Abstract
Genetic differentiation between the largely sympatric molecular forms M and S of Anopheles gambiae appears mostly limited to division 6 and part of division 5 of the X chromosome. This region is adjacent to the centromere and includes the rDNA that was used to define these forms. This localized differentiation between populations that experience gene flow strongly suggests that this region contains genes responsible for reproductive isolation. Regions adjacent to centromeres are known to experience less recombination in several species and it has recently been suggested that low recombination rates can facilitate the accumulation and maintenance of isolation genes in partially isolated populations. Therefore, we measured the recombination rate in division 5D/6 directly and estimate that it is at least 16-fold reduced across this region compared to the remainder of the X chromosome. Additionally, sequence data from four loci from field-collected mosquitoes from several West African countries show very strong differentiation between the molecular forms in division 5D/6, whereas none was observed in two loci elsewhere on the X chromosome. Furthermore, genetic variation was substantially lower in division 5D/6 compared to the two reference loci, and the inferred genealogies of the division 5D/6 genes show patterns consistent with selective sweeps. This suggests that the reduced recombination rate has increased the effect of selection on this region and that our data are consistent with the hypothesis that reduced recombination rates can play a role in the accumulation of isolation genes in the face of gene flow.
RECENT studies of genetic differentiation between closely related species have lead to the realization that incipient reproductive isolation is often a property of small genomic regions, rather than of entire genomes (e.g., Harrison 1990; Wu 2001). Support for this view comes from observations that selection can prevent gene flow of regions containing isolation genes between populations, while other regions are exchanged freely. For example, Machado et al. (2002) have shown that there is a good correspondence between the location of isolation genes and the level of genetic differentiation between Drosophila pseudoobscura and its close relatives. Similarly, Rieseberg et al. (1999) and Noor et al. (2001a) have shown a lack of gene flow for inversions containing isolation genes, but not for regions lacking such genes. Therefore, genomic regions characterized by strong genetic differentiation between incompletely isolated taxa that undergo gene flow are expected to contain isolation genes.
Differentiation between members of the Anopheles gambiae complex, which contains two of the most important vectors of malaria, appears to fit the notion of mosaic genomes. Levels of differentiation between An. gambiae and An. arabiensis vary between different chromosomal regions (Besansky et al. 2003), and different areas of the genome vary in their ability to introgress between the two species (della Torre et al. 1997; Slotman et al. 2005). In addition, several forms of An. gambiae s.s. have been defined, and differentiation between these forms appears to be limited to a few areas of the genome (Favia et al. 1997; Lanzaro et al. 1998; Gentile et al. 2001; Mukabayire et al. 2001; Wang et al. 2001; Stump et al. 2005a,b; Turner et al. 2005).
An. gambiae s.s. was subdivided into several distinct forms on the basis of the observation in West Africa that inversion karyotypes on the right arm of the second chromosome (2R) deviate strongly from Hardy–Weinberg equilibrium; i.e., a large excess of homozygotes is present (data summarized by Touré et al. 1998). On the basis of these observations several chromosomal forms of uncertain taxonomic status were proposed: Savanna, Mopti, Bamako, Forest, and Bissau (Coluzzi et al. 1985). Karyotypes that could be indicative of hybrids between forms have been observed, although it is possible that some inversion karyotypes float in the “wrong” form at low frequency (Touré et al. 1998; della Torre et al. 2002).
Attempts to identify fixed nucleotide differences between the chromosomal forms in Mali led to the definition of the molecular forms M and S, which are based on two intergenic spacer types of the X-linked rDNA (Favia et al. 1997). In Mali and Burkina Faso, the M form corresponds to the Mopti chromosomal form, whereas the S molecular form corresponds to both the Bamako and the Savanna chromosomal form. Outside of Mali and Burkina Faso, however, the close correspondence between chromosomal form and molecular forms breaks down (della Torre et al. 2001), and the Savanna chromosome type can carry the M rDNA type. Additionally, both the M and S type rDNA are present in the Forest chromosomal form, and this form can no longer be considered a single taxonomic unit (Wondji et al. 2002). Other attempts to identify fixed differences between the M and S molecular forms elsewhere in the genome have failed (Gentile et al. 2001; Mukabayire et al. 2001).
Initially, two studies using one and two microsatellite loci close to the rDNA suggested that genetic differentiation between M and S forms extends to division 6, a region adjacent to the rDNA (Wang et al. 2001; Lehmann et al. 2003). These observations instigated several recent studies that investigated this region in more detail. Stump et al. (2005a) showed that eight of nine microsatellite loci in part of division 5 and division 6, close to the centromere and the rDNA locus, are significantly differentiated, whereas other X-linked microsatellite loci are not. Barnes et al. (2005) have also recently shown significant differentiation in this region using SINE insertion polymorphisms. Finally, Stump et al. (2005b) very recently presented sequence data showing that differentiation between the M and S forms on the X chromosome is limited to divisions 5 and 6. A more extensive recent study using whole-genome DNA microarrays identified division 6 as the main of two or three small genomic areas containing fixed differences between homokaryotypic M and the S in several villages in Cameroon (Turner et al. 2005). In short, these data are consistent with severely restricted gene flow in division 6 and part of division 5, whereas gene flow is extensive elsewhere in the genome, providing evidence that isolation genes are located in this region. On the basis of these studies, it now appears that the molecular forms M and S define the reproductive units within An. gambiae, whereas there is little evidence for the relevance of the 2R karyotypes with respect to reproductive isolation.
Hybrids between the M and S molecular forms are rare (della Torre et al. 2001; Taylor et al. 2001), and very strong assortative mating has been observed between the molecular forms (Tripet et al. 2001). This indicates that premating isolation exists between the forms in nature, even though no reproductive isolation is apparent in laboratory crosses (Di Deco et al. 1980). Nonetheless, this premating isolation is incomplete and the number of observed M/S hybrids indicates that gene flow between the two molecular forms is high enough to prevent the accumulation of genetic differences by drift (Taylor et al. 2001; Tripet et al. 2001). This raises the question of how reproductive isolation between these forms was established and maintained in the face of gene flow.
Recently proposed formulations of chromosomal speciation offer novel explanations for the evolution of reproductive isolation between populations undergoing gene flow. These hypotheses are similar and propose that the reduced recombination rate associated with inversions plays a crucial role in the evolution of reproductive isolation by linking loci that prevent gene flow between the populations in both directions (Coluzzi 1982; Noor et al. 2001b; Rieseberg 2001; Navarro and Barton 2003; Butlin 2005). That low recombination rates have played a role in the isolation of the M and S molecular forms of An. gambiae was recently also argued by Stump et al. (2005b), who concluded that loci in the centromeric region of the X chromosome show the reduced nucleotide polymorphism expected in regions with lowered recombination rates.
Reduced recombination rates around the centromere have been observed in several species (Jones 1987), although exceptions, such as D. mauritiana, are known (True et al. 1996). Therefore, we measured the recombination rate across division 5D/6 directly and compared it to the rate across much of the rest of the X chromosome. We show that the recombination rate across division 5D/6 is much reduced. We also present sequence data for four introns in division 5D/6 from several countries in West and Central Africa in the M and S forms. These data indicate genetic differentiation between the M and S form in a wide geographic area, with much of the genetic variation within forms being shared across large distances. A comparison with sequence data from two other X-linked loci also suggests that the low recombination rate has affected the pattern of genetic variation in the loci located in division 5D/6, possibly by increasing the effects of selection. These results are consistent with hypotheses that reduced recombination rates could have played a facilitating role in the evolution of reproductive isolation between these sympatric taxa.
MATERIALS AND METHODS
Strains and crosses:
The S form strain Kisumu (S) was collected from Kisumu in Kenya in 1969. An M form strain (M) was collected from the village of N'Gabacoro-Droit in Mali in 2003 and was started from a single gravid female. Larvae were reared in distilled water at 27° and were fed on pulverized fish food. Adult mosquitoes were kept in 1-gal cardboard containers at 27° and 70% relative humidity and fed a 5% sugar solution. Female mosquitoes were blood fed once on hamsters before oviposition.
S females were mated with M males (SM), as well as the reciprocal (MS). SM F1 females were backcrossed to S males, resulting in SMS backcross progeny. MS F1 females were backcrossed to M males, resulting in MSM backcross progeny. The backcross progeny were genotyped for rDNA type and microsatellite loci. The microsatellite loci were previously tested on 16 females from the S and M strains, and none of the loci used in this study shared alleles between strains.
Mosquito collections:
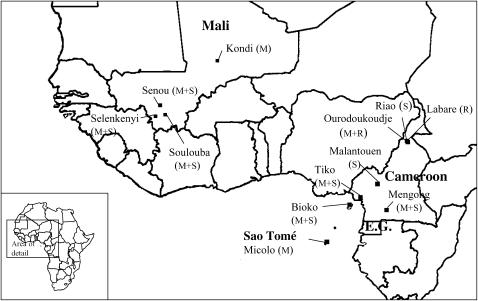
Female mosquitoes were collected from four villages in Mali, six villages in Cameroon, one site on the island of Bioko (Equatorial Guinea), as well as from a site on the island of Sao Tomé (Figure 1). In Bioko, adult female mosquitoes were collected using aspirators and human landing catches. In the remainder of the collection sites, resting female adults were collected indoors using aspirators. All collections were performed in 2002 and 2003.
Figure 1.—
Geographic location of mosquito collection sites. M, molecular form M; S, molecular form S; R, An. arabiensis.
Karyotype analysis:
Abdomens of half-gravid females from all locations except Bioko were preserved in Carnoy's fixative. The remainder of each carcass was preserved in alcohol for DNA extraction. Ovaries were prepared for chromosome analysis following della Torre (1997) and karyotypes were scored using a phase-contrast microscope according to Coluzzi et al. (1979). Karyotyped individuals were assigned to chromosomal form following Touré et al. (1998). The vast majority of the samples from Mali were assigned to the Mopti, Savanna, or Bamako chromosomal form. Karyotypes were not available for specimens from Bioko and Sao Tomé. The specimens from Cameroon were classified as the Forest chromosomal form. This form is characterized by the standard arrangement on the second chromosome and can belong to both the M and the S molecular form. Additionally, the karyotype of two S-form specimens from Malantouen, Cameroon, could be indicative of either the Savanna or the Forest chromosomal form.
Molecular analysis:
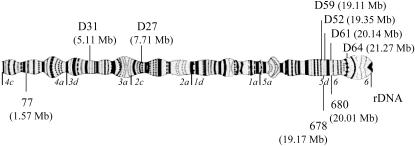
DNA was extracted using a standard extraction protocol (Post et al. 1993). A whole-genome amplification was performed on several of the field-collected samples using the GenomePhi DNA amplification kit (Amersham Bioscience, Piscataway, NJ). Species and molecular-form PCR diagnostics were performed following Fanello et al. (2002). Two of the microsatellites (AGXH77 and AGXH678) used in our study were taken from Zheng et al. (1996) and one novel microsatellite marker (680) was developed on the basis of the An. gambiae genome sequence (Holt et al. 2002). Primer sequences for locus 680 are presented in supplemental Table 5 at http://www.genetics.org/supplemental/. The locations of the microsatellite loci are indicated in Figure 2. Microsatellites were amplified using fluorescently labeled primers on a PTC-200 thermal cycler (MJ Research, Watertown, MA). PCR products were run on an ABI 3100 capillary sequencer (Applied Biosystems, Foster City, CA). The gels were analyzed using Genescan analysis software (Applied Biosystems) and Gentoyper DNA fragment analysis software (Applied Biosystems).
Figure 2.—
Position of microsatellite loci (below) and introns (above) on the polytene X chromosome. Numbers in parentheses indicate position of the loci according to the An. gambiae genome sequence (Holt et al. 2002). The X chromosome of An. gambiae is acrocentric and the centromere is located near the rDNA. Italics indicate the borders of chromosomal division. Note that borders of subdivisions are not indicated and that locus 680 is located on the border of divisions 5 and 6. Furthermore, all An. gambiae are fixed for an inversion on this chromosome with respect to other species in the An. gambiae complex, inverting divisions 1–4.
Four putative genes, here named D52, D59, D61, and D64, located close to the rDNA locus were taken from the An. gambiae genome sequence for further study (Holt et al. 2002). These genes were selected on the basis of the presence of an intron of 350–550 bp in length. For comparison, two introns of putative genes located in divisions 2 and 3, here named D31 and D27, respectively, were also included in the study (Figure 2). Primers annealing to flanking exons were designed using Primer Express version 2.0.0 (Applied Biosystems) and are presented in supplemental Table 5 at http://www.genetics.org/supplemental/. The collection site and molecular form of the specimens for which these six introns were amplified are presented in Table 1.
TABLE 1.
Sample sizes for each gene and collection site
| Gene
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D31
|
D27
|
D59
|
D52
|
D61
|
D64
|
||||||||||||||
| Species/form: | R | M | S | R | M | S | R | M | S | R | M | S | R | M | S | R | M | S | |
| Mali | |||||||||||||||||||
| Selenkenyi | 4 | 5 | — | 5 | 5 | — | 4 | 5 | — | 7 | 1 | — | 5 | 8 | — | 3 | 2 | ||
| Kondi | — | — | — | — | — | — | — | 3 | — | — | 3 | — | — | 5 | — | — | 4 | — | |
| Senou | — | — | — | — | — | — | — | 1 | 2 | — | 1 | 3 | — | 1 | 3 | — | 1 | 3 | |
| Soulouba | — | — | — | — | — | — | — | 1 | 3 | — | 2 | 4 | — | 2 | 4 | — | 2 | 3 | |
| Cameroon | |||||||||||||||||||
| Tiko | — | 4 | 5 | — | 5 | 5 | — | 5 | 3 | — | 5 | 4 | — | 5 | 4 | — | 5 | 3 | |
| Malantouen | — | — | 2 | — | — | — | — | — | 2 | — | — | 2 | — | — | 2 | — | — | 2 | |
| Labare | 2 | — | — | 2 | — | — | 1 | — | — | 1 | — | — | 3 | — | — | 3 | — | — | |
| Mengong | — | — | — | — | — | — | — | — | 1 | — | — | 1 | — | 1 | 1 | — | 1 | 2 | |
| Riao | — | — | — | — | — | — | — | — | 1 | — | — | 2 | — | — | 1 | — | — | — | |
| Ourodoukoudje | — | — | — | 1 | — | — | 2 | 1 | — | 2 | 2 | — | 2 | 1 | — | 2 | — | — | |
| Equatorial Guinea | |||||||||||||||||||
| Bioko | — | 5 | 4 | — | 6 | 4 | — | 6 | 4 | — | 6 | 4 | — | 6 | 4 | — | 4 | 4 | |
| Sao Tomé | |||||||||||||||||||
| Micolo | — | — | — | — | — | — | — | 2 | — | — | 3 | — | — | 4 | — | — | 3 | — | |
| Total | 3 | 13 | 14 | 3 | 16 | 14 | 3 | 23 | 21 | 3 | 29 | 21 | 5 | 30 | 27 | 5 | 23 | 19 | |
R, An. Arabiensis; M, molecular form M; S, molecular form S. A dash indicates unavailability of data.
PCR products were purified using the Qiaquick PCR purification kit (QIAGEN, Chatsworth, CA). PCR products were subsequently ligated into the pCR 2.1 Topo vector (Invitrogen, San Diego). Following transformation into competent cells, colonies with plasmids carrying inserts were selected for incubation. Minipreps were performed using Qiaprep spin (QIAGEN). The presence of inserts was confirmed using PCR. Sequencing reactions were performed on the purified plasmids, and sequencing reactions were run on an ABI 3100 Genetic Analyzer (Applied Biosystems). For D64, D59, D27, and D31, a single insert from each mosquito was sequenced. For D52 and D61, multiple clones were sequenced for a few individuals and two alleles were obtained for four S-form specimens for D61 and for two S- and one M-form specimen for D59.
PCR error:
PCR products were cloned before sequencing because of the presence of indels and polymorphisms. This procedure led to the incorporation of some PCR error. Three different clones were sequenced for five individuals for genes D61 and D52, and, assuming that individuals for which three different sequences were obtained are heterozygous, we observed five errors in 5376 bp of sequence. This translates into an error rate of 0.00093 substitutions/bp. This is somewhat higher than the PCR rate estimated by comparing sequences obtained directly from PCR products vs. clones (0.00061 substitutions/bp) (Kobayashi et al. 1999) and almost identical to the PCR error rate (0.0011/bp) estimated by Simard et al. (2007). If it is instead assumed that sequences that varied by a single base pair are from homozygous individuals, the error rate is 0.00107 substitution/bp, which is very similar to our previous estimate. Taking 0.00093 as our minimum estimated error rate, we expect the following number of erroneous substitutions in our total data sets: 10.7 (D31), 11.0 (D27), 16.8 (D59), 17.3 (D52), 23.1 (D61), and 11.9 (D64). However, these are expected to be present almost exclusively as singletons and therefore do not greatly influence most statistics used to analyze our data. However, data sets were also corrected for PCR error and most analyses were performed using both the corrected and the uncorrected data sets. Corrected data sets were constructed by randomly replacing a number of singletons in the data set equal to the number of expected PCR errors.
Analysis:
Sequences were aligned using the clustal W method (MegAlign, DNASTAR) and adjustments in the alignment were made on the basis of manual inspection. All exon portions of the sequences were removed prior to analyses to avoid biasing comparisons of nucleotide diversity. The amplified portion of D52 consists of two introns, interrupted by a 74-bp stretch of putative exon sequence. This exon was also removed from the analyses. The number of base pairs included in the analyses was as follows for each gene: D31, 425 bp; D27, 409 bp; D59, 411 bp; D52, 372 bp; D61, 436 bp; and D64, 306 bp.
Genealogical relationships between sequences were reconstructed on the basis of an uncorrected data set using the TCS version 1.18.mac software package (Clement et al. 2000). This program implements the statistical parsimony method developed by Templeton et al. (1992) and is particularly appropriate for population data. In contrast to traditional phylogenetic reconstruction methods, no assumptions are made about the absence of the ancestral sequence or recombination. The program also identifies the most probable ancestral sequence among the samples on the basis of coalescent theory (Donnelly and Tavaré 1986; Castelloe and Templeton 1994). All networks were reconstructed using the 95% parsimony criterion and gaps were treated as single substitutions.
Population pairwise FST values were calculated using Arlequin version 2 (Schneider et al. 1997) using both the corrected and the uncorrected data set. Each pairwise FST was tested for whether the observed value was significantly different from zero using 10,000 permutations. Gaps in the alignment were excluded from the analysis.
The average number of substitutions per site (π) was calculated using DnaSP version 4.0 (Rozas et al. 2003), following Nei (1987). To avoid biasing our estimate of genetic variation, only one randomly selected allele was included in the analysis for the few individuals where multiple clones were obtained. Using DnaSP, empirical distributions of the nucleotide diversity (π) were obtained on the basis of 10,000 coalescent simulations based on the expected heterozygosity (θ) of loci D27 and D31. On the basis of these distributions, P-values of the observed π for loci in division 5D/6 were obtained to evaluate if these were significantly different from those expected for genes D27 and D31.
Using polymorphism and divergence values provided by DnaSP, Hudson–Kreitman–Aguadé tests were performed using the program MLHKA (Wright and Charlesworth 2004). The HKA test compared the combined four loci in division 5D/6 to loci D27 and D31. The test was performed using Markov chain lengths of 100,000 and six runs of the test were performed to assure convergence. Both the corrected and the uncorrected data sets were used for this analysis.
The number of polymorphic sites, divergent sites, and the ratio of polymorphic-to-divergent sites are based on the values provided by DnaSP using the same number of sequences for all loci. These sample sizes were 13, 14, and 3 for M, S, and An. arabiensis populations, respectively, and samples from the same populations were used for all genes in most cases. These numbers were derived from the corrected data set. The ratio of polymorphic-to-divergent sites is not sensitive to which singletons were chosen for replacement, and thus the corrected data set provides a more accurate estimate.
Rm, the number of recombination events that can be parsimoniously inferred from our sample, was estimated using the “four-gamete test” method described by Hudson and Kaplan (1985), as implemented in DnaSP. The sequence data presented in this article have been submitted to GenBank under accession nos. EF056793–EF057064.
RESULTS
Recombination rate between M and S strains:
A total of 196 females and 157 males from the SMS backcross were genotyped for the rDNA and for microsatellite loci 678 and 680. In the vast majority of the cases, both locus 678 and locus 680 were successfully amplified, but in all cases a genotype was obtained for at least one locus. Additionally, 183 MSM backcross females were genotyped for rDNA type and the two microsatellite loci. In a total of 536 genotyped individuals, not a single recombination event was observed, either between the rDNA and locus 680 or between loci 678 and 680.
Although these data cannot provide an estimate of the recombination rate, it is possible to calculate the minimum recombination fraction (c) that is consistent with zero observed recombination events. In other words, what is the recombination rate at which there is a 5% change of observing zero recombination events in 536 samples? This probability can be calculated using the binomial probability function and equals 0.00560. This represents the maximum “expected” recombination fraction consistent with our results. Because of the possibility of double crossovers, the recombination fraction, which measures the fraction of observed recombinant genotypes, is not identical to the genetic or map distance m, which measures the number of crossing-over events. However, a correction is not necessary if the recombination fraction c is very low, that is, when c ≈ m. Therefore, the map distance between the rDNA and loci 678 and 680 is at most 0.560 cM.
The rDNA locus in An. gambiae is located in the heterochromatin close to the centromere of the X chromosome (Collins et al. 1989). This heterochromatin is not included in the An. gambiae genome sequence, and therefore the number of base pairs separating the rDNA locus from loci 678 and 680 is not known. However, the portion of euchromatin between locus 678 and the rDNA locus spans ∼2.97 Mb (Holt et al. 2002). Therefore, the recombination rate in this region is at most (0.560 cM/2.97 Mb) = 0.189 cM/Mb.
To compare the recombination rate of this region to that of the remainder of the X chromosome, genotypes at locus 77 were obtained for 189 MSM backcross females. This locus is located at the opposite end of the X chromosome, separated from locus 678 by ∼17.5 Mb (Figure 2). Of 189 individuals, 74 carried a recombinant genotype. Therefore, c = 74/189 = 0.392. Two map functions are used frequently to transform the observed fraction of recombinants to the map distance. Kosambi's map function (Kosambi 1944) allows for a modest amount of interference, whereas Haldane's map function does not (Haldane 1919). Therefore, Kosambi's map function results in a smaller genetic distance than Haldane's map function and its use is more conservative for our purpose. Using Kosambi's function, the map distance between locus 77 and 678 is 52.8 cM. This translates to an average recombination rate of ∼3.02 cM/Mb. Therefore, we estimate that the recombination rate in division 6 of the X chromosome is reduced at least 16-fold compared to the average recombination rate over most of the remainder of the X chromosome.
Genetic variation in intron sequences:
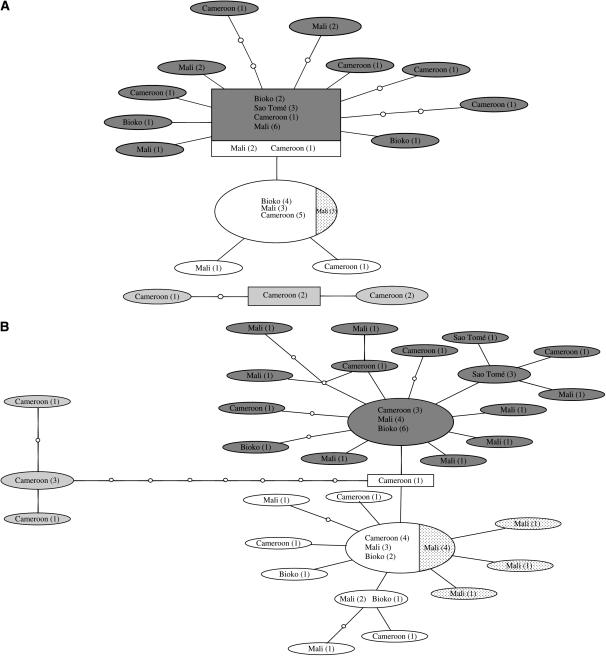
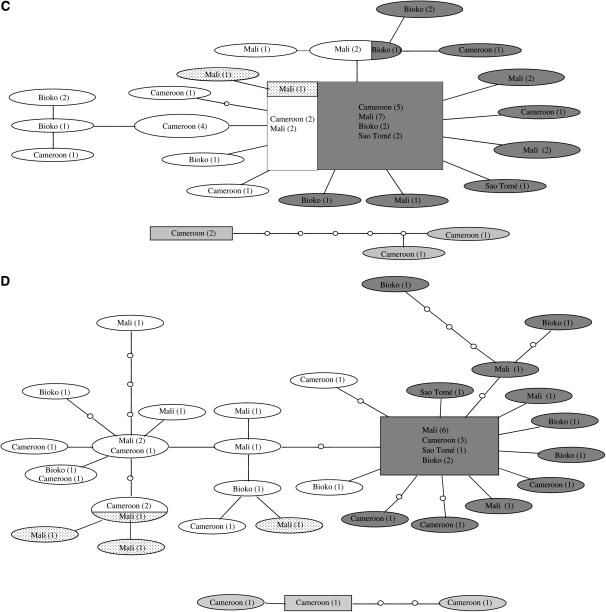
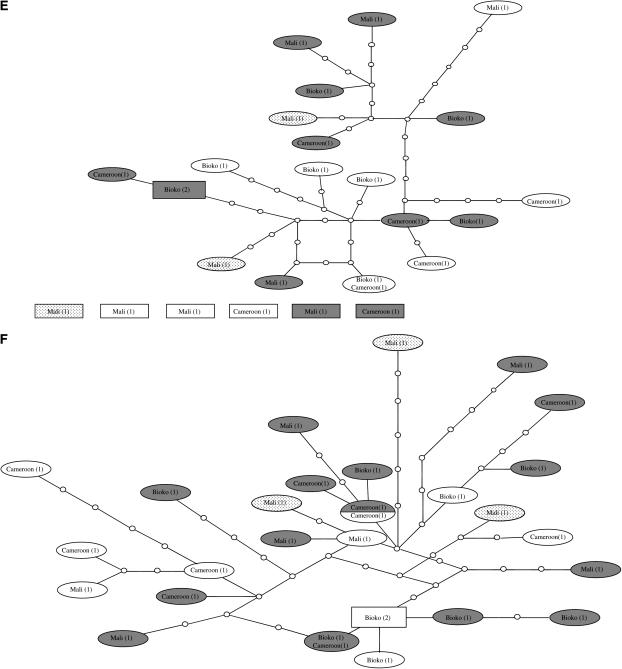
The inferred genealogies of the genes from division 5D/6 provide largely similar pictures (Figure 3, A–D). The S molecular form is clearly differentiated from the M molecular form, although three of the four genes share alleles between forms. However, this shared allele is always the most common, presumably ancestral allele. In D61 (Figure 3B), the M and S molecular forms are separated by one substitution, which may represent a fixed difference between the M and S forms. In all four genes, variation within forms is shared over a large geographic area and the most common allele is always present in Cameroon, Bioko, and Mali. Following the 95% parsimony criterion, the arabiensis alleles could be connected only to the main network for locus D61 (Figure 3B). These alleles connect to an S-form allele that is intermediate between the most common M- and S-form alleles. Therefore, the position of these alleles is not very informative regarding the possible ancestry of either form.
Figure 3.—
Networks of genealogical relationships of haplotypes of six introns in An. gambiae s.s. and An. arabiensis based on statistical parsimony (Templeton et al. 1992). Dark shading, M molecular form; no shading, S molecular form; dotted, Bamako chromosomal form; light shading, An. arabiensis. The number of haplotypes is indicated in parentheses. The putative ancestral haplotype, as calculated by TCS, is indicated by a rectangle. (A) D64, (B) D61, (C) D52, (D) D59, (E) D27, (F) D31. Nodes indicated by open circles represent missing haplotypes, i.e. haplotypes that were not sampled. Each branch between nodes represents one substitution. Intersections without an open dot are not haplotype nodes.
Pairwise FST values between the M and S molecular forms in three localities, i.e., Bioko, Tiko, and Selenkenyi were calculated if at least four sequences were available for both forms (Table 2). Results for the corrected and uncorrected data sets were highly similar and only the results based on the uncorrected data set are presented. Despite the very small sample size per population, seven of eight comparisons showed significant differentiation between forms in the genes located in division 5D/6. In contrast, no significant genetic differentiation was detected between the M and S forms in any of the three populations for genes D31 and D27. Within forms, pairwise FST values were also calculated between these three populations (i.e., a total of 36 comparisons), but no significant genetic differentiation within forms was detected. Therefore, all samples from each form were subsequently pooled. Using this complete data set, genetic differentiation was highly significant between the M and S forms in all division 5D/6 genes, ranging from 0.135 to 0.515, whereas no differentiation between forms was detected in D31 and D27.
TABLE 2.
Differentiation between the M and S molecular forms of An. gambiae
|
FST
|
||||||
|---|---|---|---|---|---|---|
| D31 | D27 | D59 | D52 | D61 | D64 | |
| Bioko | 0 | 0 | 0.187 (0.039) | 0.462 (0.015) | 0.694 (0.005) | 0.667 (0.026) |
| Tiko | 0 | 0.070 (NS) | — | 0.133 (NS) | 0.428 (0.008) | — |
| Selenkenyi | 0 | 0 | 0.567 (0.007) | 0.534 (0.007) | 0.535 (0.009) | — |
| All populations | 0 | 0.007 (NS) | 0.380 (0.000) | 0.135 (0.000) | 0.515 (0.000) | 0.396 (0.00) |
Significant P-values are indicated in parentheses. Negative FST values were converted to 0. Comparisons of populations with fewer than four sequences were not included in the analysis. NS, not significant.
The Bamako chromosomal form, which is contained within the S molecular form, has an unclear status with respect to other S-form populations (Slotman et al. 2006). We included several Bamako specimens in our study to examine their position within a genealogical network including S and M specimens from several different populations. The Bamako specimens cluster together within the S molecular form samples in all but one case. In locus D59, one of the Bamako alleles clustered with other S-form alleles. Therefore, we found no evidence that the Bamako chromosomal form is differentiated from other S forms in division 5D/6, although we did not examine this issue rigorously. The status of the Forest chromosomal forms with respect to the other chromosomal forms is also not clear. The M and S specimens of the Forest chromosomal form clustered with their respective molecular form. However, within the M and S forms, Forest specimens did not cluster (Figure 3, B and D), suggesting a lack of differentiation between Forest M and S and other M or S molecular form specimens in division 5D/6.
One feature of the inferred genealogies of all four loci, although locus D59 in the S form is an exception (Figure 3D), is the presence of one allele at high frequency, surrounded by alleles at low frequency that differ from the most common allele by one or few substitutions. This pattern could be indicative of a very strong bottleneck or a selective sweep in which one allele was driven to fixation, followed by the accumulation of several new variants (Aguadé et al. 1989). The pattern of variation in genes D27 and D31 in division 2 and 3, respectively, is very different from that in division 5D/6 genes. No alleles were found at high frequency in either D31 or D27. It should be pointed out that the following number of singletons are expected to be due to PCR error: (D64) M form 6.5; S form 5.4; (D61) M form 12.2; S form 10.9; (D52) M form 10.7; S form 6.5; (D59) M form 8.7; S form 8.0; (D31) M form 5.14; S form 5.53; (D27) M form 6.1; and S form 4.9. Given the pattern of genetic variation observed within division 5D/6 genes, the frequency of the most common allele was therefore underestimated. Inclusion of PCR error is not expected to have greatly influenced the pattern of the networks of genes D31 and D27. No high-frequency alleles were found for these two genes and most alleles are separated by several substitutions. As expected from the lack of differentiation evident from the FST values (Table 2), no clustering of alleles by form was observed. In the case of D31, six An. gambiae alleles were too diverged for connection to the network following the 95% parsimony criterion. As with the four division 5D/6 genes, little or no clustering by geography was present, indicating that much of the genetic variation is shared across the sampled geographical area. Although the TCS analysis designated an ancestral allele, this designation should be considered very tentative.
The average number of nucleotide differences per site, π, is markedly different between the four genes located in division 5D/6 and genes D31 and D27 (Table 3). In both molecular forms, the level of genetic variation was higher in D31 and D27. In the M form, π was between 5.1 and 8.5 times higher in gene D31 and between 2.4 and 4.0 times higher in gene D27 vs. genes D64, D61, D52, and D59. In the S form, π was between 3.7 and 16.1 and between 1.5 and 7.1 times higher in genes D31 and D27, respectively. Combining the data for the M and S forms does not substantially change this picture. Additionally, coalescent simulations showed that of 16 pairwise comparisons of division 5D/6 loci vs. D27 and D31, the observed value of π in division 5D/6 loci was significantly less than those expected for D27 and D31 on the basis of θ in 15 cases. In 10 of these comparisons, the P-value was <0.001. The nucleotide diversity can be corrected for the expected amount of PCR error by deducting twice the estimated PCR error rate, i.e., 0.00186. As the PCR error is expected to affect all genes equally, it does not substantially affect the comparison above. Since the PCR error is proportionally bigger in genes with low variation, inclusion of PCR error will increase the relative difference of nucleotide diversity between division 5D/6 genes and genes D27 and D31.
TABLE 3.
The observed number of nucleotide differences per site (π)
| Gene
|
||||||
|---|---|---|---|---|---|---|
| Form | D31 | D27 | D59 | D52 | D61 | D64 |
| M | 2.55 (0.50) | 1.21 (0.16) | 0.42 (0.11) | 0.30 (0.06) | 0.30 (0.06) | 0.50 (0.17) |
| S | 2.68 (0.38) | 1.13 (0.16) | 0.73 (0.10) | 0.45 (0.08) | 0.25 (0.06) | 0.16 (0.06) |
| M+S | 2.64 (0.31) | 1.19 (0.13) | 0.72 (0.08) | 0.38 (0.06) | 0.38 (0.04) | 0.47 (0.08) |
π values were multiplied by 100. Standard deviations (×100) are in parentheses.
Using the sequences from An. arabiensis, our data were also examined for signs of selection using the HKA test (Hudson et al. 1987). This test is based on the assumption that both the level of polymorphism within a species and the level of divergence between species are proportional to the neutral mutation rate (Kimura 1968, 1969). That is, the ratio of the level of polymorphism to divergence is expected to be similar for different loci under the neutral model. However, the HKA test was not significant, whether the corrected or uncorrected data set was used. A comparison of the number of divergent sites between the division 5D/6 loci and genes D31 and D27 (Table 4) suggests that the neutral mutation rate is lower for most division 5D/6 loci. However, Table 4 also indicates that the ratio of polymorphic-to-divergent sites was markedly lower for division 5D/6 than for genes D31 and D27 in seven of eight cases, suggesting that a lower neutral mutation rate does not suffice to explain the low level of polymorphism in division 5D/6.
TABLE 4.
Polymorphic sites within M and S forms, the number of divergent sites between An. gambiae and An. arabiensis, and the ratio of polymorphic-to-divergent sites
| D31 | D27 | D59 | D52 | D61 | D64 | |
|---|---|---|---|---|---|---|
| M polymorphisms | 39 | 20 | 8 | 4 | 4 | 9 |
| M divergent sites | 33.1 | 18.5 | 18.5 | 11.9 | 8.7 | 7.0 |
| M polymorphism/divergent | 1.18 | 1.08 | 0.43 | 0.34 | 0.46 | 1.29 |
| S polymorphisms | 43 | 16 | 13 | 4 | 2 | 1 |
| S divergent sites | 32.5 | 17.0 | 18.5 | 12.5 | 8.5 | 7.1 |
| S polymorphism/divergent | 1.32 | 0.94 | 0.70 | 0.32 | 0.24 | 0.14 |
Sample sizes were as follows: M form of An. gambiae: 13; S form of An. gambiae: 14; An. arabiensis: 3.
Recombination in sequence data:
Recombination events in sequence data are sometimes apparent through the presence of reticulations in networks, although these can also be caused by homoplasy, or the presence of three or more states at a single nucleotide position. A reticulation between three alleles is present in the network for locus D61 (see Figure 3B). A visual inspection of the D61 sequence alignment showed that the reticulation is caused by the presence of three character states at a single position and is therefore not evidence of recombination in this gene.
In all division 5D/6 genes, the minimum number of recombination events in our data set (Rm) was 0 for both the M and S forms, although in D69 minimum Rm was 1 when the data for the M and S populations were combined. For D27, minimum Rm was 1 and 2 for the M and S forms, respectively, and for D31, minimum Rm was 1 and 5 for the M and S forms, respectively. This confirms that recombination rates in division 6 are lower than in D27 and D31.
DISCUSSION
We have shown that recombination rates among loci of division 5D/6 of the An. gambiae genome are at least 16-fold lower than the average recombination rate over much of the remainder of the X chromosome. The estimate of the recombination rate in division 5D/6 is the highest that is consistent with zero observed recombinants; therefore the actual recombination rate in this region could be lower. Zheng et al. (1996) estimated the map distance between markers 678 and 77 to be ∼44 cM, which corresponds to 2.5 cM/Mb. Although they do not provide an estimate of the recombination rate between the rDNA and other markers, these authors did observe some recombination events between markers 412 and 678, both located in division 5D/6. However, the position of the markers 412 and 678 on their genetic map is reversed with respect to their physical position on the chromosome. These authors do not provide the genetic distance between markers 678 and 412 exactly, but it is ∼0.8 cM. These markers are 2.83 Mb apart, resulting in a recombination rate of 0.28 cM/Mb. Zheng et al. (1996) therefore also provided evidence for a relatively low recombination rate in division 5D/6.
Areas with reduced recombination typically exhibit low levels of DNA polymorphism (Aguadé et al. 1989; Begun and Aquadro 1991; Berry et al. 1991; Moriyama and Powell 1996). It is thought that this is because the increased linkage between loci in such areas can extend the effect of various types of selection on adjacent regions (Berry et al. 1991; Charlesworth et al. 1993; Gillespie 1994, 1997; Barton 1998; Comerón et al. 1999).
We also examined sequence variation in four introns in the division 5D/6 to investigate if the low recombination rate has left a footprint in the pattern of genetic variation in division 5D/6. In all four loci in division 5D/6, genetic variation was significantly less than in D31 and D27. This observation is consistent with the presence of increased linkage across division 5D/6. However, lower levels of variation could also be caused by a lower neutral mutation rate. It has been shown in several cases that a low recombination rate may reduce the neutral mutation rate (Perry and Ashworth 1999; Lercher and Hurst 2002). Pairwise HKA tests, which make use of the fact that both the level of DNA polymorphism within populations and the level of divergence between species are correlated to the neutral mutation rate, did not indicate significant differences between division 5D/6 and genes D31 and D27. However, the ratio of segregating to diverging sites was lower in the division 5D/6 genes in seven of eight cases, indicating that the division 5D/6 loci have relatively low levels of DNA polymorphism in An. gambiae. Therefore, although it appears that the genes in division 5D/6 may experience a lower neutral mutation rate, our data also suggest that positive selection may have played a role in reducing variation in this region. This would be consistent with a recent study by Stump et al. (2005b) who used the HKA test to show that the level of polymorphism in a sample of genes from divisions 5 and 6 is reduced relative to other regions of the X chromosome in An. gambiae.
Additionally, the network analyses of all four division 5D/6 introns in the M form and two of the four division 5D/6 introns in the S form indicate the presence of a single high-frequency allele with several low-frequency variants clustered around it. This is the pattern expected after a very strong bottleneck or selective sweep, when a single allele has become (almost) fixed followed by the accumulation of some new variants. This is true regardless of whether PCR error was incorporated, as in this case this would have led to an underestimation of the frequency of the most common allele. Whereas the effect of a selective sweep is local, a bottleneck would affect the level of genetic variation across the genome. Although the above pattern of variation could also fit a neutral model, both genes D31 and D27 showed a strikingly different pattern of variation. No high-frequency alleles were found for these genes, i.e., most alleles were sampled only once, and the alleles did not cluster around a single variant, suggesting that selective sweeps may have occurred in division 5D/6.
Several studies have presented evidence that An. gambiae s.s. has undergone a range expansion recently (Donelly et al. 2001; Lehmann et al. 2003). Coluzzi (1982) suggested that the expansion of this highly antropophilic species could have been linked to the expansion of human populations in sub-Saharan Africa during the agrarian revolution 10,000–4000 years ago, although this area has also been subject to large periodic droughts (Reader 1997). The fact that genetic variation in our samples was shared across a large geographic area is consistent with a recent population expansion of An. gambiae. Regardless of whether a recent range expansion took place, however, the difference in the pattern of genetic variation between division 5D/6 genes and D31 and D27 indicates that the lack of genetic variation and the presence of a single high-frequency allele in all but one case in division 5D/6 loci is not likely the result of the demographic history of An. gambiae.
That low recombination rates might facilitate speciation was realized some time ago (Felsenstein 1981), and more specifically it has been argued that the low recombination rates associated with inversions facilitates speciation (Coluzzi 1982; Noor et al. 2001b; Rieseberg 2001; Navarro and Barton 2003; Butlin 2005). In its most recent form, a low recombination rate is hypothesized to facilitate the accumulation of reproductive isolation genes between sympatric populations undergoing extensive gene flow (Noor et al. 2001b; Rieseberg 2001; Navarro and Barton 2003; Butlin 2005). Although formulated for inversions, this hypothesis could also apply to other regions of the genome that experience low recombination rates.
The strongest evidence for the importance of inversions, i.e., a low recombination rate, in speciation comes from a Drosophila literature survey compiled by Noor et al. (2001b). Most Drosophila species separated by fixed inversions are sympatric, whereas virtually all species that are not separated by inversions are allopatric. Noor et al. (2001a) and Rieseberg et al. (1999) observed that inversions that are fixed between closely related species often preferentially contain isolation genes. Additionally, Brown et al. (2004) showed that sterility of hybrids between the sympatric species D. pseudoobscura pseudoobscura and D. persimilis was almost exclusively associated with inversions, whereas sterility of hybrids between the allopatric D. pseudoobscura bogotana and D. persimilis was not.
Previous studies have shown that the molecular forms M and S are highly differentiated in division 5D/6 close to the centromere (Favia et al. 1997; Wang et al 2001; Lehmann et al. 2003; Barnes et al. 2005; Stump et al. 2005a; Turner et al. 2005). Our results confirm that highly significant differentiation between the M and S forms is present over a wide area in West and Central Africa in at least an ∼3-Mb region adjacent to the centromere on the X chromosome and that gene flow between the forms in this region is therefore severely restricted. In contrast, no differentiation between the M and the S form was present at loci D31 and D27.
On the basis of these studies, and in particular the study by Turner et al. (2005) who performed a whole-genome scan for fixed differences between molecular forms M and S, it is expected that division 5D/6 contains genes involved in reproductive isolation between the M and S molecular forms. Stump et al. (2005b), on the basis of reduced levels of nucleotide polymorphism and evidence of positive selection on genes in division 5/6, also proposed that a reduced recombination rate in the centromere region has played a role in the isolation of the M and S forms of An. gambiae. Our data, showing that the recombination rate in division 5D/6 is in fact much reduced, as well as a similar reduction in genetic variation in this region, provide additional evidence for the role of recombination in speciation in An. gambiae. This provides us with a framework for understanding the genetic differentiation and reproductive isolation between two sympatric forms of the most important malaria vector worldwide.
On the basis of the An. gambiae genome sequence, ∼75 genes, which are expected to include genes responsible for isolation between the M and S forms, are located between gene D59 and the centromere. This number may actually be larger since Stump et al. (2005a,b) found evidence of differentiation between the M and S forms extending to division 5B. If this is accurate, the number of candidate genes is at least 200. It should be pointed out that the number of genes in the area that is differentiated is important not only because these genes are candidate speciation genes, but also because a role of low recombination rates in facilitating speciation is more plausible if more genes are linked. This is because the probability of linkage between loci that prevent gene flow between populations obviously increases with the number of linked genes. In fact, the typically lower number of genes in centromere regions has been used to argue against these regions as potential sites for the accumulation of isolation genes (Navarro and Barton 2003). However, it is not known how many linked genes are required for the scenarios discussed here to be plausible.
Regions of low recombination differentiate more rapidly between recently isolated populations than other regions (Stephan and Mitchell 1992; Begun and Aquadro 1993). Presumably, this is because selective sweeps tend to fix different alleles in different populations. Therefore, the question arises whether the differentiation between the M and S forms in division 5D/6 could be the direct result of low recombination rates, rather than of the presence of isolation genes. However, we think this is extremely unlikely. Such a scenario would require the almost complete absence of gene flow between forms and the available evidence consistently indicates hybridization between the M and S forms (Taylor et al. 2001; Tripet et al. 2001; della Torre et al. 2005).
Acknowledgments
We are grateful to A. Spielman for providing samples from Bioko and D. Charlwood for providing samples from Sao Tomé. In addition, we thank F. Tripet, D. J. Begun, S. Vicario, J. Marshall, and several anonymous reviewers for providing criticism on earlier versions of the manuscript. We also thank C. Meneses for technical assistance. This work was supported by National Institutes of Health grant AI 40308 to G.C.L. and by World Health Organization Special Programme for Research and Training in Tropical Diseases (Tropical Disease Research) grant A40354 to G.C.L. and M.S.
References
- Aguadé, M., N. Miyashita and C. H. Langley, 1989. Reduced variation in the yellow-achaete-scute region in natural populations of Drosophila melanogaster. Genetics 122: 607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, M. J., N. F. Lobo, M. B. Coulibaly, N'F Sagnon, C. Costantini et al., 2005. SINE insertion polymorphism on the X chromosome differentiates Anopheles gambiae molecular forms. Insect Mol. Biol. 14: 353–363. [DOI] [PubMed] [Google Scholar]
- Barton, N. H., 1998. The effect of hitchhiking on neutral genealogies. Genet. Res. 72: 123–133. [Google Scholar]
- Begun, D. J., and C. F. Aquadro, 1991. Molecular population genetics of the distal portion of the X chromosome in Drosophila: evidence for genetic hitchhiking of the yellow-achaete region. Genetics 129: 1147–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun, D. J., and C. F. Aquadro, 1993. African and North American populations of Drosophila melanogaster are very different at the DNA level. Nature 365: 548–550. [DOI] [PubMed] [Google Scholar]
- Berry, A. J., J. W. Ajioka and M. Kreitman, 1991. Lack of polymorphism on the Drosophila fourth chromosome resulting from selection. Genetics 129: 1111–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besansky, N. J., J. Krzywinski, T. Lehmann, F. Simard, M. Kern et al., 2003. Semipermeable species boundaries between Anopheles gambiae and Anopheles arabiensis: evidence from multilocus DNA sequence variation. Proc. Natl. Acad. Sci. USA 100: 10818–10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, K. M., L. M. Burk, L. M. Henagan and M. F. Noor, 2004. A test of the chromosomal rearrangement model of speciation in Drosophila pseudoobscura. Evolution 58: 1856–1860. [DOI] [PubMed] [Google Scholar]
- Butlin, R. K., 2005. Recombination and speciation. Mol. Ecol. 14: 2621–2635. [DOI] [PubMed] [Google Scholar]
- Castelloe, J., and A. R. Templeton, 1994. Root probabilities for intraspecific gene trees under neutral coalescent theory. Mol. Phylogenet. Evol. 3: 102–113. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., M. T. Morgan and D. Charlesworth, 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134: 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement, M., D. Posada and K. A. Crandall, 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9: 1657–1659. [DOI] [PubMed] [Google Scholar]
- Collins, F. H., S. M. Paskewitz and V. Finnerty, 1989. Ribosomal RNA genes of the Anopheles gambiae species complex. Adv. Dis. Vector Res. 6: 1–28. [Google Scholar]
- Coluzzi, M., 1982. Spatial distribution of chromosomal inversions and speciation in Anopheline mosquitoes, pp 113–153 in Mechanisms of Speciation, edited by C. Barigozzi. Alan R. Liss, New York. [PubMed]
- Coluzzi, M., A. Sabatini, V. Petrarca and M.A. Di Deco, 1979. Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Trans. R. Soc. Trop. Med. Hyg. 73: 483–497. [DOI] [PubMed] [Google Scholar]
- Coluzzi, M., V. Petrarca and M. A. Di Deco, 1985. Chromosomal inversion intergradation and incipient speciation in Anopheles gambiae. Boll. Zool. 52: 45–63. [Google Scholar]
- Comerón, J. M., M. Kreitman and M. Aguadé, 1999. Natural selection on synonymous sites is correlated with gene length and recombination in Drosophila. Genetics 151: 239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- della Torre, A., 1997. Polytene chromosome preparation from Anopheline mosquitoes, pp. 329–336 in Molecular Biology of Insect Disease Vectors, edited by J. M. Crampton, C. B. Beard and C. Louis. Chapman & Hall, London.
- della Torre, A., L. Merzagora, J. R. Powell and M. Coluzzi, 1997. Selective introgression of paracentric inversions between two sibling species of the Anopheles gambiae complex. Genetics 146: 239–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- della Torre, A., C. Fanello, M. Akogbeto, J. Dossou-Yovo, G. Favia et al., 2001. Molecular evidence of incipient speciation within Anopheles gambiae s.s in West Africa. Insect Mol. Biol. 10: 9–18. [DOI] [PubMed] [Google Scholar]
- della Torre, A., C. Costantini, N. J. Besansky, A. Caccone, V. Petrarca et al., 2002. Speciation within Anopheles gambiae: the glass is half full. Science 298: 115–117. [DOI] [PubMed] [Google Scholar]
- della Torre, A., Z. Tu and V. Petrarca, 2005. A review/update on the distribution and genetic differentiation of Anopheles gambiae s.s. molecular forms. Insect Biochem. Mol. Biol. 35: 755–769. [DOI] [PubMed] [Google Scholar]
- Di Deco, M. A., V. Petrarca, F. Villani and M. Coluzzi, 1980. Polimorfismo cromosomico da inversioni paracentriche ed eccesso degli eterocariotypi in ceppi di Anopheles allevati in laboratorio. Parassitologia 22: 304–306. [Google Scholar]
- Donelly, M. J., M. C. Licht and T. Lehmann, 2001. Evidence for recent population expansion in the evolutionary history of the malaria vectors Anopheles arabiensis and Anopheles gambiae. Mol. Biol. Evol. 18: 1353–1364. [DOI] [PubMed] [Google Scholar]
- Donnelly, P., and S. Tavaré, 1986. The ages of alleles and a coalescent. Adv. Appl. Prob. 18: 1–19. [Google Scholar]
- Fanello, C., F. Santolamazza and A. della Torre, 2002. Simultaneous identification of species and molecular forms of the Anopheles gambiae complex by PCR-RFLP. Med. Vet. Entomol. 16: 461–464. [DOI] [PubMed] [Google Scholar]
- Favia, G., A. della Torre, M. Bagayoko, A. Lanfrancotti, N'F. Sagnon et al., 1997. Molecular identification of sympatric chromosomal forms Anopheles gambiae and further evidence of their reproductive isolation. Insect Mol. Biol. 6: 377–383. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J., 1981. Skepticism towards anta Rosalia, or why are there so few kinds of animals? Evolution 35: 124–138. [DOI] [PubMed] [Google Scholar]
- Gentile, G., M. Slotman, V. Ketmaier, J. R. Powell and A. Caccone, 2001. Attempts to molecularly distinguish cryptic taxa in Anopheles gambiae s.s. Insect Mol. Biol. 10: 25–32. [DOI] [PubMed] [Google Scholar]
- Gillespie, J. H., 1994. Alternatives to the neutral theory, pp. 1–17 in Non-neutral Evolution: Theories and Molecular Data, edited by B. Golding. Chapman & Hall, New York.
- Gillespie, J. H., 1997. Junk ain't what junk does: neutral alleles in a selected context. Gene 205: 291–299. [DOI] [PubMed] [Google Scholar]
- Haldane, J. B. S., 1919. The combination of linkage values, and the calculation of distance between the loci of linked factors. J. Genet. 8: 299–309. [Google Scholar]
- Harrison, R. G., 1990. Hybrid zones: windows on evolutionary process, pp. 69–128 in Oxford Surveys in Evolutionary Biology, Vol. 7, edited by D. Futuyma and J. Antonovics. Oxford University Press, Oxford.
- Holt, R. A., G. M. Subramanian, A. Halpern, G. G. Sutton, R. Charlab et al., 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298: 129–149. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., and N. L. Kaplan, 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 147: 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., M. Kreitman and M. Aguadé, 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, G. H., 1987. Chiasmata, pp. 213–244 in Meiosis, edited by P. B. Moens. Academic Press, New York.
- Kimura, M., 1968. Evolutionary rate at the molecular level. Nature 217: 624–626. [DOI] [PubMed] [Google Scholar]
- Kimura, M., 1969. The number of heterozygous nucleotide sites maintained in a finite population due to the steady flux of mutations. Genetics 61: 893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, N., K. Tamura and T. Aotsuka, 1999. PCR error and molecular population genetics. Biochem. Genet. 37: 317–321. [DOI] [PubMed] [Google Scholar]
- Kosambi, D. D., 1944. The estimation of map distances from recombination values. Ann. Eugen. 12: 172–175. [Google Scholar]
- Lanzaro, G. C., Y. T. Touré, J. Carnahan, L. Zheng, G. Dolo et al., 1998. Complexities in the genetic structure of Anopheles gambiae populations in West Africa as revealed by microsatellite DNA analysis. Proc. Natl. Acad. Sci. USA 95: 14260–14265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann, T., M. Licht, N. Elissa and B. T. A. Maega, 2003. Population structure of Anopheles gambiae in Africa. J. Hered. 94: 133–147. [DOI] [PubMed] [Google Scholar]
- Lercher, M. J., and L. D. Hurst, 2002. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 18: 337–340. [DOI] [PubMed] [Google Scholar]
- Machado, C. A., R. M. Kliman, J. A. Markert and J. Hey, 2002. Inferring the history of speciation from multilocus DNA sequence data: the case of Drosophila pseudoobscura and its close relatives. Mol. Biol. Evol. 19: 472–488. [DOI] [PubMed] [Google Scholar]
- Moriyama, E. N., and J. R. Powell, 1996. Intraspecific nuclear DNA variation in Drosophila. Mol. Biol. Evol. 13: 261–277. [DOI] [PubMed] [Google Scholar]
- Mukabayire, O., J. Caridi, X. Wang, Y. T. Touré, M. Coluzzi et al., 2001. Patterns of DNA sequence variation in chromosomally recognized taxa of Anopheles gambiae: evidence from rDNA and single-copy loci. Insect Mol. Biol. 10: 33–46. [DOI] [PubMed] [Google Scholar]
- Navarro, A., and N. H. Barton, 2003. Accumulating postzygotic isolation in parapatry: a new twist on chromosomal speciation. Evolution 57: 447–459. [DOI] [PubMed] [Google Scholar]
- Nei, M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York.
- Noor, M. A. F., K. L. Grams, L. A. Bertucci, Y. Almendarez, J. Reiland et al., 2001. a The genetics of reproductive isolation and the potential for gene exchange between Drosophila pseudoobscura and D. persimilis via backcross hybrid males. Evolution 55: 512–521. [DOI] [PubMed] [Google Scholar]
- Noor, M. A. F., K. L. Grams, L. A. Bertucci and J. Reiland, 2001. b Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. USA 98: 12084–12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, J., and A. Ashworth, 1999. Evolutionary rate of a gene affected by chromosomal position. Curr. Biol. 9: 987–989. [DOI] [PubMed] [Google Scholar]
- Post, R. J., P. K. Flook and A. L. Millest, 1993. Methods for the preservation of insects for DNA studies. Biochem. Syst. Ecol. 21: 85–92. [Google Scholar]
- Reader, J., 1997. Africa: A Biography of the Continent. Vintage, New York.
- Rieseberg, L. H., 2001. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 16: 351–358. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L. H., J. Whitton and K. Gardner, 1999. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics 152: 713–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sánchez-DelBarrio, X. Messequer and R. Rozas, 2003. DnaSP, DNA polymorphism analysis by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Schneider, S., J.-M. Kueffer, D. Roessli and L. Excoffier, 1997. Arlequin, Version 2.0. Genetics and Biometry Laboratory, University of Geneva, Geneva.
- Simard, F., M. Licht, N. J. Besansky and T. Lehmann, 2007. Polymorphism at the defensin locus in the Anopheles gambiae complex: testing different selection hypotheses. Infect. Genet. Evol. (in press). [DOI] [PMC free article] [PubMed]
- Slotman, M., A. della Torre, M. Calzetta and J. R. Powell, 2005. Differential introgression of chromosomal regions between Anopheles gambiae and An. arabiensis. Am. J. Trop. Med. Hyg. 73: 326–335. [PubMed] [Google Scholar]
- Slotman, M. A., M. M. Mendez, A. della Torre, G. Dolo, Y. T. Touré et al., 2006. Genetic differentiation between the Bamako and Savana chromosomal forms of Anopheles gambiae as indicated by amplified fragment length polymorphism analysis. Am. J. Trop. Med. Hyg. 74: 641–648. [PubMed] [Google Scholar]
- Stephan, W., and S. J. Mitchell, 1992. Reduced levels of DNA polymorphisms and fixed between-population differences in the centromeric region of Drosophila ananassae. Genetics 132: 1039–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stump, A. D., J. A. Shoener, C. Constantini, N'F. Sagnon, N. J. Besansky, 2005. a Sex-linked differentiation between incipient species of Anopheles gambiae. Genetics 169: 1509–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stump, A. D., M. C. Fitzpatrick, N. F. Lobo, S. Traoré, N'F Sagnon et al. 2005. b Centromere-proximal differentiation and speciation in Anopheles gambiae. Proc. Natl. Acad. Sci. USA 102: 15930–15935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, C., Y. T. Touré, J. Carnahan, D. E. Norris, G. Dolo et al., 2001. Gene flow among populations of the malaria vector, Anopheles gambiae, in Mali, West Africa. Genetics 157: 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton, A. R., K. A. Crandall and C. F. Sing, 1992. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132: 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touré, Y. T., V. Petrarca, S. Traoré, A. Coulibaly, H. M. Maiga et al., 1998. The distribution and inversion polymorphism of chromosomally recognized taxa of the Anopheles gambiae complex in Mali, West-Africa. Parassitologia 40: 477–511. [PubMed] [Google Scholar]
- Tripet, F., Y. T. Touré, C. E. Taylor, D. E. Norris, G. Dolo et al., 2001. DNA analysis of transferred sperm reveals significant levels of gene flow between molecular forms of Anopheles gambiae. Mol. Ecol. 10: 1725–1732. [DOI] [PubMed] [Google Scholar]
- True, J. R., J. M. Mercer and C. C. Laurie, 1996. Differences in crossover frequency and distribution among three sibling species of Drosophila. Genetics 142: 507–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, T. L., M. W. Hahn and S. V. Nuzhdin, 2005. Genomic islands of speciation in Anopheles gambiae. PloS Biol. 3: 1572–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R., L. Zheng, Y. T. Touré, T. Dandekar and F. Kafatos, 2001. When genetic distance matters: measuring genetic differentiation at microsatellite loci in whole genome scans of recent and incipient mosquito species. Proc. Natl. Acad. Sci. USA 98: 10769–10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wondji, C., F. Simard and D. Fontenille, 2002. Evidence for genetic differentiation between the molecular forms M and S within the Forest chromosomal form of Anopheles gambiae in an area of sympatry. Insect Mol. Biol. 11: 11–19. [DOI] [PubMed] [Google Scholar]
- Wright, S. I., and B. Charlesworth, 2004. The HKA test revisited: a maximum-likelihood-ratio test of the standard neutral model. Genetics 168: 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C.-I., 2001. The genic view of speciation. J. Evol. Biol. 14: 851–865. [Google Scholar]
- Zheng, L. B., M. O. Benedict, A. J. Cornel, F. H. Collins and F. C. Kafatos, 1996. An integrated genetic map of the African human malaria vector mosquito, Anopheles gambiae. Genetics 136: 941–952. [DOI] [PMC free article] [PubMed] [Google Scholar]