E. Tomkinson, and T. Ellenberger. 2004. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature 432:473-478. [DOI] [PubMed] [Google Scholar]
- 94.Raghunathan, S., A. G. Kozlov, T. M. Lohman, and G. Waksman. 2000. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Biol. 7:648-652. [DOI] [PubMed] [Google Scholar]
- 95.Raghunathan, S., C. S. Ricard, T. M. Lohman, and G. Waksman. 1997. Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength X-ray diffraction on the selenomethionyl protein at 2.9-A resolution. Proc. Natl. Acad. Sci. USA 94:6652-6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Richard, D. J., S. D. Bell, and M. F. White. 2004. Physical and functional interaction of the archaeal single-stranded DNA-binding protein SSB with RNA polymerase. Nucleic Acids Res. 32:1065-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Robbins, J. B., M. C. Murphy, B. A. White, R. I. Mackie, T. Ha, and I. K. Cann. 2004. Functional analysis of multiple single-stranded DNA-binding proteins from Methanosarcina acetivorans and their effects on DNA synthesis by DNA polymerase BI. J. Biol. Chem. 279:6315-6326. [DOI] [PubMed] [Google Scholar]
- 98.Robinson, N. P., and S. D. Bell. 2005. Origins of DNA replication in the three domains of life. FEBS J. 272:3757-3766. [DOI] [PubMed] [Google Scholar]
- 99.Robinson, N. P., I. Dionne, M. Lundgren, V. L. Marsh, R. Bernander, and S. D. Bell. 2004. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell 116:25-38. [DOI] [PubMed] [Google Scholar]
- 100.Sakurai, S., K. Kitano, H. Yamaguchi, K. Hamada, K. Okada, K. Fukuda, M. Uchida, E. Ohtsuka, H. Morioka, and T. Hakoshima. 2005. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 24:683-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Reference deleted.
- 102.Sandman, K., and J. N. Reeve. 2005. Archaeal chromatin proteins: different structures but common function? Curr. Opin. Microbiol. 8:656-661. [DOI] [PubMed] [Google Scholar]
- 103.Seybert, A., D. J. Scott, S. Scaife, M. R. Singleton, and D. B. Wigley. 2002. Biochemical characterisation of the clamp/clamp loader proteins from the euryarchaeon Archaeoglobus fulgidus. Nucleic Acids Res. 30:4329-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Seybert, A., M. R. Singleton, N. Cook, D. R. Hall, and D. B. Wigley. 2006. Communication between subunits within an archaeal clamp-loader complex. EMBO J. 25:2209-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Seybert, A., and D. B. Wigley. 2004. Distinct roles for ATP binding and hydrolysis at individual subunits of an archaeal clamp loader. EMBO J. 23:1360-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shechter, D., C. Y. Ying, and J. Gautier. 2004. DNA unwinding is an Mcm complex-dependent and ATP hydrolysis-dependent process. J. Biol. Chem. 279:45586-45593. [DOI] [PubMed] [Google Scholar]
- 107.Shechter, D. F., C. Y. Ying, and J. Gautier. 2000. The intrinsic DNA helicase activity of Methanobacterium thermoautotrophicum delta H minichromosome maintenance protein. J. Biol. Chem. 275:15049-15059. [DOI] [PubMed] [Google Scholar]
- 108.Shin, J. H., B. Grabowski, R. Kasiviswanathan, S. D. Bell, and Z. Kelman. 2003. Regulation of minichromosome maintenance helicase activity by Cdc6. J. Biol. Chem. 278:38059-38067. [DOI] [PubMed] [Google Scholar]
- 109.Shin, J. H., Y. Jiang, B. Grabowski, J. Hurwitz, and Z. Kelman. 2003. Substrate requirements for duplex DNA translocation by the eukaryal and archaeal minichromosome maintenance helicases. J. Biol. Chem. 278:49053-49062. [DOI] [PubMed] [Google Scholar]
- 110.Singleton, M. R., R. Morales, I. Grainge, N. Cook, M. N. Isupov, and D. B. Wigley. 2004. Conformational changes induced by nucleotide binding in Cdc6/ORC from Aeropyrum pernix. J. Mol. Biol. 343:547-557. [DOI] [PubMed] [Google Scholar]
- 111.Starai, V. J., H. Takahashi, J. D. Boeke, and J. C. Escalante-Semerena. 2004. A link between transcription and intermediary metabolism: a role for Sir2 in the control of acetyl-coenzyme A synthetase. Curr. Opin. Microbiol. 7:115-119. [DOI] [PubMed] [Google Scholar]
- 112.Takayama, Y., Y. Kamimura, M. Okawa, S. Muramatsu, A. Sugino, and H. Araki. 2003. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 17:1153-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vivona, J. B., and Z. Kelman. 2003. The diverse spectrum of sliding clamp interacting proteins. FEBS Lett. 546:167-172. [DOI] [PubMed] [Google Scholar]
- 114.Wadsworth, R. I., and M. F. White. 2001. Identification and properties of the crenarchaeal single-stranded DNA binding protein from Sulfolobus solfataricus. Nucleic Acids Res. 29:914-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Warbrick, E. 2000. The puzzle of PCNA's many partners. Bioessays 22:997-1006. [DOI] [PubMed] [Google Scholar]
- 116.Weinreich, M., C. Liang, and B. Stillman. 1999. The Cdc6p nucleotide-binding motif is required for loading mcm proteins onto chromatin. Proc. Natl. Acad. Sci. USA 96:441-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.White, M. F., and S. D. Bell. 2002. Holding it together: chromatin in the Archaea. Trends Genet. 18:621-626. [DOI] [PubMed] [Google Scholar]
- 118.Woese, C. R., and G. E. Fox. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. USA 74:5088-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xie, Y., and J. N. Reeve. 2004. Transcription by an archaeal RNA polymerase is slowed but not blocked by an archaeal nucleosome. J. Bacteriol. 186:3492-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yu, X., M. S. VanLoock, A. Poplawski, Z. Kelman, T. Xiang, B. K. Tye, and E. H. Egelman. 2002. The Methanobacterium thermoautotrophicum MCM protein can form heptameric rings. EMBO Rep. 3:792-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang, R., and C. T. Zhang. 2003. Multiple replication origins of the archaeon Halobacterium species NRC-1. Biochem. Biophys. Res. Commun. 302:728-734. [DOI] [PubMed] [Google Scholar]
- 122.Zhuang, Z., B. L. Yoder, P. M. Burgers, and S. J. Benkovic. 2006. The structure of a ring-opened proliferating cell nuclear antigen-replication factor C complex revealed by fluorescence energy transfer. Proc. Natl. Acad. Sci. USA 103:2546-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]