Abstract
A hallmark of positive selection (adaptive evolution) in protein-coding regions is a dN/dS ratio >1, where dN is the number of nonsynonymous substitutions/nonsynonymous sites and dS is the number of synonymous substitutions/synonymous sites. Zonadhesin is a male reproductive protein localized on the sperm head, comprising many domains known to be involved in cell-cell interaction or cell adhesion. Previous studies have shown that VWD domains (homologous to the D domains of the von Willebrand factor) are involved directly in binding to the female zona pellucida (ZP) in a species-specific manner. In this study, we sequenced 47 coding exons in 12 primate species and, by using maximum-likelihood methods to determine sites under positive selection, we show that VWD2, membrane/A5 antigen mu receptor, and mucin-like domains in zonadhesin are rapidly evolving and, thus, may be involved in binding to the ZP in a species-specific manner in primates. In addition, polymorphism data from 48 human individuals revealed significant polymorphism-to-divergence heterogeneity and a significant departure from equilibrium-neutral expectations in the frequency spectrum, suggesting balancing selection and positive selection occurring in zonadhesin (ZAN) within human populations. Finally, we observe adaptive evolution in haplotypes segregating for a frameshift mutation that was previously thought to indicate that ZAN was a potential pseudogene.
Many steps of the mammalian fertilization cascade exhibit species specificity.1 For example, the sperm first bind in a species-specific manner with the zona pellucida (ZP), the extracellular coat of the egg.2 In general, when eggs and sperm come from heterospecific species, the initial binding to the ZP does not occur3 as efficiently as in homospecific mixtures. Once bound, the acrosomal reaction is triggered, involving exocytosis of the acrosome, creating an acrosomal shroud. Next, the sperm penetrates the egg ZP, enters the perivitelline space between the ZP and the plasma membrane, and then fuses with the plasma membrane.
Several male reproductive proteins are thought to be located in the acrosome and sperm head, and several dozen have been proposed to be involved in the species-specific binding to eggs. Some of these molecules have been shown to possess high levels of divergence between closely related species.4–6 For example, sperm-associated proteins, protamines 1 and 2, may influence sperm morphology and thereby increase competitiveness to fertilize eggs. Additionally, the female reproductive genes ZP2 and ZP3 are shown to be subject to adaptive evolution,7 and particular sites in these genes have been identified both to be under positive selection and to be involved in species-specific egg-sperm interaction. Several models have been proposed to explain the elevated levels of evolution in reproductive proteins.8,9 Many of these models have overlapping predictions and are considered a subset of sexual-selection theory. For example, sperm competition exists because of sperm competing to fertilize the egg first, and proteins in spermatozoa may be evolving to increase this ability. Sexual conflict arises when sperm cells are too abundant and the egg experiences fitness loss.10,11 Sexual selection, in which one egg preferentially binds with a sperm that carries a particular allele,12 has also been proposed to drive the rapid evolution of reproductive genes. Finally, cryptic female choice13 may also select for sperm that are efficient at fertilization, so that the female’s sons will produce sperm that are also efficient fertilizers.
Despite the abundance of available literature about sperm proteins, which proteins bind to the ZP of the egg remains elusive and controversial. ZP proteins have been shown to be subject to positive selection, suggesting that new candidates could be identified through correlated changes in the sperm receptors. The male receptors may evolve rapidly as well, to maintain molecular interaction with the female proteins. In this study, we characterize primate zonadhesin (ZAN [MIM 602372]), a large multiple-domain protein that resides on the anterior part of the sperm head and acts as a receptor to the ZP matrix of the egg.14 The human ZAN protein exists as six splice variants among exons 41–43, ranging in length from 2,600 to 2,724 codons. Many of these variants are derived from testis EST data. Previously characterized as part of this protein are membrane/A5 antigen mu receptor (MAM) domains, a mucin-like domain, and von Willebrand factor D (VWD) domains. Each of these domains is described as occurring in a variety of cell-cell interactions.15 For example, the large carbohydrate-rich domain extends beyond most other cell-surface glycoproteins, indicating its role in inhibiting or promoting cell adhesion.16 The MAM domains are also involved in cell-adhesion interactions and occur in a large number of membrane proteins, such as meprins. VWD domains, which are also adhesive proteins, have been shown to be the most functionally important domains, in terms of ZP binding.17 The ZAN molecule is thought to undergo posttranslational modification through proteolysis18 and glycosylation at the many potential sites of N- and O-glycosylation. This produces several forms of ZAN with differing capabilities of ZP interactions. In pig, for example, these forms exist not in testis but after the sperm leave the testis, consisting of complexes of fragments of the VWD domains.
In addition to the species-specific–binding VWD and cell-adhesion domains contained therein, previous analyses have hinted at positive selection promoting the divergence of ZAN. In alignments between pig, mouse, human, and rabbit, 2% of the sites were determined to have dN/dS ratios of 3.6,5 where dN is the number of nonsynonymous substitutions/nonsynonymous sites and dS is the number of synonymous substitutions/synonymous sites. However, the maximum-likelihood methods used are not robust with the small sample used in this study reported elsewhere,19 and there may be different regions under selection in such diverse mammals. Therefore, we used several different statistics to characterize the evolution of ZAN from a large primate data set. Lastly, a frameshift mutation in the human zonadhesin gene (ZAN) was detected at moderately high frequency, which was interpreted as potential evidence of the gene being a pseudogene.20 Here, we present statistical tests, using both polymorphism and divergence data that indicate that ZAN has been subjected to adaptive evolution in primates and, in particular, that haplotypes carrying the frameshift mutation have recently been under directional selection in the human lineage.
Material and Methods
Polymorphism Survey
A total of 48 human individual genomic DNA samples, 25 of African American descent and 23 of European descent, were obtained from the Coriell Institute for Medical Research. Primers for all 47 coding-sequence (CDS) exons (UTR exons excluded at the 5′ end of ZAN) were designed using Primer3.21 The universal primer M13 tails were added to each genome primer sequence. The PCR for each exon consisted of ∼15 ng of template DNA and a 1-min extension time at 72°C, and annealing-temperature gradients were performed to determine the optimal temperature at which to obtain the cleanest PCR product. Each product was diluted five times with water and was sequenced directly with the use of ABI Big Dye Terminator Reaction mix on a 3100 machine, with the M13 universal primers.
Divergence Survey
The following species were obtained from the Coriell Institute for Medical Research in this study: Pan troglodytes (chimpanzee), Pan paniscus (pygmy chimpanzee), Homo sapiens (human), Pongo pygmaeus (orangutan), Erythrocebus patas (red guenon), Macaca mulatta (rhesus monkey), Macaca nigra (celebes crested macaque), Macaca nemestrina (pig-tailed macaque), Lagothrix lagotricha (common woolly monkey), Ateles geoffroyi (black-handed spider monkey), Saguinus labiatus (red-chested mustached tamarin), and Callithrux jacchus (white-tufted-ear marmoset). To examine divergence among species, we performed PCR, as detailed above, of each of the 47 exons of ZAN from each species. Sequences were deposited in GenBank, under accession numbers DQ383284–DQ383294.
Polymorphism Analyses
Raw chromatograms were used in the alignment process using Phred and Phrap.22,23 A ZAN reference sequence (complete CDS [GenBank accession number AY046055]) was used in the analysis to facilitate exon/PCR alignments and haplotype reconstruction. The alignments were viewed and edited with Consed,24 and SNPs were identified using Polyphred25 and were manually verified by eye. The widely used Bayesian methods implemented in PHASE26 were used to infer haplotypes. To examine departures from neutrality, we calculated Tajima’s D,27 Fu and Li’s D,28 the McDonald and Krietman (MK) test,29 and Fay and Wu’s H,30 using DNAsp.31 Chimpanzee ZAN was used as the outgroup species for the MK and Fay and Wu tests. Significance for Fay and Wu’s H statistic was determined by coalescent simulations using a recombination parameter, R, estimated from the data by use of Hudson’s method.32 To look for specific regions in ZAN that might be significant for Fay and Wu’s H, we calculated H for each exon (PCR product), using Fay’s program, Hstat, locally on a Unix platform. The tests based on the frequency spectrum (Fay and Wu’s H, Tajima's D, and Fu and Li’s D) detect levels of variation that are inconsistent with the expectation of a neutral equilibrium model, but care must be taken to account for demographic effects.33 The MK29 test compares levels of synonymous and nonsynonymous variation within and between species, and departures from neutrality are considered robust to demographic effects.33
We ran DNA slider, using chimpanzee as the outgroup, to test for significance of heterogeneity of the ratio of polymorphic sites to fixed differences in ZAN.34,35 For this study, a window size of 10 variable sites was used. Against the resulting heterogeneity, four tests were conducted in DNA slider as follows. The runs statistic (KR) summarizes the number of runs of contiguous polymorphisms and contiguous fixed differences. The greater the heterogeneity, the fewer runs will be made. The Kolmogorov-Smirnov statistic (DKS) gives the absolute difference between the observed number of polymorphisms and the expected cumulative number. This test is most powerful at detecting a division within the gene, with one region having high polymorphism and the other low. The maximum sliding G statistic (Gmax) computes a 2×2 table comparing the frequencies of polymorphisms and fixed differences within and outside a window of size n, where n is determined to be the smallest window size with an expected number of polymorphisms greater than or equal to five, a number based on G tests. This computation is repeated for every possible window size from Nmin to Nmax, and the largest G statistic is the maximum sliding G statistic. This statistic is most powerful at detecting one or two peaks of elevated polymorphism. The fourth test, the mean sliding G statistic (Gmean) is similar to the one described above, except that the mean of G is computed. This test is best for finding wider peaks of elevated polymorphism, in comparison with the Gmax, which is better for narrower peaks.
Divergence Analyses
A series of likelihood-ratio tests were performed to determine the mode of evolution of ZAN, with the use of codeml from the software package Phylogenetic Analysis by Maximum Likelihood (PAML).36 These likelihood models allow specific sites to be tested for positive selection and are more sensitive than the models that average dN/dS values over the entire sequence.37 Alignments of the 12 species of primates were done with Phrap and were viewed with Consed. Protein sequences were obtained using Bioedit.38 To run PAML, we first inferred a phylogeny of all the primate species in this study with exons spliced together, using the maximum-likelihood method, as implemented in DNAml, from the software package Phylogeny Inference Package (PHYLIP).39 The tree was consistent with the known phylogenetic relationships of these species. For all PAML analyses, sites with missing data were ignored. The first test compared a model assuming a single dN/dS ratio for all lineages (branches) with a free-ratio model in which each lineage has a different dN/dS. This latter model determines whether certain branches in the phylogeny experience different selective pressures and, therefore, have different dN/dS values. The remaining tests all allowed dN/dS to vary among sites and are as follows: M0 allows one value of dN/dS at all sites; M1a (nearly neutral model) allows two categories—1 and a value estimated between 0 and 1; M2a (positive-selection model) allows three categories—1, a value estimated between 0 and 1, and >1; and M3 (discrete model) allows three categories. M7 and M8 assume a beta distribution limited to the interval between 0 and 1 and are identical, except that M8 allows an additional category, where dN/dS can be >1. The M7 test, in comparison with the M8 test, is considered to be more effective in determining potential sites of positive selection. For our analysis, we compared M0 and M3, M1a and M2a, and M7 and M8. Finally, we also compared M8 with the model M8a, where the additional class was set at 1.5 For each comparison, the significance was evaluated by taking the negative of twice the difference in the log likelihood obtained from the two nested models and comparing it with a χ2 distribution. Degrees of freedom were obtained from the difference of the numbers of parameters in each model. The five splice variants of ZAN were analyzed separately, with variant 3 omitted because there was a single base-pair insertion present in all the primate species except human in exon 42, making the remaining codons unalignable. The gorilla ZAN contained several stop codons and was also omitted from the PAML analysis.
Exon/Intron Phylogenies
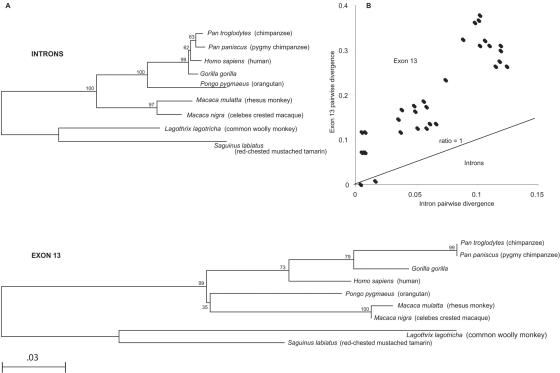
Introns 7, 8, 15, 16, 18, and 20—totaling 980 bp—and all 1,515 bp of exon 13 were used to reconstruct gene trees for nine primate species. The introns were chosen since the sequence was available for the same nine primate species mentioned above. Exon 13 was chosen because it contained three potential sites predicted to be subject to positive selection (table 1). Genetic distances were calculated using the Tamura-Nei two-parameter model, and the trees were generated using the neighbor-joining method employed by Molecular Evolutionary Genetics Analysis (MEGA) software.40
Table 1. .
Overview of Location of Sites of Positive Selection Determined by PAML
| Location (bp/codon) |
Exon | Probability That dN/dS>1 | dN/dS ± SE |
| 774/258 | 7 | .70 | 1.58 ±. 71 |
| 1,029/343 | 9 | .70 | 1.58 ±. 71 |
| 1,206/402 | 10 | .92 | 1.93 ±. 63 |
| 1,230/410 | 10 | .91 | 1.91 ± .64 |
| 2,667/889 | 13 | .79 | 1.72 ± .73 |
| 2,844/948 | 13 | .73 | 1.63 ± .72 |
| 2,928/976 | 13 | .91 | 1.90 ± .64 |
| 4,932/1,644 | 25 | .83 | 1.78 ± .68 |
| 4,935/1,645 | 25 | .87 | 1.85 ± .66 |
| 5,094/1,698 | 26 | .85 | 1.81 ± .69 |
| 5,502/1,834 | 29 | .83 | 1.78 ± .68 |
| 5,739/1,913 | 30 | .75 | 1.66 ± .72 |
| 5,784/1,928 | 31 | .73 | 1.62 ± .72 |
| 6,153/2,051 | 32 | .80 | 1.73 ± .78 |
| 6,531/2,177 | 34 | .70 | 1.57 ± .71 |
| 6,546/2,182 | 34 | .72 | 1.61 ± .71 |
| 7,869/2,623 | 44 | .72 | 1.61 ± .71 |
| 8,124/2,708 | 46 | .71 | 1.60 ± .74 |
To determine whether exon 13 was significantly more divergent than introns, we calculated the average pairwise divergence between all pairwise comparisons for both exon 13 and the introns, using the Tamura-Nei two-parameter model. We determined SEs by bootstrap analysis, using MEGA3. For comparison, we plotted pairwise distance between all possible comparisons for exon 13 and plotted it against all pairwise distance between the introns. This analysis is analogous to a dN/dS plot, since the introns are in the same genomic location as exon 13 and, theoretically, should be equivalent to the neutral mutation rate in this region.
Results
We sequenced all 47 CDS exons of the male reproductive gene ZAN in 48 human individuals and 12 primate species. A total of 8,677 bp of exon were sequenced, and ∼13,869 bp of intron were sequenced per individual. The majority of primers designed were in introns, to span the entire exon. Numerous indels were detected in the intron alignment and were ignored in analyses.
ZAN Divergence
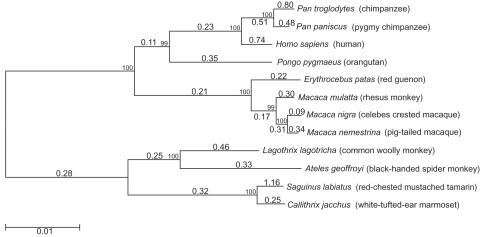
For the divergence studies, we sequenced 47 exons in ZAN for 12 primate species. We then analyzed the sequence data to detect variation in dN/dS ratios between sites, using PAML.36 The five splice variants of human ZAN showed significance for varying dN/dS ratios among branches (table 2 and fig. 1), with dN/dS ratios ranging in values from 0.17 to 1.16. The significant varying degree of dN/dS values in the primate lineages suggests different rates of evolution among lineages and is surprising, given the larger number of df (20) for this analysis.
Table 2. .
Results of PAML Model Comparisons of Six Different Variants of ZAN
| −2Δl Model Comparisonsb |
Parameter Estimatesunder M8 |
||||||||||
| Variant | N | No. of Codons |
Tree Lengtha |
dN/dS | Free Ratio vs. M0 |
M0 vs. M3 | M1 vs. M2 | M7 vs. M8 | ω (p1) | β (p0) | Positively Selected Sitesc |
| 1 | 12 | 2,724 | .69 | .30 | 41.7d | 135.4e | 4.4 | 7.5fe | 2.7 (.03) | .31, .86 (.96) | 402, 410, 934, 976, 1644, 1645, 1698, 1834, 1913, 1928, 2051 |
| 2 | 12 | 2,668 | .67 | .29 | 44.8e | 130.8e | 4.1 | 7.1f | 2.8 (.02) | .30, .83 (.97) | 402, 410, 934, 976, 1644, 1645, 1698, 1834, 1913, 1928, 2051 |
| 4 | 12 | 2,624 | .68 | .30 | 42.7e | 134.2e | 4.8 | 7.9f | 2.8 (.03) | .30, .81 (.96) | 402, 410, 889, 934, 976, 1644, 1645, 1698, 1834, 1913, 1928, 2051, 2621 |
| 5 | 12 | 2,600 | .68 | .30 | 45.8e | 129.7e | 5.2 | 8.7f | 2.3 (.05) | .43, 1.38 (.94) | 402, 410, 889, 934, 976, 1644, 1645, 1698, 1834, 1913, 1928, 2051 |
| 6 | 12 | 2,724 | .68 | .32 | 38.9d | 144.9e | 4.4 | 7.3f | 2.12 (.06) | .38, 1.20 (.93) | 258, 343, 402, 410, 889, 948, 976, 1644, 1645, 1698, 1834, 1913, 1928, 2051, 2177, 2182, 2623, 2708 |
In substitutions per codon.
−2Δl is the negative of twice the difference between the nested models.
Posterior probabilities for sites are as follows: bold font indicates >.90, underlined font indicates >.80, and regular font indicates >.70.
P<.01.
P<.001.
P<.05.
Figure 1. .
Neighbor-joining tree of all 47 exons of ZAN for 12 primate species used in this study. The larger-font numerals are dN/dS ratios for each of the lineages determined by PAML, determined from the free-ratio model. The smaller-font numbers located at branch nodes are bootstrap values.
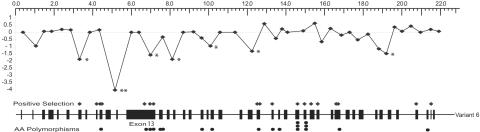
Since ZAN has several alternatively spliced forms, we analyzed variation in the dN/dS ratios between sites for each of the six spliced forms. These methods allow for the detection of specific sites that have been subject to positive selection.33,41 Several sites of positive selection were identified in variants 1–5, with dN/dS averaging 2.54 across selected sites from M8 (table 2). Sites of positive selection in this study were derived from variant 6 because it contains the most exons that can be aligned between species. Variant 6 contains 18 potential sites under positive selection dispersed throughout the gene (table 2 and fig. 2). Interestingly, the sites predicted to be subjected to positive selection between primates fall in regions similar to the sites of amino acid polymorphisms within humans (fig. 2). Two sites are in exon 25, located within VWD2, with posterior probabilities of 0.83 and 0.87 (tables 1 and 2). Exons 31 and 32, making up part of VWD3, each have one potential site, with probabilities of 0.73 and 0.80, respectively. Exon 10, encoding the beginning of the MAM3 domain, shows two sites of positive selection with probabilities >0.90. Also coding for MAM domains are exons 7 and 9, each containing one site, with probabilities of 0.70.
Figure 2. .
Indication of points of significance of Fay and Wu's H in ZAN plotted against the exons of variant 6 (black boxes). The diamonds indicate the values of H per exon and sites of positive selection in the exons determined by PAML. Two asterisks (**) indicate P<.01, and one asterisk (*) indicates P<.05.
Exon 13—which, at 1,515 bp, is the largest exon in ZAN—displays three potential sites under positive selection, one of which has a probability of 0.91. The other two sites have probabilities of 0.79 and 0.73. No known domains have been inferred in the region of exon 13, which falls between the MAM and the VWD domains. Functional studies that would identify the roles of key peptides within ZAN have not been performed, and no known domains exist within exon 13 of this protein. Elevated positive selection in exon 13 possibly indicates that the region immediately upstream of the VWD domains may play some role in the recognition or binding process with the ZP.
The exon 13 tree was determined to be 4.4 times longer than that of the intron tree (fig. 3A). The average pairwise divergence between the introns and exon 13 showed, by bootstrap analysis, that exon 13 was significantly more divergent than the introns (P<.05) (fig. 3B). This was not observed for any other exons. Since intron divergence between human and chimpanzee in ZAN was as expected for neutral evolution (∼1%42), this is consistent with adaptive evolution in exon 13. Additionally, the average dN/dS ratio for exon 13 was 2.1. However, we note that increased rate of exon compared with intron divergence is not a test of adaptive evolution but rather is a demonstration that the exon is rapidly evolving. For example, introns could contain conversed regulatory sequences. However, in the case of ZAN, the rate of intron divergence is similar to dS (another proxy for neutral rate of evolution) and to other estimates for the neutral rate in these primates.
Figure 3. .
A, Neighbor-joining phylogenetic trees of exon 13 and introns 7, 8, 15, 16, 18, and 20, generated using the Tamura-Nei distance method. Nine of the 12 primate species were used in this tree, since the exon 13 sequences of 3 species were not obtained. B, Plot of all pairwise distance comparisons of exon 13 and the introns. Each point represents the comparison of two species, and the diagonal represents the point at which each of the comparisons are equal to 1.
ZAN Polymorphism
A total of 19 nonsynonymous polymorphisms (amino acid changes) were detected using DNAsp, of which 5 were radical changes involving charge and hydrophobicity. Interestingly, the sites of nonsynonymous polymorphisms were in the same regions of sites predicted to be subject to adaptive evolution between species. Neither Tajima’s D,27 Fu and Li’s D,28 nor the MK test29 was significant for any of the overall African American or European American populations (table 3). Fay and Wu's H30 was significant for the overall population (P<.05) and for the African American population (P<.001) but not for the European American population (table 3). The H test with the African American sample remained significant when compared with coalescent simulations, including with an admixture model (P<.001). We determined Fay and Wu’s H statistic for each exon (fig. 2). Seven sites of significance were observed, and these are distributed evenly throughout the gene. This pattern is roughly similar to that for sites of positive selection determined by PAML. The most significant point of Fay and Wu’s H is located at 5,127 bp, with a value of −3.9 (P<.01), and the weakest is located at 10,108 bp (H statistic −0.93; P<.05). Because of hitchhiking, the actual target of positive selection and the point of highest significance for H may not necessarily coincide exactly. Where recombination is normal or high, the effect of hitchhiking can be limited to a region of <1 kb.30
Table 3. .
Population Statistics of Human Populations in ZAN
| Population | No. of Alleles |
π | Tajima’s D | Fu and Li's D |
Fay and Wu's H |
| African American | 50 | .006 | −.769 | −.91 | −13.98a |
| European | 46 | .00056 | .753 | −.19 | −1.49 |
| Combined | 96 | .00051 | −.627 | −1.58 | −13.17b |
P<.001.
P<.01.
A frameshift in exon 30,20 which caused the protein to truncate 20 aa downstream of the mutation, occurred in the human population at a frequency of 76%. Nineteen individuals were heterozygous for this allele, and 27 were homozygous. This frameshift was not present in any of the other primate species in this study. On the basis of the high presence of the frameshift mutation, we calculated Fay and Wu’s H30 on the haplotypes with and without the frameshift mutation (table 4). In all cases, those haplotypes with the frameshift mutation had a significantly negative Fay and Wu’s H, whereas the H for those without the frameshift did not depart from equilibrium-neutral expectations. This is particularly interesting since the European American population was not significant with all haplotypes but was significant when only those haplotypes with the frameshift mutation were considered. This suggests that there has been adaptive evolution to increase the frequency of the frameshift mutation. Interestingly, there are nine substitutions separating the clade containing the frameshift mutation from those without it, one of which is an amino acid–altering substitution.
Table 4. .
Fay and Wu’s H Based on Presence of Frameshift
| Sample and Presence of Frameshifta |
Sample Size (N) |
θb | H | Pc |
| Combined: | ||||
| Without | 23 | 7.8 | −5.3 | .07 |
| With | 73 | 8.9 | −6.3 | <.00001 |
| European American: | ||||
| Without | 16 | 5.5 | −7.3 | .46 |
| With | 27 | 6.8 | −7.9 | .01 |
| African American: | ||||
| Without | 6 | 13.2 | −3.2 | .17 |
| With | 44 | 11.5 | −16.2 | <.00001 |
Population and haplotypes analyzed with or without the frameshift mutation.
Per gene.
Probability of departure from equilibrium-neutral expectation.
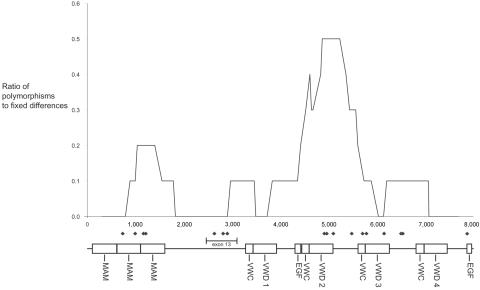
We conducted four tests in DNA slider and found them all to be significant for heterogeneity present in ZAN. After Bonferroni correction,43 all but the K runs test remained significant (table 5). Natural selection can cause greater heterogeneity in the polymorphism-to-divergence ratio than expected under the neutral model. Near a balanced polymorphism, the area will have a greater number of neutral polymorphisms and fewer fixed differences and, thus, greater polymorphism-to-divergence ratios,35 similar to the pattern seen in figure 4 at ∼1,500 and ∼5,000 bp. It is clear that the KS statistic, best at seeing a division between two genes—one low and one high in polymorphism—does not apply to ZAN. The opposite can be said of an adaptive substitution (positive selection)—there will be fewer polymorphisms and more fixed differences than expected under a neutral model. Evidence of a selective sweep is present between 2,500 and 3,000 bp spanning exon 13. Three sites of positive selection are observed, indicating fixed amino acid substitutions between species and no polymorphisms within species. This suggests that rapid fixation of a positively selected point removed linked neutral variation. Nonetheless, the results of DNA slider indicate that there have been differing evolutionary forces and constraints across ZAN, creating significant heterogeneity.44
Table 5. .
Levels of Significance of the Four Tests Run against ZAN Polymorphism-to-Divergence Heterogeneity for Eight Different Recombination Parameters[Note]
|
P |
||||
| Recombination Parameter |
Gmax ⩽17.96 | Runs ⩽17 | KS ⩾.029 | Gmean ⩾6.53 |
| 2.00 | .007 | .023 | .006 | .001 |
| 4.00 | .003 | .017 | .007 | .001 |
| 6.00 | .004 | .022 | .007 | .002 |
| 8.00 | .005 | .021 | .012 | .002 |
| 10.80 | .009 | .023 | .007 | .003 |
| 12.00 | .001 | .019 | .007 | .001 |
| 14.00 | .003 | .015 | .009 | .002 |
| 16.00 | .01 | .022 | .007 | .004 |
Note.— The problem of multiple comparisons was corrected by applying the Bonferroni correction, then comparing it with the highest P value for each of the recombination parameters. All but the runs statistic remain significant.
Figure 4. .
Ratio of polymorphism to fixed differences in ZAN exons 1–47, with chimpanzee used as the outgroup. Also indicated are the domains found by SMART. The diamonds show sites of positive selection found by PAML for variant 6.
Discussion
ZAN demonstrates great variation among species in domain arrangements and numbers.15,17 For example, mouse and pig ZAN genes exhibit 25 and 4 VWD domains, respectively. Functionally, these domains are proteolysed, glycosylated, and assembled into different structures that may mediate species-specific adhesion to ZP. Pig ZAN fragments p45 and p105 (VWD1 and VWD2–D3 domains, respectively) were observed to bind in a species-specific manner to the ZP of the pig but not to the ZP of other species.16 Two goals of this study were to determine specific regions in primate ZAN that similarly may be subject to positive selection, to maintain interaction with the female adaptively evolving ZP,7 and to investigate levels and patterns of human polymorphism.
ZAN is potentially subject to several forces of natural selection. Balancing selection is consistent with the pattern of high peaks34 evident at ∼5,000 bp (fig. 4). In addition, predicted sites of positive selection occur in the same region. This pattern of balancing selection and positive selection in the same gene has been documented in the major histocompatibility complex, in which functional diversity is maintained in the peptide-binding regions.45,46 This could indicate that regions (domains or exons) in ZAN have a heterozygotic advantage and that positive selection plays a role in the evolution of ZAN-binding properties. Most of ZAN has a low polymorphism-to-divergence ratio, indicating that a large fraction of this gene is under purifying selection, presumably to maintain function of certain regions. VWD2, a domain involved in ZP binding in other species, contains two potential sites of positive selection. In addition, this domain contains the conserved motif CGLCG, which is important for oligomerization and proper function of von Willebrand domains.18 VWD3 contains two sites of positive selection contained within exons 31 and 32. However, the frameshift in the human population in exon 30 truncates the protein within exon 30. This frameshift occurs at a high frequency (76%), with 27 individuals homozygous for this condition. There have been several genes proposed to be involved in the binding of the ZP—for example, sp5647 and PKDREJ.48 Perhaps the signal pathway to the acrosomal reaction is redundant, with several genes able to bind, which will ensure that the process will not fail; in this case, knocking out one gene will not affect the process.49 Therefore, ZAN could be part of the gene family involved in the binding process to the ZP and subject to partial functional redundancy. The truncated protein may indicate one particular variant of ZAN contributing only domains upstream of exon 30 to the ZP-binding process. Or, the variation of posttranslational modifications of ZAN mRNA between species would also lend to certain domains contributing to the ZP-binding process, as suggested by the fact that no truncated proteins were found among the other primate species in this study.
In contrast with the interpretation of the frameshift mutation causing ZAN to be a pseudogene, calculating Fay and Wu’s H on the haplotypes with or without the frameshift mutation demonstrated that only those with the frameshift mutation had a significantly negative value of H (table 4). Although applying Fay and Wu’s H statistic to a subset of the data could result in false positives, this form of subdividing the data for explorative analysis can yield interesting insights into gene evolution.50 There are two possible explanations for significantly negative H values for the alleles with the frameshift mutation. First, it may be that there has been positive selection for the frameshift mutation or the linked amino acid–altering polymorphism. Interestingly, the linked amino acid polymorphism is in a region with several sites predicted to have been subjected to positive selection between primates. Alternatively, reducing the number of samples for the analysis of Fay and Wu’s H could have reduced the power of the test. Arguing against this possibility is the observation that only the European American population was significant when the sample size was reduced to include only the haplotypes with the frameshift mutation. From these observations, we conclude that there has been positive selection to increase the frequency of the frameshift mutation or of one of the linked polymorphisms.
It is extremely interesting to observe positive selection increasing the frequency of a frameshift mutation. Frameshifts are typically considered to be deleterious, resulting in the gene becoming a nonfunctional pseudogene.20 For example, a large multigene family encodes olfactory receptors, with some loci containing alleles segregating as potential pseudogenes on the basis of the presence of frameshift mutations.51 Our results indicate that the mere presence of a frameshift mutation may not be sufficient to indicate a pseudogene. Consistent with this observation, several rapidly evolving genes do show alterations of the stop codon, which adds additional amino acid to the protein. There have been other examples of selection acting on frameshift mutations in humans,52 including the caspase-12 gene, in which a loss of function has been hypothesized to confer sepsis resistance.53 Obviously, functional analyses will be necessary to determine if the frameshift mutations encode proteins with perhaps novel functions or if they have become nonfunctional.
Exon 10 was shown to have two sites with probabilities >0.90 for positive selection, in addition to sites in exons 7 and 9 with probabilities of 0.70 (table 1). These exons contain domains homologous to the MAM domains in previously characterized ZAN proteins. These domains, however, are thought to have no binding function in ZAN because some studies (in rabbit54) show that these regions are removed before sperm reach the female reproductive tract. However, posttranslational modification of the precursor mRNA may depend on the species.17
MAM domains are suggested to have roles in some cell-surface molecules and are functionally critical in these cell-cell interactions.55 Therefore, whereas the function of MAM domains in ZAN is unclear, positive selection observed in these regions among primates in this study may indicate some species-specific binding of ZAN MAM domains in the reproductive process or roles in binding to the ZP. Moreover, it has been proposed that MAM domains in other primate species are positively selected,56 and the authors suggest that the variability of posttranslational proteins is enhanced by positive selection, given the taxon differences in the predicted posttranslational modifications.
Exon 13 contains three sites of positive selection, with probabilities of 0.79, 0.73, and 0.91. These points are not within any domain structure detected by SMART (fig. 4); however, exon 13 contains 29 small amino acid repeats of motifs PTEK (P/T/S/L)(T/S)(V/I) and PTE (E/V)(P/T)(T/V), which give it a mucin-like domain structure similar to the mucin repeats in mouse, PTE (E/V)(P/T)(T/V), and the 53 repeats of pig, consisting of PTE (K/R)(P/T)T(V/I).16 Repetitive motifs rich in proline and threonine are characteristics of mucin domains, and the structural properties of such domains inhibit or promote cell adhesion. The ZAN mucin in primates could function similarly, perhaps by inhibiting inappropriate spermatozoa or by promoting adhesion of the correct sperm in the oviductal isthmus.57 The tree length for exon 13 was 4.4 times greater than that for the introns (fig. 3A). The morpheus gene family shows similar topology in which the branch lengths are longer in exon 2 than in the introns and exon 2 was subjected to extreme positive selection.58 In addition, the difference of the overall mean of pairwise comparisons between exon 13 and the introns was significant (0.203±0.029 and 0.065±.006, respectively), indicating that exon 13 is evolving faster than the neutral rate of introns. Plotting pairwise distance comparisons of exon 13 and the introns shows that the majority of points lie above the diagonal, indicating that the exon 13/intron ratio is >1 (fig. 3B). This further supports that exon 13 is adaptively evolving and could be a primary binding region of ZAN to the ZP of the egg or a part of another species-specific role in the reproductive process.
There is indication of positive selection dispersed throughout ZAN from two independent tests—the maximum-likelihood test employed by PAML and the Fay and Wu H statistic calculated overall or for each exon (PCR product). Regardless of where the targets of selection may occur, it is clear that ZAN contains significant incidences of high frequency–derived variants from hitchhiking, a strong indication of positive selection. It is our interpretation that each of the potential binding domains—VWD2 and MAM—and the mucin-like domain within exon 13 fall within the recombination effect of hitchhiking resulting from positive selection.
The analysis of the human polymorphism data demonstrates two striking features. First, the signal of recent selection, as indicated by the Fay and Wu H test, is significant only in the African population. This is in contrast to several recent genomewide scans for adaptive evolution59–62 in which the signal of selection is more prevalent in non-African populations. A second observation was apparent selection for a haplotype with a frameshift mutation, which was observed in both the European and African populations. The selective pressure driving the frameshift mutation to high frequency remains unknown but suggests that the gene is still functional.
Results of this study lend support to models of sperm competition and sexual conflict driving the divergence of reproductive proteins8 and male-female interactions under which positive selection is expected in male reproductive proteins. Sexual conflict is consistent with the significant variance of dN/dS ratios among primate lineages (fig. 1), given that the primate species in this study exhibit a wide variety of mating systems, including monogamy, polygyny, multimale-multifemale, and dispersed. It follows that, with these different mating systems, there will be varying degrees of selective pressures and, therefore, differing rates of molecular evolution on reproductive genes. We were, however, unable to find any correlation between levels of female promiscuity and the corresponding lineage dN/dS ratios (fig. 1), in which it is expected that dN/dS ratios will increase as the number of partners per ovulatory period increase.6 This is due, in part, to the relatively unknown number of mating partners of some of the species in our study and to our small sample size.
Primates may be expected to exhibit lower incidences of positive selection than do broadcast spawning invertebrates for whom closely related species can be in the same vicinity during mating. Primate mating systems involve an additional level of prezygotic isolation, which reduces the need for molecular barriers to fertilization. Selection favoring strong species barriers, as opposed to the prezygotic barriers of the mammalian mating systems, may lower the probabilities of sites under positive selection. Despite this, sites of positive selection were observed in the present study for primate ZAN.
In conclusion, ZAN is a multiple-domain protein consisting of three MAM domains, five VWD domains, and one large mucin-like domain. Evidence of positive selection from multiple approaches occurs in these domains, suggesting species-specific binding to the ZP or other species-specific processes in the reproductive cascade. The changes in the MAM domains observed in this study, along with species-dependent posttranslational modifications, potentially change the binding properties of ZAN. We found two sites of positive selection in VWD2, a primary domain involved in binding to the ZP. In addition, the large mucin-like domain that codes for cell-surface glycoproteins was found to have three sites of positive selection and extreme elevated levels of divergence compared with the neutral introns. In human populations, a detrimental frameshift occurs at a frequency of 76% in exon 30. Whereas this may indicate a pseudogene, we interpret that, because of the increase in the frameshift haplotype frequency, it may be positively selected and is potentially a transcribed gene in the spermatozoa. Possible scenarios are unidentified variants in ZAN, which splices among exon 30, resulting in translation of prior or succeeding exons. Finally, this frameshift is potentially linked to amino acid–altering polymorphisms and to sites of positive selection between primates in the region that may be under selection.
Acknowledgments
We thank members of the Swanson lab, Josh Akey, and two anonymous reviewers, for advice and discussion. Support was provided by National Institutes of Health grants HD42563 and HD42454 and by National Science Foundation grants DEB-0213171 and DEB-0410112.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for ZAN reference sequences [accession numbers DQ383284–DQ383294 and AY046055])
- Hstat, http://www.genetics.wustl.edu/jflab/htest.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ZAN) [PubMed]
- SMART, http://smart.embl-heidelberg.de/
References
- 1.Vacquier VD (1998) Evolution of gamete recognition proteins. Science 281:1995–1998 10.1126/science.281.5385.1995 [DOI] [PubMed] [Google Scholar]
- 2.Wassarman PM, Jovine L, Litscher ES (2001) A profile of fertilization in mammals. Nat Cell Biol 3:E59–E64 10.1038/35055178 [DOI] [PubMed] [Google Scholar]
- 3.Wassarman PM (1999) Mammalian fertilization: molecular aspects of gamete adhesion, exocytosis, and fusion. Cell 96:175–183 10.1016/S0092-8674(00)80558-9 [DOI] [PubMed] [Google Scholar]
- 4.Wyckoff GJ, Wang W, Wu CI (2000) Rapid evolution of male reproductive genes in the descent of man. Nature 403:304–309 10.1038/35002070 [DOI] [PubMed] [Google Scholar]
- 5.Swanson WJ, Nielsen R, Yang Q (2003) Pervasive adaptive evolution in mammalian fertilization proteins. Mol Biol Evol 20:18–20 [DOI] [PubMed] [Google Scholar]
- 6.Dorus S, Evans PD, Wyckoff GJ, Choi SS, Lahn BT (2004) Rate of molecular evolution of the seminal protein gene SEMG2 correlates with levels of female promiscuity. Nat Genet 36:1326–1329 10.1038/ng1471 [DOI] [PubMed] [Google Scholar]
- 7.Swanson WJ, Yang Z, Wolfner MF, Aquadro CF (2001) Positive Darwinian selection drives the evolution of several female reproductive proteins in mammals. Proc Natl Acad Sci USA 98:2509–2514 10.1073/pnas.051605998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swanson WJ, Vacquier VD (2002) Rapid evolution of reproductive proteins. Nat Rev Genet 3:137–144 10.1038/nrg733 [DOI] [PubMed] [Google Scholar]
- 9.McCartney MA, Lessios HA (2004) Adaptive evolution of sperm binding tracks egg incompatibility in neotropical sea urchins of the genus Echinometra. Mol Biol Evol 21:732–745 10.1093/molbev/msh071 [DOI] [PubMed] [Google Scholar]
- 10.Gavrilets S (2000) Rapid evolution of reproductive barriers driven by sexual conflict. Nature 403:886–889 10.1038/35002564 [DOI] [PubMed] [Google Scholar]
- 11.Frank SA (2000) Sperm competition and female avoidance of polyspermy mediated by sperm-egg biochemistry. Evol Ecol Res 2:613–625 [Google Scholar]
- 12.Palumbi SR (1999) All males are not created equal: fertility differences depend on gamete recognition polymorphisms in sea urchins. Proc Natl Acad Sci USA 96:12632–12637 10.1073/pnas.96.22.12632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eberhard WG (1996) Female control: sexual selection by cryptic female choice. Princeton University Press, Princeton, NJ [Google Scholar]
- 14.Lea IA, Sivashanmugam P, O’Rand MG (2001) Zonadhesin: characterization, localization, and zona pellucida binding. Biol Reprod 65:1691–2001 10.1095/biolreprod65.6.1691 [DOI] [PubMed] [Google Scholar]
- 15.Gao Z, Garbers DL (1998) Species diversity in the structure of zonadhesin, a sperm-specific membrane protein containing multiple cell adhesion molecule-like domains. J Biol Chem 273:3415–3421 10.1074/jbc.273.6.3415 [DOI] [PubMed] [Google Scholar]
- 16.Hardy DM, Garbers DL (1995) A sperm membrane protein that binds in a species-specific manner to the egg extracellular matrix is homologous to von Willebrand factor. J Biol Chem 270:26025–26028 10.1074/jbc.270.44.26025 [DOI] [PubMed] [Google Scholar]
- 17.Bi M, Hickox JR, Winfrey VP, Olson GE, Hardy DM (2003) Processing, localization and binding activity of zonadhesin suggest a function in sperm adhesin to the zona pellucida during exocytosis of the acrosome. Biochem J 375:477–488 10.1042/BJ20030753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hickox JR, Bi M, Hardy DM (2001) Heterogeneous processing and zona pellucida binding activity of pig zonadhesin. J Biol Chem 276:41502–41509 10.1074/jbc.M106795200 [DOI] [PubMed] [Google Scholar]
- 19.Anisimova M, Bielawski JP, Yang Z (2001) Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol Biol Evol 18:1585–1592 [DOI] [PubMed] [Google Scholar]
- 20.Hillier LW, Fulton RS, Fulton LA, Graves TA, Pepin KH, Wagner-McPherson C, Layman D, et al (2003) The DNA sequence of human chromosome 7. Nature 424:157–164 10.1038/nature01782 [DOI] [PubMed] [Google Scholar]
- 21.Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386 [DOI] [PubMed] [Google Scholar]
- 22.Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194 [PubMed] [Google Scholar]
- 23.Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8:175–185 [DOI] [PubMed] [Google Scholar]
- 24.Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202 [DOI] [PubMed] [Google Scholar]
- 25.Nickerson DA, Tobe VO, Taylor SL (1997) PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res 25:2745–2751 10.1093/nar/25.14.2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDonald JH, Kreitman M (1991) Adaptive protein evolution at the Adh locus in Drosophila. Nature 351:652–654 10.1038/351652a0 [DOI] [PubMed] [Google Scholar]
- 30.Fay JC, Wu CI (2000) Hitchhiking under positive Darwinian selection. Genetics 155:1405–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174–175 10.1093/bioinformatics/15.2.174 [DOI] [PubMed] [Google Scholar]
- 32.Hudson RR (1987) Estimating the recombination parameter of a finite population model without selection. Genet Res 50:245–250 [DOI] [PubMed] [Google Scholar]
- 33.Nielsen R (2005) Molecular signatures of natural selection. Annu Rev Genet 39:197–218 10.1146/annurev.genet.39.073003.112420 [DOI] [PubMed] [Google Scholar]
- 34.McDonald JH (1998) Improved tests for heterogeneity across a region of DNA sequence in the ratio of polymorphism to divergence. Mol Biol Evol 15:377–384 [DOI] [PubMed] [Google Scholar]
- 35.McDonald JH (1996) Detecting non-neutral heterogeneity across a region of DNA sequence in the ratio of polymorphism to divergence. Mol Biol Evol 13:253–260 [DOI] [PubMed] [Google Scholar]
- 36.Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13:555–556 [DOI] [PubMed] [Google Scholar]
- 37.Yang Z, Nielsen R (2002) Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol 19:908–917 [DOI] [PubMed] [Google Scholar]
- 38.Hall TA (1999) Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98NT. Nucleic Acids Symp Ser 41:95–98 [Google Scholar]
- 39.Felsenstein J (1989) PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164–166 [Google Scholar]
- 40.Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163 10.1093/bib/5.2.150 [DOI] [PubMed] [Google Scholar]
- 41.Yang Z, Bielawski JP (2000) Statistical methods for detecting molecular adaptation. Trends Ecol Evol 15:496–503 10.1016/S0169-5347(00)01994-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mikkelsen TS, Hillier LW, Eichler EE, Zody MC, Jaffe DB, Yang SP, Enard W, et al (2005) Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437:69–87 10.1038/nature04072 [DOI] [PubMed] [Google Scholar]
- 43.Bonferroni CE (1936) Teoria statistica delle classi e calcolo delle probabilità. Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze 8:3–62 [Google Scholar]
- 44.Hasselmann M, Beye M (2004) Signatures of selection among sex-determining alleles of the honey bee. Proc Natl Acad Sci USA 101:4888–4893 10.1073/pnas.0307147101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garrigan D, Hedrick PW (2001) Class I MHC polymorphism and evolution in endangered California Chinook and other Pacific salmon. Immunogenetics 53:483–489 10.1007/s002510100352 [DOI] [PubMed] [Google Scholar]
- 46.Hughes AL, Nei M (1988) Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335:167–170 10.1038/335167a0 [DOI] [PubMed] [Google Scholar]
- 47.Bookbinder LH, Cheng A, Bleil JD (1995) Tissue- and species-specific expression of sp56, a mouse sperm fertilization protein. Science 269:86–89 [DOI] [PubMed] [Google Scholar]
- 48.Hughes J, Ward CJ, Aspinwall R, Butler R, Harris PC (1999) Identification of a human homologue of the sea urchin receptor for egg jelly: a polycystic kidney disease-like protein. Hum Mol Genet 8:543–549 10.1093/hmg/8.3.543 [DOI] [PubMed] [Google Scholar]
- 49.Snell WJ, White JM (1996) The molecules of mammalian fertilization. Cell 85:629–637 10.1016/S0092-8674(00)81230-1 [DOI] [PubMed] [Google Scholar]
- 50.Wang E, Ding YC, Flodman P, Kidd JR, Kidd KK, Grady DL, Ryder OA, Spence MA, Swanson JM, Moyzis RK (2004) The genetic architecture of selection at the human dopamine receptor D4 (DRD4) gene locus. Am J Hum Genet 74:931–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Menashe I, Man O, Lancet D, Gilad Y (2003) Different noses for different people. Nat Genet 34:143–144 10.1038/ng1160 [DOI] [PubMed] [Google Scholar]
- 52.Wang X, Grus WE, Zhang J (2006) Gene losses during human origins. PLoS Biol 4:e52 10.1371/journal.pbio.0040052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xue Y, Daly A, Yngvadottir B, Liu M, Coop G, Kim Y, Sabeti P, Chen Y, Stalker J, Huckle E, Burton J, Leonard S, Rogers J, Tyler-Smith C (2006) Spread of an inactive form of caspase-12 in humans is due to recent positive selection. Am J Hum Genet 78:659–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lea IA, Sivashanmugam P, O’Rand MG (2001) Zonadhesin: characterization, localization, and zona pellucida binding. Biol Reprod 65:1691–1700 10.1095/biolreprod65.6.1691 [DOI] [PubMed] [Google Scholar]
- 55.Takagi S, Hirata T, Agata K, Mochii M, Eguchi G, Fujisawa H (1991) The A5 antigen, a candidate for the neuronal recognition molecule, has homologies to complement components and coagulation factors. Neuron 7:295–307 10.1016/0896-6273(91)90268-5 [DOI] [PubMed] [Google Scholar]
- 56.Herlyn H, Zischler H (2005) Identification of a positively evolving putative binding region with increased variability in posttranslational motifs in zonadhesin MAM domain 2. Mol Phylogenet Evol 37:62–72 10.1016/j.ympev.2005.04.001 [DOI] [PubMed] [Google Scholar]
- 57.Gagneux P, Varki A (1999) Evolutionary considerations in relating oligosaccharide diversity to biological function. Glycobiology 9:747–755 10.1093/glycob/9.8.747 [DOI] [PubMed] [Google Scholar]
- 58.Johnson ME, Viggiano L, Bailey JA, Abdul-Rauf M, Goodwin G, Rocchi M, Eichler EE (2001) Positive selection of a gene family during the emergence of humans and African apes. Nature 413:514–519 10.1038/35097067 [DOI] [PubMed] [Google Scholar]
- 59.Carlson CS, Thomas DJ, Eberle MA, Swanson JE, Livingston RJ, Rieder MJ, Nickerson DA (2005) Genomic regions exhibiting positive selection identified from dense genotype data. Genome Res 15:1553–1565 10.1101/gr.4326505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kelley JL, Madeoy J, Calhoun JC, Swanson W, Akey JM (2006) Genomic signatures of positive selection in humans and the limits of outlier approaches. Genome Res 16:980–989 10.1101/gr.5157306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang ET, Kodama G, Baldi P, Moyzis RK (2006) Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc Natl Acad Sci USA 103:135–140 10.1073/pnas.0509691102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Voight BF, Kudaravalli S, Wen X, Pritchard JK (2006) A map of recent positive selection in the human genome. PLoS Biol 4:e72 10.1371/journal.pbio.0040072 [DOI] [PMC free article] [PubMed] [Google Scholar]