Abstract
Magnesium chelatase inserts Mg2+ into protoporphyrin IX and is the first unique enzyme of the chlorophyll biosynthetic pathway. It is a heterotrimeric enzyme, composed of I- (40 kDa), D- (70 kDa) and H- (140 kDa) subunits. The I- and D-proteins belong to the family of AAA+ (ATPases associated with various cellular activities), but only I-subunit hydrolyses ATP to ADP. The D-subunits provide a platform for the assembly of the I-subunits, which results in a two-tiered hexameric ring complex. However, the D-subunits are unstable in the chloroplast unless ATPase active I-subunits are present. The H-subunit binds protoporphyrin and is suggested to be the catalytic subunit. Previous studies have indicated that the H-subunit also has ATPase activity, which is in accordance with an earlier suggested two-stage mechanism of the reaction. In the present study, we demonstrate that gel filtration chromatography of affinity-purified Rhodobacter capsulatus H-subunit produced in Escherichia coli generates a high- and a low-molecular-mass fraction. Both fractions were dominated by the H-subunit, but the ATPase activity was only found in the high-molecular-mass fraction and magnesium chelatase activity was only associated with the low-molecular-mass fraction. We demonstrated that light converted monomeric low-molecular-mass H-subunit into high-molecular-mass aggregates. We conclude that ATP utilization by magnesium chelatase is solely connected to the I-subunit and suggest that a contaminating E. coli protein, which binds to aggregates of the H-subunit, caused the previously reported ATPase activity of the H-subunit.
Keywords: ATPases associated with various cellular activities (AAA+), bchH, chlorophyll, magnesium chelatase, protoporphyrin IX, Rhodobacter capsulatus
Abbreviations: AAA+, ATPases associated with various cellular activities; ATP[S], adenosine 5′-[γ-thio]triphosphate; D, magnesium chelatase D-subunit; DTT, dithiothreitol; EM, electron microscopy; I, magnesium chelatase I-subunit; LM, ‘light’ membrane-bound fraction; S, soluble fraction; TFA, trifluoroacetic acid
INTRODUCTION
Magnesium chelatase (EC 6.6.1.1) catalyses the insertion of Mg2+ into protoporphyrin IX, the first committed step in chlorophyll biosynthesis [1]. Early characterizations of magnesium chelatase employed intact chloroplasts to assay enzyme activity [2–4]. It was shown that magnesium chelatase required the porphyrin and metal ion substrates, but it also had a requirement for ATP. Further characterization of magnesium chelatase was hampered by a complete loss of activity after plastid breakage. The first work on magnesium chelatase from broken chloroplasts showed that enzyme activity required both a soluble (S) and a ‘light’ membrane-bound fraction (LM) [5]. It was also shown that the time course of enzyme activity was not linear, but exhibited a lag phase before the enzymatic product, Mg-protoporphyrin IX, was detected [6]. The lag phase could be eliminated if the S and LM extracts were mixed and pre-incubated in the presence of ATP before protoporphyrin was added. Through a series of experiments where the pre-incubation and the reaction were performed at different concentrations, Walker and Weinstein [7] found that ATP[S] (adenosine 5′-[γ-thio]triphosphate) could replace ATP in the activation stage during pre-incubation, but not in the chelation stage of the reaction. From this observation they concluded that magnesium chelation proceeds via a two-stage ATP-dependent mechanism.
It is now known that the magnesium chelatase is composed of three heterogeneous subunits (I, D and H) with average molecular masses of 40, 70 and 140 kDa respectively. Today, recombinant proteins of all three subunits are available from Rhodobacter sphaeroides [8–10], Rhodobacter capsulatus [11], Chlorobium vibrioforme [12], Synechocystis PCC6803 [13,14] and Acidophilum rubrum [15]. X-ray structural analysis revealed that the R. capsulatus I-subunit belongs to the large family of AAA+ (ATPases associated with various cellular activities) proteins [16]. The N-terminal half of the D-subunit shows high sequence similarity to the I-subunit, while the C-terminal half includes a metal ion co-ordination motif [MIDAS (metal-ion-dependent adhesion site)] characteristic of integrin I domains [16]. The N-terminal similarity to the I-subunit strongly suggests that the D-subunit is also an AAA+ protein, although no ATPase activity has been detected from this subunit [17–19]. AAA+ proteins form oligomeric ring structures, and indeed from negative-stained EM (electron microscopy) studies it was shown that the I-subunits from R. capsulatus [20] and Synechocystis PCC6803 [21] form hexameric and heptameric rings respectively. In addition, a two-tiered hexameric ring structure of an ID complex was demonstrated recently (J. Lundqvist, E. Axelsson, A. Hansson, D. Birch, M. Hansson, S. Al-Karadaghi and R. D. Willows, unpublished work). Typically AAA+ proteins are chaperones that can bind and hydrolyse ATP. During the ATP hydrolytic cycle, AAA+ oligomeric ring structures go through a conformational change that transfers mechanical energy to the substrate protein or DNA [22–24]. Today it is well established that ATP hydrolysis can be performed by the magnesium chelatase I-subunit from various organisms [17–19,25–27].
The availability of recombinant magnesium chelatase systems has afforded detailed in vitro investigations showing that the oligomerization of I- and D-subunits is responsible for the observed lag phase. Pre-incubation of I- and D-subunits with Mg2+ and ATP eliminates the lag phase [9,10,14,28]. As earlier found by Walker and Weinstein [7], this initial step does not require hydrolysis of ATP, as ADP and non-hydrolysable ATP analogues (but not AMP) can substitute for ATP [18,25,29]. It is clear, however, that the overall magnesium chelatase reaction requires ATP hydrolysis. Therefore ATPase activity assigned to the H-subunit has been interesting, since this subunit also binds the porphyrin substrate and is regarded as the catalytic subunit [9,11,14,30]. The ATPase activity of the H-subunit of R. sphaeroides and C. vibrioforme was 0.90 and 1.8 nmol/min per mg of protein respectively [17,19]. Otherwise, the H-subunit of Synechocystis PCC6803 has been most intensively studied. In one report, no ATPase activity could be detected [19], whereas 0.95±0.34 nmol/min per mg was reported in another study [18]. In addition, the binding of the porphyrin substrate analogue deuteroporphyrin IX to the H-subunit of Synechocystis, but not R. sphaeroides, was stimulated 4-fold by ATP [30]. In the present study we have investigated the ATPase activity of R. capsulatus H-subunit and showed that the ATPase activity could be separated from magnesium chelatase activity. Therefore we suggest that the H-subunit participates in the magnesium chelatase reaction without hydrolysing ATP. Instead, ATP utilization by magnesium chelatase is connected to activities of the I-subunit.
EXPERIMENTAL
Production of the magnesium chelatase H-subunit
Plasmid pET15bBchH [11] contains the R. capsulatus bchH gene cloned into the pET15b vector (Novagen). The bchH gene encodes the magnesium chelatase H-subunit, and when expressed from pET15bBchH it obtains a His tag in its N-terminus. Plasmid pET15bBchH was transformed into Escherichia coli BL21 (DE3) pLys (Novagen), and the His-tagged H-subunit was produced and purified as described in [11] using an Ni2+-chelating column. During the expression and purification procedure, the cultures, the pellets, lysates and purification fractions were protected from light where possible in order to avoid photo-oxidation.
Gel filtration chromatography
A Sephacryl S-200 HR 16/60 (Amersham Biosciences) column was equilibrated with a buffer consisting of 50 mM Tricine/NaOH (pH 8.0), 15 mM MgCl2, 6% (v/v) glycerol and 4 mM DTT (dithiothreitol) at a flow rate of 1 ml/min. His-tagged H-subunit (400 μl; 10 μg/μl) purified on an Ni2+-chelating column was loaded in the dark on to the Sephacryl column and 500 μl fractions were collected. When necessary, eluted fractions were pooled and concentrated using YM-10 Centricon (Amicon). The column was calibrated with the following molecular mass standards: BSA (66 kDa), alcohol dehydrogenase (150 kDa), β-amylase (200 kDa) and apoferritin (443 kDa).
Magnesium chelatase activity measurements
Recombinant R. capsulatus H-protein was thawed quickly and kept on ice. During the entire assay procedure it was kept in darkness in order to prevent photo-oxidation and bleaching of protein and endogenously bound porphyrin. All assays were performed in 100 μl volumes with 25 μg of H-subunit, 4.5 μg of I-subunit and 10 μg of D-subunit. The reaction contained: 50 mM Tricine/NaOH (pH 8.0), 15 mM MgCl2, 1 mM DTT, 10 mM phosphocreatine, creatine kinase (2 units per reaction), 4 mM ATP and 1 μM protoporphyrin IX. Protoporphyrin IX was first mixed with a small amount of 1 M NaOH and then dissolved in DMSO. The concentration of protoporphyrin IX was determined in 2.7 M HCl using the ϵ408 (molar absorption coefficient) of 262 mM−l·cm−l. The final concentration of DMSO in assays was not greater than 0.5%. Assays were incubated for a maximum of 90 min at 32 °C, and the reaction was terminated with the addition of 900 μl of acetone/water/13.4 M ammonia mixture (80:20:1, by vol.), followed by centrifugation to remove precipitants. For Mg-protoporphyrin IX detection, the supernatants were analysed on a FluroMax-2 spectrophotometer (Jobin Yvon Spex; Horiba) with excitation set at 418 nm and emission detected between 550 and 650 nm. Excitation and emission slit widths were set to 10 nm. A peak at 590 nm indicated the presence of Mg-protoporphyrin IX. A standard curve was produced using authentic Mg-protoporphyrin IX (Porphyrin Products, Logan, UT, U.S.A.) acetone mix.
ATPase activity measurements
ATPase activity was measured by hydrolysis of [8-14C]ATP (specific radioactivity 2.1 GBq/mmol; PerkinElmer). The reaction contained: 50 mM Tricine/NaOH (pH 8.0), 15 mM MgCl2, 6% (v/v) glycerol, 1 mM DTT, 80 μM ATP, 20 μM [8-14C]ATP and 7 μg of protein in a total volume of 10 μl. During incubation at 32 °C, 1 μl samples were removed at 2 h and quenched by addition of 1 μl of 2 M formic acid. The ATP and ADP components were then separated by TLC (polyethyleneimine–cellulose F; Merck), using 1 M formic acid and 0.7 M LiCl as the mobile phase. A STORM phosphoimager system (Amersham Biosciences) was used for detection and quantification.
MS
Protein bands of interest were cut from SDS/polyacrylamide gels and destained in 75 μl of 50 mM NH4HCO3 and 50% (v/v) ethanol for 60 min. The liquid was removed and 50 μl of ethanol was added to shrink the gel pieces for 15 min. The liquid was removed and the gel pieces were dried in a SpeedVac concentrator (Savant, Farmingdale, NY, U.S.A.). The gel pieces were rehydrated in 10 μl of digestion buffer consisting of 25 mM NH4HCO3 and 12.5 μg/μl of sequencing-grade trypsin (Promega, Madison, WI, U.S.A.) on ice for 45 min. Digestion buffer without trypsin (10 μl) was added to cover the pieces and to keep them wet during enzyme cleavage. The samples were incubated at 37 °C overnight and the supernatant was collected. To further extract the peptides from the gel pieces, 10 mM NH4HCO3, 50% ethanol and 0.5% (w/v) TFA (trifluoroacetic acid) were added and incubated for 60 min at 37 °C. The gel pieces were spun down and the supernatant was collected and pooled with the supernatant from the overnight incubation. The samples were dried using a SpeedVac concentrator and were then redissolved in 5 μl of 0.1% TFA. A 0.5 μl sample was spotted directly on to a stainless-steel MALDI-target plate and the solvent was allowed to completely evaporate. Then, 0.5 μl of a matrix solution containing 5 mg/ml α-cyano-4-hydroxycinnamic acid, 50% (v/v) acetonitrile, 0.1% (w/v) TFA and 5 mM NH4H2PO4 [31] was added and allowed to dry. MALDI–TOF (matrix-assisted laser-desorption ionization–time-of-flight) MS spectra were recorded automatically using a 4700 Proteomics Analyzer (Applied Biosystems, Framingham, CA, U.S.A.) mass spectrometer in positive reflector mode. The MS spectra were internally calibrated using two trypsin autoproteolysis peptides with the m/z values 842.51 and 2211.097 Da. Protein identification was performed using the GPS Explorer software, with an in-house installed Mascot search engine (Matrix Science, London, U.K.) [32] searching the Swiss-Prot database. Parameters specified in the search were: taxa, E. coli or other bacteria; missed cleavages, 1; peptide mass tolerance, ±50 p.p.m. A protein was considered to be identified when the level of confidence exceeded 99%.
EM
Carbon-coated 400 mesh copper grids (Ax-labs, Vedbæk, Denmark) were first washed for 1 min with a drop of 50 mM Tricine/NaOH (pH 8.0), 15 mM MgCl2 and 1 mM DTT. Excess liquid was removed by brief contact with filter paper. Ni2+ affinity-purified H-subunit was diluted to 0.01 mg/ml with the wash buffer. A drop of the protein solution was applied to the grid and incubated for 1 min in darkness, and excess sample was removed. The grid was immediately stained with filtered 2% (w/v) uranyl acetate for 30 s and excess stain was removed. EM was performed using a 120 kV Philips CM-10 microscope at ×55000 magnification. Using a single particle reconstruction of R. capsulatus H-subunit as a guide (N. Sirijovski, J. Lundqvist, M. Rosenbäck, S. Al-Karadaghi, R. D. Willows and M. Hansson, unpublished work), we searched for areas in the micrographs that contained stain-excluding particles equal to or greater than the monomeric size.
Other methods
SDS/PAGE [10% (w/v) acrylamide] was performed as described by Fling and Gregerson [33] with the Tris–Tricine buffer system of Schägger and von Jagow [34]. Proteins on SDS/PAGE were visualized by staining with colloidal Coomassie Brilliant Blue G-250 [35]. Amino acid sequence alignments were performed using the ClustalW algorithm [36]. The ScanProsite tool was used to search for ATPase moieties [37].
RESULTS
H-subunits do not contain conserved ATPase moieties
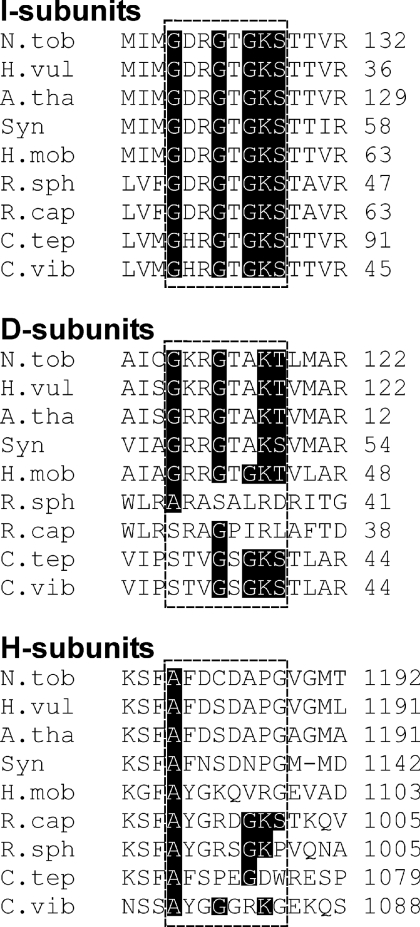
An abundance of sequence and crystallographic data analysis [38–42] have shown that the best conserved motif for binding of ATP or GTP is the Walker A consensus ([AG]XXXXGK[ST]), which is also known as the P-loop [38,40]. An alignment of magnesium chelatase I-subunits shows the conserved P-loop motif characteristic of AAA+ proteins (G[DH]RGTGKS) (Figure 1). ATP hydrolysis by this AAA+ protein is strongly supported by biochemical and structural investigations [16–19,25–27]. We performed a careful search for the P-loop signature in a representative set of nine magnesium chelatase D- and H-subunits (Figure 1). Concerning the D-subunit, only the He. mobilis protein contained the full P-loop motif (GRRGTGKT). An incomplete P-loop motif was found in N. tabacum, A. thaliana, Ho. vulgare and Synechocystis PCC 6803 (G[KR]RGTAK[TS]). R. capsulatus and R. sphaeroides completely lacked the motif, and C. tepidum and C. vibrioforme contain only traces of the motif (GKS). The relative position of motif in the I- and D-subunits is conserved within their AAA+ domains. It can be speculated that the semi-conserved motif in the D-subunits is due to a gene duplication event followed by divergence over the course of evolution without selective pressure to maintain the P-loop motif in the D-subunit.
Figure 1. Alignments showing the best candidates for a P-loop motif ([AG]XXXXGK[ST]) in the three polypeptides of magnesium chelatase.
The P-loop motif is conserved in the I-subunit sequences. The motif is also found in the individual polypeptides of Heliobacillus mobilis D-subunit and the R. capsulatus H-subunit. The analysis suggests that it is unlikely that ATPase activity is a general feature of the D- and H-subunits. Only 15 amino acid residues are shown from an alignment of the entire polypeptides. Sources: R. capsulatus (GenBank® accession number Z11165), R. sphaeroides (AF017642 and AJ010302), Chlorobium tepidum (AY005135), C. vibrioforme (Z83933), Nicotiana tabacum (AF014052, AF014053 and Y10022), Arabidopsis thaliana (AF083555, X91411 and Z68495), Hordeum vulgare (AY039003, DQ125942 and DQ529207), Synechocystis PCC 6803 [13] and H. mobilis (AF080002).
Regarding the H-subunit, no traces of a conserved site with a P-loop signature could be found by analysis of amino acid sequence alignments. However, when analysing individual polypeptides a perfect motif was found in the polypeptide of R. capsulatus (AYGRDGKS) (Figure 1). Of the eight other organisms included in the alignment, R. sphaeroides contained a sequence most reminiscent of a P-loop (AYGRSGKP). Only the first alanine residue in the motif is conserved in all eight polypeptides. It is likely that this motif is a semi-conserved artefact between two closely related organisms, since not all proteins that contain the P-loop motif are ATP binding, e.g. human ferritin light chain and chymotrypsin. This analysis did not strengthen the argument that ATPase activity is a general feature of the H-subunit, although it could explain ATP hydrolysis by the R. capsulatus enzyme.
Light exposure increases the heterogeneity of affinity-purified H-subunit
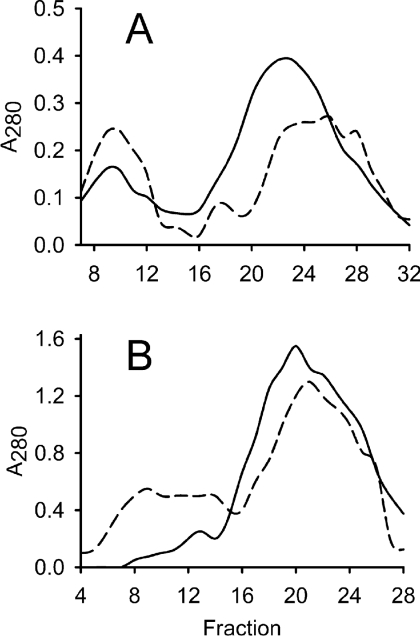
When excited by light, porphyrins generate ROS (reactive oxygen species). The magnesium chelatase H-subunit is brownish upon purification due to bound protoporphyrin IX. It has been suggested previously that light-induced inactivation of the H-subunit via the bound protoporphyrin could account for the rapid inhibition of bacteriochlorophyll biosynthesis in R. capsulatus during transition from anaerobic photosynthetic growth to aerobic heterotrophic growth [43]. We analysed the effect of light treatment on the H-subunit in vitro. A system for producing R. capsulatus H-subunit in E. coli has been developed including a one-step affinity-purification method [11]. In the present study, we produced affinity-purified H-subunit in the dark, although some light exposure could not be avoided. Two 40 mg aliquots of this protein were analysed. One aliquot was kept in the dark and on ice as a control. The other identical sample was also kept on ice, but exposed to ordinary room light of 15 μE·m−2·s−1 from a 58 W fluorescent light tube for 30 min. These samples were analysed by gel filtration and in both cases a peak of high molecular mass was obtained in addition to a peak of lower molecular mass (Figure 2A). Interestingly, light treatment resulted in an increased amount of protein of high molecular mass and a decreased amount of low-molecular-mass protein. Fractions 9 and 22 corresponded to approximate molecular masses of 400 and 150 kDa respectively. Fractions 7–12 from the respective analysis were pooled and selected to represent the high-molecular-mass fraction. Fractions 18–27 were pooled and represented the low-molecular-mass fraction. In the dark control, the H-subunit dominated both the high- and the low-molecular-mass peaks according to SDS/PAGE analyses (Figure 3, lanes 2 and 3). This was also supported by MS performed on non-trypsin-treated material. The high- and low-molecular-mass protein fractions gave masses of 132830 and 132581 Da respectively, which was close to the predicted molecular mass of 132524 Da for the His-tagged R. capsulatus H-subunit.
Figure 2. Gel filtration of affinity-purified H-subunit.
Elution of protein was followed by absorbance (A) at 280 nm. The column was calibrated with apoferritin (443 kDa, peak at fraction 9), β-amylase (200 kDa, fraction 18), alcohol dehydrogenase (150 kDa, fraction 22) and BSA (66 kDa, fraction 29). (A) Affinity-purified H-subunit (40 mg) was exposed to light before loading on to a Sephacryl S-200 HR 16/60 column (dashed line). An equal sample, kept in darkness, was used as a control (solid line). The first (fractions 7–12) and second (fractions 18–27) peaks of each analysis were selected to represent the high- and low-molecular-mass fractions respectively and used in further analysis. (B) Pooled and concentrated fractions from low-molecular-mass H-subunit obtained by gel filtration were exposed to light and a second round of gel filtration was performed (dashed line). An identical sample was kept in the dark as a control (solid line). The experiment shows that light causes de novo formation of a high-molecular-mass fraction.
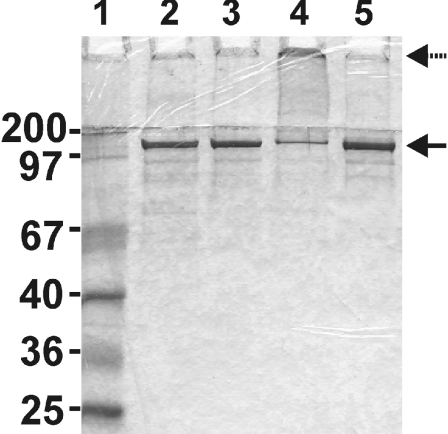
Figure 3. Coomassie-stained SDS/PAGE of gel filtration fractions.
Fractions from a gel filtration experiment with light-treated and dark-treated His-tagged R. capsulatus H-subunit were analysed by SDS/PAGE; 10 μg of protein was loaded. Lane 1, size marker; lane 2, pooled fractions 7–12 of the dark-treated sample in Figure 2(A); lane 3, pooled fractions 18–27 of the dark-treated sample; lane 4, pooled fractions 7–12 of the light-exposed sample; lane 5, pooled fractions 18–27 of the light-exposed sample. The solid and dashed arrows indicate His-tagged H-subunit and high-molecular-mass aggregates retarded in the stacking gel respectively.
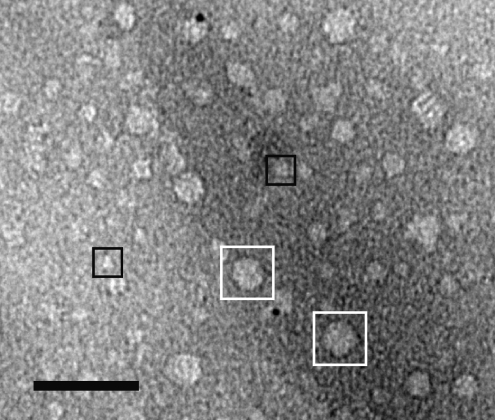
The SDS/PAGE profile of the light-treated material was different from that of the dark control. The amount of H-subunit corresponding to the monomeric size was reduced in the high-molecular-mass fraction and instead a significant amount of protein was retarded in the stacking gel (Figure 3, lane 4). The observation suggested that the H-subunits form large aggregates upon light exposure. The existence of aggregates could be demonstrated by negative-staining EM (Figure 4). In a parallel study we could easily pick over 5000 single particles that correspond to monomeric H-subunit and a three-dimensional model was produced (N. Sirijovski, J. Lundqvist, M. Rosenbäck, S. Al-Karadaghi, R. D. Willows and M. Hansson, unpublished work). The H-subunit is approx. 100 Å (1 Å=0.1 nm) across. Figure 4 illustrates a region of a micrograph containing a few monomeric H-subunit particles and a large number of particles of up to 300 Å across. In order to further analyse the light-dependent formation of aggregates, a low-molecular-mass fraction from a dark control was split into two equal samples. One sample was exposed to light as described above, while the other sample was kept in the dark as a control. Upon gel filtration, a new high-molecular mass-fraction was obtained at the expense of the low-molecular-mass fraction in the light-treated sample, whereas the low-molecular-mass proteins remained in the dark-treated control (Figure 2B). The experiment demonstrated that the protoporphyrin-binding H-subunit is very sensitive to light and that even brief exposure causes novel aggregation of H-subunits.
Figure 4. Negative-staining electron micrograph of BchH.
The micrograph shows a section containing both aggregated BchH particles (examples marked with white boxes) and monomeric sized BchH particles (black boxes). Scale bar, 50 nm.
Separation of magnesium chelatase and ATPase activity
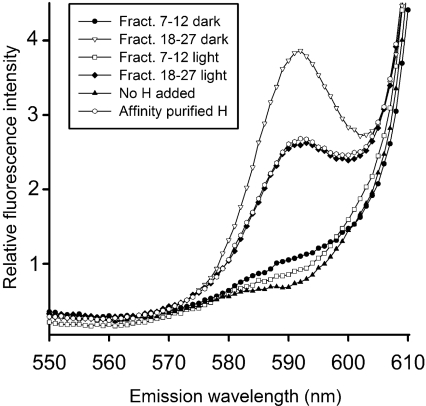
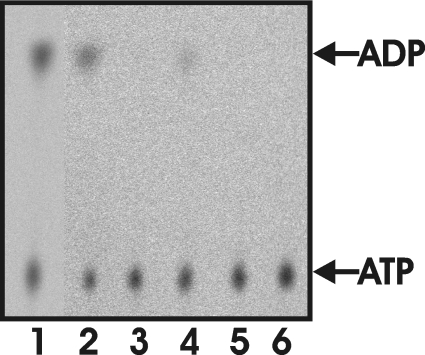
Bioinformatic analyses of the magnesium chelatase subunits call the ATPase activity of the H-subunit in question. However, it is clear that affinity-purified H-subunit is active in magnesium chelatase activity measurements (Figure 5), but also hydrolysed ATP to ADP (Figure 6, lane 1). The ATPase activity was 0.65 nmol of ADP formed/min per mg of protein, which is in the same range of the activity determined for the H-subunit of R. sphaeroides, C. vibrioforme and Synechocystis PCC6803 [17–19]. Further purification of the affinity-purified H-subunit by gel filtration chromatography resulted in a high- and a low-molecular-mass fraction (Figure 2A). When assayed for magnesium chelatase activity (Figure 5) and ATPase activity (Figure 6), it was found that the high-molecular-mass fraction had ATPase activity, but could not contribute to magnesium chelatase activity. In contrast, the low-molecular-mass peak had no ATPase activity, but contributed to magnesium chelatase activity. Light treatment of the affinity-purified H-subunit did not increase the ATPase activity. When protein from the dark-treated low-molecular-mass peak was pooled and re-loaded on to the gel filtration column after light or dark treatment, it was evident that light directly induces the formation of the high-molecular-mass peak (Figure 2B). However, in this case no ATPase activity was detected in the de novo high-molecular-mass peak. Considering these data, it is unlikely that ATP hydrolysis is an intrinsic activity of the H-subunit that is activated by light. Instead, the ATPase activity is more likely to originate from a contaminating E. coli protein that is present in the affinity-purified preparation of H-subunit. In order to identify such a contaminating ATPase, we separated proteins with SDS/PAGE and employed MS of trypsin-digested material to identify low-abundance proteins, which were seen in the region between 25 and 100 kDa. All bands formed by low-abundance proteins on the gel were identified as fragments of the H-subunit. The fragments were often internal regions of the H-polypeptide rather than C-terminal truncations with a retained His tag. Therefore it is likely that fragments were formed after the elution from the affinity column and suggest the presence of proteases. Concerning the contaminating ATPase, we conclude that it is present at a very low level in our preparation. We considered the possibility that the H-subunit fragments in the MS analysis could mask the contaminating ATPase. Therefore we performed careful analyses of residual peaks not identified as fragments of the H-subunit. Although it was difficult to obtain good sequence coverage, we did identify two peptides from a band of approx. 70 kDa in the high-molecular-mass fraction that corresponded to regions within the ClpA protein from E. coli (Swiss-Prot accession number P0ABH9). The observed mass peaks were of 1513.82 and 2722.27 Da and match with the internal ClpA polypeptides HPHAVLLLDEIEK (theoretical mass 1513.830 Da) and NEVTGANVLVAIFSEQESQAAYLLR (2722.40 Da). ClpA catalyses protein unfolding and limited protein remodelling. Together with its proteolytic partner ClpP, it participates in ATP-dependent intracellular protein degradation [44]. Since ClpA is a highly active ATPase and binds to hydrophobic surface patches not normally exposed in native proteins, it is a possible candidate to be the contaminating ATPase in our high-molecular-mass fractions.
Figure 5. Magnesium chelatase activity of gel filtration fractions of light- and dark-exposed His-tagged H-subunit.
The amount of formed Mg-protoporphyrin IX was analysed by spectrofluorimetry using a λex of 418 nm. Mg-protoporphyrin IX forms a peak at 590 nm. Light- and dark-treated H-subunits from pooled fractions 7–12 and 18–27 in Figure 2(A) were used in the analysis.
Figure 6. ATPase activity of gel filtration fractions as analysed by TLC and visualized by autoradiography.
Light- and dark-exposed His-tagged H-subunits fractionated with gel filtration were tested for their ability to hydrolyse ATP into ADP. After 2 h the assay were stopped and the nucleotides were separated with TLC. Lane 1, affinity-purified His-tagged H-subunit (activity 0.65 nmol of ADP formed/min per mg of protein); lane 2, pooled fractions 7–12 of the dark-treated sample in Figure 2(A) (0.61 nmol/min per mg); lane 3, pooled fractions 18–27 of the dark-treated sample (<0.05 nmol/min per mg); lane 4, pooled fractions 7–12 of the light-exposed sample (0.30 nmol/min per mg); lane 5, pooled fractions 18–27 of the light-exposed sample (<0.05 nmol/min per mg); lane 6, negative control with no protein (<0.05 nmol/min per mg).
DISCUSSION
Here, we present data that strongly question previous reports [17–19] that the magnesium chelatase H-subunit possesses intrinsic ATPase activity. Instead we suggest that the ATPase activity is attributed to a low-abundance contaminating E. coli protein that persists during affinity purification of the H-subunit. The one-step affinity-purification method employing nickel immobilized on a column to purify His-tagged H-proteins has been used in all studies where ATPase activity of H-subunit has been analysed [17–19,30]. We suggest that the contaminating ATPase could be a native E. coli chaperone involved in refolding or degradation of aggregated protein. A contaminating chaperone, E. coli GroEL, has previously been seen during expression and purification of the R. sphaeroides D-subunit [9]. However, in contrast with our study, the contaminating GroEL protein dominated the fractions that could contribute to magnesium chelatase activity and the recombinant D-subunit could not be detected.
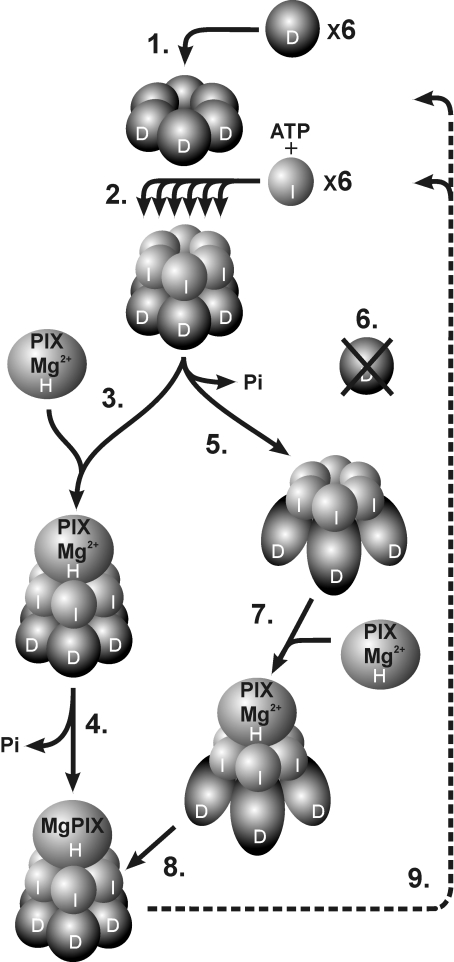
To us it is clear that the two-stage model for the magnesium chelatase reaction as originally suggested by Walker and Weinstein [7] is better explained without hydrolysis of ATP by the H-subunit. Instead, both the activation step and the catalytic step relate to the use of ATP by the I-subunit. The activation step, first observed by Walker and Weinstein, is now referred to as an oligomerization step involving the I- and the D-subunits. In this process, a hexameric D-ring provides a platform for the stepwise assembly of the I-subunits into a double hexameric ring (Figure 7) (E. Axelsson, J. Lundqvist, A. Sawicki, S. Nilsson, I. Schröder, S. Al-Karadaghi, R. D. Willows and M. Hansson, unpublished work). This hypothesis is based on EM structural analysis showing that D-subunits can form oligomeric ring structures in the absence of nucleotide and that a two-tiered hexameric ID complex is formed in the presence of ATP (J. Lundqvist, E. Axelsson, A. Hansson, D. Birch, M. Hansson, S. Al-Karadaghi and R. D. Willows, unpublished work). Since this ID-structure has only been characterized at low resolution and it is yet to be determined whether different nucleotides such as ADP and ATP[S] result in alternative structures, we cannot rule out the possibility that hydrolysed ATP is present within the structure. The ability of I-subunit to bind to D-subunit in the absence of ATP hydrolysis has been shown in independent studies. In one experiment, the I-subunit of Synechocystis was treated with N-ethylmaleimide, which covalently modifies cysteine residues. This treatment resulted in an inactive I-subunit both with respect to ATP hydrolysis and magnesium chelatase activity. However, in the presence of ATP, ADP or a non-hydrolysable ATP analogue, the modified I-subunit could still associate with the D-subunit [18]. In another study, semi-dominant barley mutants, deficient in the gene encoding the I-subunit, were characterized at the DNA level and the mutations were introduced into the recombinant system for expression of R. capsulatus I-subunit [26]. These modified I-subunits were unable to hydrolyse ATP and were devoid of magnesium chelatase activity. They could still form oligomeric complexes with themselves, with wild-type I-protein and with D-protein if ATP or ADP was present [25,26]. However, the function of the I-subunit as a chaperone of the D-subunit was apparently lost, since no D-subunit could be detected in chloroplast extracts of barley I-mutants. This begs the question of when ATP is hydrolysed in the catalytic cycle, which has so far been narrowed down to the second stage of the magnesium chelatase reaction suggested by Walker and Weinstein [7]. It has been proposed that the D-subunit blocks the ATPase activity of the I-subunit until the ID complex interacts with an H-subunit–protoporphyrin complex [16]. The transient interaction with the H-subunit releases the inhibition, triggering ATP hydrolysis as well as insertion of Mg2+ into protoporphyrin. However, in the view of an absence of D-subunit in barley I-mutants, it seems that binding of I to D is not enough to maintain D in the cell, but is dependent on the ATPase activity of the I-subunit. Thus ATP hydrolysis should be important for a step after the formation of the ID complex, but before interaction with the H-subunit (Figure 7). It should be noted that the D-subunit is stably maintained in barley mutants with deletions of the entire gene encoding the H-subunit [45]. It has been shown for several AAA+ proteins that a significant conformational change occurs during the ATP hydrolytic cycle [23,46–48]. In some cases ATP hydrolysis is required for substrate recognition and in others it is not. If ATP hydrolysis occurs after the holoenzyme is formed, then a conformational change can be expected to provide the mechanical force that probably remodels the H-subunit and drives the magnesium chelatase reaction. Alternatively, if ATP hydrolysis occurs prior to the formation of the holoenzyme, then a conformational change in the I-hexamer would be expected to expose essential amino acid residues in the ID complex that are only then recognized by the H-subunit, resulting in magnesium chelation. This kind of mechanism is found in the phage shock protein F, which is an AAA+ protein that regulates transcription in response to stress [49]. This chaperone binds and hydrolyses NTP in order to adopt an alternative conformation so as to expose inaccessible motifs required to interact with its target α54 and form the holoenzyme. Future structural studies of the magnesium chelatase involving various ATP analogues will aid in clarifying the role and timing of ATP hydrolysis in the first step of chlorophyll biosynthesis.
Figure 7. Model suggesting progressive subunit assembly of magnesium chelatase in relation to ATP hydrolysis.
(1) Six D-subunits form a stable hexameric ring in an ATP-independent process. (2) Using the hexameric D-ring structure as a platform, six I-subunits are assembled stepwise into a hexameric ring. The formation of the two-tiered complex is nucleotide-dependent. (3) Two alternative models are shown, which differ with respect to the timing of ATP hydrolysis. In one model the catalytic H-subunit binds to an ID complex where the ATP hydrolytic capacity of the I-subunit is blocked by the D-subunit. (4) The transient interaction with the H-subunit triggers ATP hydrolysis and insertion of Mg2+ into protoporphyrin. (5) In the alternative model, ATP hydrolysis by the I-subunits converts the D-subunits into a stable form and possibly exposes key residues required for the recognition of H-subunits. (6) Without the ATP-dependent activity of the I-subunits the D-subunits are degraded. (7) The catalytic H-subunit interacts with the activated ID complex and (8) Mg2+ is inserted into protoporphyrin without ATP hydrolysis at this step. (9) After catalysis, the IDH complex disassociates to the level of the D-hexamer, which remains for a new round of catalysis. At present, it is unknown whether the H-subunit docks to the I- or D-side of the complex, although docking to the I-subunits is shown in the Figure. PIX, protoporphyrin.
Acknowledgments
This work was made possible thanks to generous support from the Swedish Research Council to M. H. and S. A.-K. and the Australian Research Council (grant no. A09905713) to R. D. W. A fellowship from the Sven and Lilly Lawski Foundation to N. S. is gratefully acknowledged.
References
- 1.Willows R. D., Hansson M. Mechanism, structure and regulation of magnesium chelatase. In: Kadish K. M., Smith K., Guilard R., editors. The Porphyrin Handbook II, Vol. 13. Amsterdam: Elsevier Science; 2003. pp. 1–47. [Google Scholar]
- 2.Fuesler T. P., Castelfranco P. A., Wong Y. S. Formation of magnesium-containing chlorophyll precursors from protoporphyrin IX, δ-aminolevulinic acid, and glutamate in isolated, photosynthetically competent, developing chloroplasts. Plant Physiol. 1984;74:928–933. doi: 10.1104/pp.74.4.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuesler T. P., Wright L. A., Castelfranco P. A. Properties of magnesium chelatase in greening etioplasts. Plant Physiol. 1981;67:246–249. doi: 10.1104/pp.67.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castelfranco P. A., Weinstein J. D., Schwarcz S., Pardo A. D., Wezelman B. E. The magnesium insertion step in chlorophyll biosynthesis. Arch. Biochem. Biophys. 1979;192:592–598. doi: 10.1016/0003-9861(79)90130-9. [DOI] [PubMed] [Google Scholar]
- 5.Walker C. J., Weinstein J. D. In vitro assay of the chlorophyll biosynthetic enzyme magnesium chelatase: resolution of the activity into soluble and membrane bound fractions. Proc. Natl. Acad. Sci. U.S.A. 1991;88:5789–5793. doi: 10.1073/pnas.88.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker C. J., Weinstein J. D. Further characterization of the magnesium chelatase in isolated developing cucumber chloroplasts: substrate specificity, regulation, intactness and ATP requirements. Plant Physiol. 1991;95:1189–1196. doi: 10.1104/pp.95.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker C. J., Weinstein J. D. The magnesium-insertion step of chlorophyll biosynthesis is a two-stage reaction. Biochem. J. 1994;299:277–284. doi: 10.1042/bj2990277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibson L. C. D., Willows R. D., Kannangara C. G., von Wettstein D., Hunter C. N. Magnesium-protoporphyrin chelatase of Rhodobacter sphaeroides: reconstitution of activity by combining the products of the bchH, -I, and -D genes expressed in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1995;92:1941–1944. doi: 10.1073/pnas.92.6.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willows R. D., Gibson L. C. D., Kanangara C. G., Hunter C. N., von Wettstein D. Three separate proteins constitute the magnesium chelatase of Rhodobacter sphaeroides. Eur. J. Biochem. 1996;235:438–443. doi: 10.1111/j.1432-1033.1996.00438.x. [DOI] [PubMed] [Google Scholar]
- 10.Gibson L. C. D., Jensen P. E., Hunter C. N. Magnesium chelatase from Rhodobacter sphaeroides: initial characterization of the enzyme using purified subunits and evidence for a BchI–BchD complex. Biochem. J. 1999;337:243–251. [PMC free article] [PubMed] [Google Scholar]
- 11.Willows R. D., Beale S. I. Heterologous expression of the Rhodobacter capsulatus BchI, -D, and -H genes that encode magnesium chelatase subunits and characterization of the reconstituted enzyme. J. Biol. Chem. 1998;273:34206–34213. doi: 10.1074/jbc.273.51.34206. [DOI] [PubMed] [Google Scholar]
- 12.Petersen B. L., Jensen P. E., Gibson L. C., Stummann B. M., Hunter C. N., Henningsen K. W. Reconstitution of an active magnesium chelatase enzyme complex from the bchI, -D, and -H gene products of the green sulfur bacterium Chlorobium vibrioforme expressed in Escherichia coli. J. Bacteriol. 1998;180:699–704. doi: 10.1128/jb.180.3.699-704.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen P. E., Gibson L. C., Henningsen K. W., Hunter C. N. Expression of the chlI, chlD, and chlH genes from the cyanobacterium Synechocystis PCC6803 in Escherichia coli and demonstration that the three cognate proteins are required for magnesium-protoporphyrin chelatase activity. J. Biol. Chem. 1996;271:16662–16667. doi: 10.1074/jbc.271.28.16662. [DOI] [PubMed] [Google Scholar]
- 14.Jensen P. E., Gibson L. C. D., Hunter C. N. Determinants of catalytic activity with the use of purified I, D and H subunits of the magnesium protoporphyrin IX chelatase from Synechocystis PCC6803. Biochem. J. 1998;334:335–344. doi: 10.1042/bj3340335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masuda T., Inoue K., Masuda M., Nagayama M., Tamaki A., Ohta H., Shimada H., Takamiya K.-I. Magnesium insertion by magnesium chelatase in the biosynthesis of zinc bacteriochlorophyll a in an aerobic acidophilic bacterium Acidiphilum rubrum. J. Biol. Chem. 1999;274:33594–33600. doi: 10.1074/jbc.274.47.33594. [DOI] [PubMed] [Google Scholar]
- 16.Fodje M. N., Hansson A., Hansson M., Olsen J. G., Gough S., Willows R. D., Al-Karadaghi S. Interplay between an AAA module and an integrin I domain may regulate the function of magnesium chelatase. J. Mol. Biol. 2001;311:111–122. doi: 10.1006/jmbi.2001.4834. [DOI] [PubMed] [Google Scholar]
- 17.Hansson M., Kannangara C. G. ATPases and phosphate exchange activities in magnesium chelatase subunits of Rhodobacter sphaeroides. Proc. Natl. Acad. Sci. U.S.A. 1997;94:13351–13356. doi: 10.1073/pnas.94.24.13351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen P. E., Gibson L. C., Hunter C. N. ATPase activity associated with the magnesium-protoporphyrin IX chelatase enzyme of Synechocystis PCC6803: evidence for ATP hydrolysis during Mg2+ insertion, and the MgATP-dependent interaction of the ChlI and ChlD subunits. Biochem. J. 1999;339:127–134. [PMC free article] [PubMed] [Google Scholar]
- 19.Petersen B. L., Kannangara C. G., Henningsen K. W. Distribution of ATPase and ATP-to-ADP phosphate exchange activities in magnesium chelatase subunits of Chlorobium vibrioforme and Synechocystis PCC6803. Arch. Microbiol. 1999;171:146–150. doi: 10.1007/s002030050692. [DOI] [PubMed] [Google Scholar]
- 20.Willows R. D., Hansson A., Birch D., Al-Karadaghi S., Hansson M. EM single particle analysis of the ATP-dependent BchI complex of magnesium chelatase: an AAA+ hexamer. J. Struct. Biol. 2004;146:227–233. doi: 10.1016/j.jsb.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 21.Reid J. D., Siebert C. A., Bullough P. A., Hunter C. N. The ATPase activity of the ChlI subunit of magnesium chelatase and formation of a heptameric AAA+ ring. Biochemistry. 2003;42:6912–6920. doi: 10.1021/bi034082q. [DOI] [PubMed] [Google Scholar]
- 22.Vale R. D. AAA proteins: Lords of the ring. J. Cell Biol. 2000;150:F13–F19. doi: 10.1083/jcb.150.1.f13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogura T., Wilkinson A. J. AAA+ superfamily ATPases: common structure – diverse function. Genes Cells. 2001;6:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- 24.Neuwald A. F., Aravind L., Spouge J. L., Koonin E. V. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 25.Lake V., Olsson U., Willows R. D., Hansson M. ATPase activity of magnesium chelatase subunit I is required to maintain subunit D in vivo. Eur. J. Biochem. 2004;271:2182–2188. doi: 10.1111/j.1432-1033.2004.04143.x. [DOI] [PubMed] [Google Scholar]
- 26.Hansson A., Willows R. D., Roberts T. H., Hansson M. Three semidominant barley mutants with single amino acid substitutions in the smallest magnesium chelatase subunit form defective AAA+ hexamers. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13944–13949. doi: 10.1073/pnas.212504499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petersen B. L., Kannangara C. G., Henningsen K. W. ATPase and phosphate exchange activities in Mg-chelatase subunits of Chlorobium and Synechocystis. Photosynth. Mech. Eff. Proc. Int. Congr. 11th. 1998;4:3241–3244. [Google Scholar]
- 28.Papenbrock J., Gräfe S., Kruse E., Hanel F., Grimm B. Mg-chelatase of tobacco: identification of a Chl D cDNA sequence encoding a third subunit, analysis of the interaction of the three subunits with the yeast two-hybrid system, and reconstitution of the enzyme activity by co-expression of recombinant CHL D, CHL H and CHL I. Plant J. 1997;12:981–990. doi: 10.1046/j.1365-313x.1997.12050981.x. [DOI] [PubMed] [Google Scholar]
- 29.Jensen P. E., Reid J. D., Hunter C. N. Modification of cysteine residues in the ChlI and ChlH subunits of magnesium chelatase results in enzyme inactivation. Biochem. J. 2000;352:435–441. [PMC free article] [PubMed] [Google Scholar]
- 30.Karger G. A., Reid J. D., Hunter C. N. Characterization of the binding of deuteroporphyrin IX to the magnesium chelatase H subunit and spectroscopic properties of the complex. Biochemistry. 2001;40:9291–9299. doi: 10.1021/bi010562a. [DOI] [PubMed] [Google Scholar]
- 31.Smirnov I. P., Zhu X., Taylor T., Huang Y., Ross P., Papayanopoulos I. A., Martin S. A., Pappin D. J. Suppression of α-cyano-4-hydroxycinnamic acid matrix clusters and reduction of chemical noise in MALDI–TOF mass spectrometry. Anal. Chem. 2004;76:2958–2965. doi: 10.1021/ac035331j. [DOI] [PubMed] [Google Scholar]
- 32.Perkins D. N., Pappin D. J., Creasay D. M., Cottrell J. S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 33.Fling S. P., Gregerson D. S. Peptide and protein molecular weight determination by electrophoresis using a high-molarity Tris buffer system without urea. Anal. Biochem. 1986;155:83–88. doi: 10.1016/0003-2697(86)90228-9. [DOI] [PubMed] [Google Scholar]
- 34.Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 35.Neuhoff V., Arold N., Taube D., Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis. 1988;9:255–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- 36.Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gattiker A., Gasteiger E., Bairoch A. ScanProsite: a reference implementation of a PROSITE scanning tool. Appl. Bioinformatics. 2002;1:107–108. [PubMed] [Google Scholar]
- 38.Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Möller W., Amons R. Phosphate-binding sequences in nucleotide-binding proteins. FEBS Lett. 1985;186:1–7. doi: 10.1016/0014-5793(85)81326-0. [DOI] [PubMed] [Google Scholar]
- 40.Saraste M., Sibbald P. R., Wittinghofer A. The P-loop: a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 41.Koonin E. V. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J. Mol. Biol. 1993;229:1165–1174. doi: 10.1006/jmbi.1993.1115. [DOI] [PubMed] [Google Scholar]
- 42.Dever T. E., Glynias M. J., Merrick W. C. GTP-binding domain: three consensus sequence elements with distinct spacing. Proc. Natl. Acad. Sci. U.S.A. 1987;84:1814–1818. doi: 10.1073/pnas.84.7.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willows R. D., Lake V., Roberts T. H., Beale S. I. Inactivation of Mg chelatase during transition from anaerobic to aerobic growth in Rhodobacter capsulatus. J. Bacteriol. 2003;185:3249–3258. doi: 10.1128/JB.185.11.3249-3258.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoskins J. R., Singh S. K., Maurizi M. R., Wickner S. Protein binding and unfolding by the chaperone ClpA and degradation by the protease ClpAP. Proc. Natl. Acad. Sci. U.S.A. 2000;97:8892–8897. doi: 10.1073/pnas.97.16.8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olsson U., Sirijovski N., Hansson M. Characterization of eight barley xantha-f mutants deficient in magnesium chelatase. Plant Physiol. Biochem. 2004;42:557–564. doi: 10.1016/j.plaphy.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 46.Hanson P. I., Roth R., Morisaki H., Jahn R., Heuser J. E. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- 47.Rouiller I., DeLaBarre B., May A. P., Weis W. I., Brunger A. T., Milligan R. A., Wilson-Kubalek E. M. Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nat. Struct. Biol. 2002;9:950–957. doi: 10.1038/nsb872. [DOI] [PubMed] [Google Scholar]
- 48.Song H. K., Hartmann C., Ramachandran R., Bochtler M., Behrendt R., Moroder L., Huber R. Mutational studies on HslU and its docking mode with HslV. Proc. Natl. Acad. Sci. U.S.A. 2000;97:14103–14108. doi: 10.1073/pnas.250491797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bordes P., Wigneshweraraj S. R., Schumacher J., Zhang X., Chaney M., Buck M. The ATP hydrolysing transcription activator phage shock protein F of Escherichia coli: identifying a surface that binds α54. Proc. Natl. Acad. Sci. U.S.A. 2003;100:2278–2283. doi: 10.1073/pnas.0537525100. [DOI] [PMC free article] [PubMed] [Google Scholar]