Abstract
The many reports of trans interactions between homologous as well as nonhomologous loci in a wide variety of organisms argue that such interactions play an important role in gene regulation. The yellow locus of Drosophila is especially useful for investigating the mechanisms of trans interactions due to its ability to support transvection and the relative ease with which it can be altered by targeted gene replacement. In this study, we exploit these aspects of yellow to further our understanding of cis as well as trans forms of enhancer–promoter communication. Through the analysis of yellow alleles whose promoters have been replaced with wild-type or altered promoters from other genes, we show that mutation of single core promoter elements of two of the three heterologous promoters tested can influence whether yellow enhancers act in cis or in trans. This finding parallels observations of the yellow promoter, suggesting that the manner in which trans interactions are controlled by core promoter elements describes a general mechanism. We further demonstrate that heterologous promoters themselves can be activated in trans as well as participate in pairing-mediated insulator bypass. These results highlight the potential of diverse promoters to partake in many forms of trans interactions.
THE regulation of a gene can be a complicated process, involving many layers of organization. In addition to being controlled by intricate intragenic interactions, the expression of a gene can be influenced by neighboring genetic elements found both in linear and three-dimensional proximity. Such elements can include other genes, insulators, sequences involved in the maintenance of chromatin states (such as the Polycomb and Trithorax response elements), and heterochromatin (reviewed by Capelson and Corces 2004; Cernilogar and Orlando 2005; Loden and van Steensel 2005; Shearwin et al. 2005). Of special interest is the potential of a gene to be influenced by the enhancers and promoters of a neighboring gene. Although mechanisms exist to constrain enhancers to their own promoter, P-element-mediated enhancer traps and transient transfection of cell lines demonstrated early on that enhancers have the capacity to interact with heterologous promoters (O'Kane and Gehring 1987; Bellen et al. 1989; Kermekchiev et al. 1991). Here we examine enhancer–promoter specificity, investigating why intergenic interactions occur in some situations but not in others.
One way to study enhancer–promoter interactions is to dissect the process by which an enhancer chooses between multiple promoters. This approach has revealed that enhancer choice can be modulated by the identity, relative strengths, and/or accessibilities of the promoters (Li and Noll 1994; Cai and Levine 1995; Scott and Geyer 1995; Merli et al. 1996; Krebs and Dunaway 1998; Ohtsuki et al. 1998; Sharpe et al. 1998) as well as the distance between an enhancer and its potential target promoters (Dillon et al. 1997; Kmita et al. 2002). Studies have also highlighted the role of transcription factors (reviewed by Kadonaga 2004), chromatin proteins (reviewed by McBryant et al. 2006), and a diverse array of chromosomal elements, such as promoter-targeting sequences, tethering elements, and facilitators, in enabling enhancers and promoters to interact across large distances (Sipos et al. 1998; Zhou and Levine 1999; Calhoun et al. 2002; Calhoun and Levine 2003; Lin et al. 2003; Chen et al. 2005; reviewed by Dorsett 1999; Sipos and Gyurkovics 2005). Still other studies have focused on the structure of the promoter itself, revealing that the promoter preference displayed by some enhancers may reflect the presence of specific arrays of core promoter elements (Ohtsuki et al. 1998; Butler and Kadonaga 2001; Conte et al. 2002) such as the TATA box, initiator (Inr; Smale and Baltimore 1989), and downstream promoter element (DPE), which bind TFIID (Burke and Kadonaga 1996; Burke and Kadonaga 1997; reviewed by Smale 2001; Smale and Kadonaga 2003).
Most studies concerning gene expression assume that enhancer–promoter interactions occur between elements located in cis on the same chromosome. However, in Drosophila, where homologous chromosomes are paired in somatic cells (reviewed by McKee 2004), such interactions can also occur in trans in a manner that is sensitive to the degree of pairing. These and other pairing-dependent phenomena represent transvection effects and have been reported at greater than a dozen loci in Drosophila (reviewed by Pirrotta 1999; Wu and Morris 1999; Duncan 2002; Kennison and Southworth 2002; see also Coulthard et al. 2005). Coupled with the growing evidence for related trans interactions in mice (Rassoulzadegan et al. 2002; Herman et al. 2003; Baarends et al. 2005; Turner et al. 2005), humans (Ashe et al. 1997; Thatcher et al. 2005; Bacher et al. 2006; Xu et al. 2006), plants (reviewed by Grant-Downton and Dickinson 2004), nematodes (Bean et al. 2004), and fungi (reviewed by Galagan and Selker 2004; see also Pratt et al. 2004; Shiu et al. 2006; Vyas et al. 2006), transvection effects argue that an understanding of gene regulation will require an understanding of trans- as well as cis-regulatory mechanisms. Below, we describe transvection at the Drosophila yellow locus, with a focus on mechanisms that determine whether a gene will hold its enhancers in cis or release them to act on a promoter in trans (Geyer et al. 1990; Morris et al. 1998, 1999a,b, 2004).
The yellow gene, required for the dark pigmentation of larval and adult cuticular structures, is ideal for gene expression studies as it is nonessential, structurally simple, and produces mutant phenotypes that are easily scored. It is located on the X chromosome and has a single promoter and transcription unit under the control of tissue-specific enhancers (Geyer and Corces 1987; Biessmann and Mason 1988; Martin et al. 1989; Gause et al. 1998; Wittkopp et al. 2002). Disruption of the promoter or coding region results in overall reduced pigmentation, while disruption of the enhancers results in corresponding tissue-specific mutant phenotypes. Interestingly, heterozygosity for some combinations of alleles produces levels of pigmentation that are greater than that observed in females homozygous for either allele (Geyer et al. 1990; Morris et al. 1999a). These examples of intragenic complementation are believed to result from transvection.
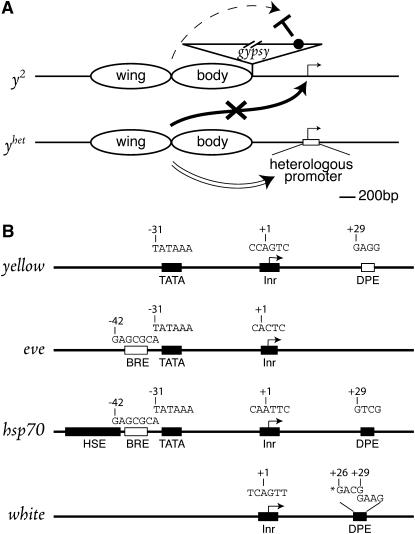
In general, the yellow alleles that participate in transvection are of two distinct classes, the first of which consists of alleles whose wing and/or body enhancers are disabled (Geyer et al. 1990; Morris et al. 1999a). A good example of this class is y2, in which a gypsy retrotransposon has inserted 700 bp upstream of the promoter, separating it from the wing and body enhancers (Figure 1; Geyer and Corces 1987). As gypsy contains an insulator that blocks enhancer–promoter interactions (Holdridge and Dorsett 1991; Jack et al. 1991; Geyer and Corces 1992; reviewed by Kuhn and Geyer 2003; Brasset and Vaury 2005), y2 flies have mutant yellow pigmentation of the wings and body. In contrast, because the gypsy insulator does not block the intronic bristle enhancer (Figure 1), y2 flies have wild-type dark bristles. Transvection occurs when an allele of this first class is heterozygous for an allele of the second class, which typically contains a mutation of the promoter region (Geyer et al. 1990; Morris et al. 1999a). One representative of this second class is y1#8, which carries a 780-bp deletion of the promoter region and gives a completely mutant phenotype (Figure 1; Geyer et al. 1990). Strikingly, females heterozygous for y2 and y1#8 are darkly pigmented as a result of the enhancers of y1#8 activating the promoter of y2 in trans (Geyer et al. 1990; Morris et al. 1998, 2004).
Figure 1.—
Enhancer action in trans. Deletion (Δ) of the promoter in y1#8 releases the wing and body enhancers (ovals) to activate (solid arrow) the promoter of the paired y2 allele. Trans-activation of the y2 promoter is detectable due to the disruption of cis-activation (dashed arrow blocked by boldface T) by the gypsy retrotransposon at −700 relative to the start site of transcription. The upstream regulatory region, promoter, first exon (solid rectangle), and 5′ intronic region of yellow, including the bristle enhancer, are drawn approximately to scale. Although the wing and body enhancers are drawn as nonoverlapping, these two elements may not be cleanly separable (Gause et al. 1998). The gypsy retrotransposon, which is 7 kbp in length, is not drawn to scale. The gypsy insulator (solid circle) is located at the 5′-end of the gypsy retrotransposon. Scale is indicated in bottom right-hand corner (200 bp).
Although the yellow wing and body enhancers are able to act in trans, they show a strong bias toward the promoter lying immediately downstream in cis, gaining the capacity to act in trans only when that promoter has been compromised (Geyer et al. 1990; Morris et al. 1999a,b). For example, the wing and body enhancers are not released when y2 is heterozygous with the y1 null allele, whose causative lesion is a simple A-to-C change in the translation initiation codon and which maintains an intact promoter. That enhancer release may depend on disruption of promoter function has also been proposed for transvection at several other loci (for example, Martinez-Laborda et al. 1992; Hendrickson and Sakonju 1995; Casares et al. 1997; Sipos et al. 1998) and, while this mechanism may not be applicable in all situations (Goldsborough and Kornberg 1996; Casares et al. 1997; Sipos et al. 1998), it suggests that at least one basis for enhancer–promoter fidelity is cis-preference (reviewed by Duncan 2002). At yellow, our understanding of cis-preference rests on observations that mutations of the core promoter elements cause the promoter of yellow to release the upstream wing and body enhancers to trans action. Thus far, three core promoter elements have been recognized at yellow: a TATA box, an Inr, and a potential DPE, and mutational analyses have demonstrated that at least two of them, the TATA box and Inr, are critical for cis-preference (Morris et al. 1999b, 2004). Importantly, demonstration of cis-preference at yellow involved introduction of mutations into the yellow promoter at its natural chromosomal location by P-element-mediated targeted gene replacement (Gloor et al. 1991; Keeler et al. 1996). As gene replacement allows the manipulation of a gene in its natural genomic context, it precludes complications arising from position effect. Accordingly, all the mutations analyzed in this study were generated through gene replacement.
Of the models proposed to explain cis-preference, two are of particular relevance. In the first, promoter competency signals cis-linked enhancers to interact exclusively with the promoter and, if compromised, releases the enhancers to act in trans. In this view, we suggested that promoter competency could reflect transcriptional strength or the integrity of the promoter as defined by the status of its core promoter elements. In the second model, the identity of the promoter, as determined by its sequence or core promoter elements, entrains an enhancer that would otherwise be able to act in trans. To distinguish between these two models, new alleles containing promoters from other genes were generated and tested for their ability to maintain cis-preference. In these hybrid alleles, the yellow promoter from position −63 to +130, including the TATA box and Inr but not the translation initiation codon at +172, was substituted by targeted gene replacement with 193 bp of the analogous regions of promoters from three other genes (Figure 2). If strict promoter identity were key for cis-preference, the heterologous promoters should fail, releasing enhancers to trans action (Morris et al. 2004).
Figure 2.—
Intact heterologous promoters maintain cis-preference. (A) Trans-activation is not detected (X) when heterologous promoters (open rectangle) maintain cis-preference (double-lined arrow) of wing and body enhancers. The first exon and intronic bristle enhancer are not depicted for the sake of simplicity. yhet is the yellow allele carrying the eve, hsp70, or white promoter; all other symbols are as in Figure 1. (B) Core promoter elements of eve, hsp70, and white promoters (drawn to scale). The sequence of core promoter elements, previously reported (solid rectangles; see text) and putative (open rectangles), are indicated above each element except for HSE (CTCGAATGTTCGCGAAA). TATA, TATA box; Inr, initiator; BRE, TFIIB binding element; DPE, downstream promoter element; HSE, heat-shock element. The two sequences mutated in the white promoter are indicated separately (*).
The three new yellow alleles that were generated, yeve, yhsp70, and yw, contained sequences from the promoters of the even-skipped (eve), heat-shock protein 70 (hsp70), and white (w) genes of Drosophila. In terms of core elements, these promoters share similarities with, but also differ from, the yellow promoter as well as each other (Figure 2B). Specifically, the eve promoter has a TATA box and Inr (Macdonald et al. 1986) as well as a potential TFIIB recognition element (BRE) (Lagrange et al. 1998), all of which were included in yeve. The hsp70 promoter has a TATA box, Inr, DPE (Burke and Kadonaga 1997), and a potential BRE, which were all included in yhsp70. In addition, it contains several heat-shock-responsive elements (Amin et al. 1987), of which only one was included in the hybrid allele. The white promoter has the least number of recognizable core promoter elements, containing only an Inr and a DPE (Kutach and Kadonaga 2000), both of which were included in yw. Significantly, cis-preference of the wing and body enhancers was maintained by all three heterologous promoters (Figure 2A), a surprising result, given the disparity, in terms of strength and composition, between the various promoters. In sum, this finding ruled out a strict role for promoter identity in the maintenance of cis-preference and strongly indicated that promoter competency is a key factor (Morris et al. 1999b, 2004).
In this report, we describe three studies that have furthered our understanding of enhancer–promoter interactions. First, we tested the generality of the model in which cis-preference is controlled by promoter competency by determining whether, reminiscent of the situation at yellow, core promoter elements are important contributors to the capacity of the eve, hsp70, and white promoters to maintain cis-preference. These experiments included quantitative real-time transcript analyses, enabling a more meaningful study of whether the competency of the promoter to maintain cis-preference reflects its transcriptional strength or overall integrity. Second, we ascertained whether the capacity to participate in trans interactions may be a general property of promoters by determining whether the heterologous promoters can be activated in trans by yellow enhancers. Finally, we addressed whether heterologous promoters can participate in a second form of transvection, which involves insulator bypass.
MATERIALS AND METHODS
Scoring of pigmentation:
Pigmentation was scored as previously described (Morris et al. 1998). Females were aged 3–4 days posteclosion and scored on a scale of 1–5 for both wing and body pigmentation, where “body” refers to the posterior border of the adult abdominal bands. A score of 1 represents a null or nearly null phenotype, while a score of 5 represents wild-type or nearly wild-type pigmentation. For consistency, experimental females were scored by comparison to control females of genotypes whose wing and body pigmentation had been previously defined: y1/y1#8 (1, 1), y2/y1 (1, 1–2), y82f29/y1#8 (3, 3), y2/y1#8 (4, 4), and y+/y1 (5, 5), where y+ is an allele that was generated by targeted gene replacement of the yh12w+ allele (see below) using confirmed wild-type yellow sequence. Comparisons were performed on females aged for the same number of days. Slight variation for females scored at the same value was occasionally observed and indicated as a range in the score. Complementation was noted to have occurred between two alleles if the wing or body pigmentation was at least one point darker than the pigmentation of females homozygous for either allele or, in the case of bypass studies, hemizygous for either allele. Hemizygous females carry the allele of interest heterozygous with Df(1)y− ac− w118, which is a deficiency, noted in the text as Df(1), that deletes the yellow gene. Flies were cultured at 25° ± 1° as previously described (Morris et al. 1998). Crosses were done with three females mated to three males in vials that were brooded at least every other day. Approximately 50–150 females were scored in two independent crosses for each experimental genotype.
Targeted gene replacement:
Targeted gene replacement was carried out as previously described (Gloor et al. 1991; Keeler et al. 1996; Morris et al. 1999b, 2004). Briefly, we targeted our gene replacement events to the yh12w+ allele, which contains a mini-w+-bearing P element inserted at +4 relative to the transcription start site of the X-linked yellow gene. The chromosome containing the yh12w+ allele also carries w1118. In general, to induce gene replacement, a plasmid source of transposase is co-injected with another plasmid containing a full-length copy of the yellow gene carrying the desired changes in and around the promoter region. Following injection, P-element mobilization by the transposase results in a double-strand break and a gap at the site of mobilization. In males, which have only one X chromosome, repair of the gap frequently uses yellow sequences from the injected plasmid. Incorporation of the desired changes into cells destined to become the germline allows recovery of new yellow alleles in the progeny of the injected embryos. Plasmid sources of template and transposase were used at concentrations of 1.0 and 0.25 mg/ml, respectively (Keeler et al. 1996; Morris et al. 1999b). Approximately 700 embryos were injected per promoter construct, with an average hatch rate of 40% and an average conversion rate of 0.4% (of total embryos injected). Candidate lines were screened by single-fly PCR using primers flanking the promoter region (y2425F, GAGCCTCCTGGCCTTACAATTTAC; y3298R, ATTTAACTTCCACTTACCATCACGCC) and/or the insertion site of the gypsy insulator (y2039F, TGCTCAAAATCACCTGCCAATAAC; y2256R, CATGCAGACAAAAATCCAGAAA). Fly lines giving products of the appropriate size were confirmed by sequence analysis of the PCR products. In addition, Southern analysis of genomic DNA digested separately with HindIII–BamHI and PstI and hybridized with a full-length yellow probe was performed to confirm the integrity of the yellow gene.
Plasmid construction:
Templates used in targeted gene replacement were full-length (7.8-kbp) yellow genes generated from the following plasmids. The pUC8yKB plasmid is a pUC8 plasmid with the 0.5-kbp KpnI–BamHI fragment from the wild-type yellow gene, including the entire promoter region. The pUC8y1KB plasmid is the same as pUC8yKB, except that it carries an A-to-C change in the translation initiation codon. The pUC8ySB plasmid includes the 3.1-kbp SalI–BamHI fragment, which includes the entire promoter region as well as the yellow enhancers. Again, pUC8y1SB is the same as pUC8ySB, except that it carries the A-to-C change in the translation initiation codon. Versions of pUC8yKB, pUB8y1KB, pUC8ySB, and pUC8y1SB were constructed with the eve, hsp70, and white promoters inserted in place of the yellow promoters and were named by adding “-eve,” “-hsp70,” and “-w,” respectively, as suffixes to the base plasmid names (Morris et al. 2004).
Mutations were introduced into the heterologous promoters by PCR, where one of the two primers was designed to carry the desired sequence change. For yw-DPE26 and yw-DPE29, primers w-DPE26-R (CACAGCACTTTGTGTTTAATTGATGGCGTAAACCGCTTGGAGCTTATGAACGAAACCGCT) and w-DPE29-R (CACAGCACTTTGTGTTTAATTGATGGCGTAAACCGCTTGGAGATGAGTCACGAAACCGCT) were used with primer w-cla-F (CGTTTTCATTTTATCGATCGCTGC) on pUC8ySB-w. The resulting PCR products were digested with ClaI–DraIII and cloned into pUC8yKB-w and pUC8y1KB-w. These plasmids were then digested with KpnI–BamHI, and the resulting KpnI–BamHI fragments were cloned into pUC8ySB. For the yeve-tata and yeve-tata-1 alleles, primers Eve-TATAF (TTTTATCGATGACGGCGGCCATTTGCCTGCAGAGCGCAGCGGCCCGGGAGGGGCGGGG) and y3070R (CAAGGATCCACCCTTTGTCCTGGAAC) were used on the plasmids pUC8ySB-eve and pUC8y1SB-eve. For the yhsp70-tata and yhsp70-tata-1 alleles, primers Hsp70-TATAF (TTTTATCGATCGCCTCGAATGTTCGCGAAAAGAGCGCCGGAGCCCGGGTAGAGGCGCT) and y3070R were used on the plasmids pUC8ySB-hsp70 and pUC8y1SB-hsp70. These four PCR products were digested with ClaI–BamHI and cloned into pUC8ySBLS4, which is a version of pUC8ySB with a ClaI site engineered at −69 relative to the transcription start site (Morris et al. 2004).
The final stage of cloning involved the pBSX plasmid, which is a modified Bluescript plasmid in which the KpnI site had been replaced by an XbaI site. The pBSXyBG plasmid is pBSX carrying the 4.7-kbp BamHI–BglII fragment of yellow. This fragment contains the majority of the coding region of yellow, which is not found in the SalI–BamHI fragment. The pUC8ySB derivatives described above were digested with SalI and BamHI, and then the SalI–BamHI fragments were cloned into pBSXyBG to generate full-length yellow templates.
The constructs containing the gypsy insulator were made by first using Asp718 to digest the three pBSX plasmids, containing the eve, hsp70, and white promoters (Morris et al. 2004), and MV108, a plasmid containing the full-length yellow gene as well as a NotI site located at −700 relative to the transcription start site. The Asp718 fragments from the first three plasmids, which contain the entire promoter region of each construct, were then cloned into MV108, now lacking its promoter region. The gypsy insulator fragment, cloned between direct repeats of loxP sites, was excised with NotI from MV144, a plasmid with a full-length yellow gene and a gypsy insulator. The gypsy insulator fragment was then cloned into the MV108 derivatives containing the heterologous promoters. Plasmids were a gift from P. Geyer (personal communication; Parnell et al. 2003), whose assistance contributed to the design of our cloning strategy.
Excision of the gypsy insulator:
Excision of the gypsy insulator was accomplished by the following crosses. Females of the genotype yhet-700gin/yhet-700gin, where het represents eve, hsp70, or white, were crossed to males of the genotype y1 w67c2/Y; nocSco/CyO[cre, mini-w+]. Resulting male progeny of the genotype yhet-700gin/Y; +/CyO[cre, mini-w+] were crossed to FMA3/Y females, where FMA3 is an attached-X chromosome. Resulting Cy+ male progeny with pigmentation darker than that of the originating yhet-700gin/yhet-700gin parent were individually crossed to FMA3/Y females. Males from these crosses were then crossed to y1 females, and the resulting heterozygous female progeny were scored for pigmentation. For heterozygotes bearing the eve and white promoters, deletion of the gypsy insulator resulted in the restoration of pigmentation to levels corresponding to scores of (5, 5) and (4, 4), respectively. Deletion of the gypsy insulator was confirmed in these lines by single-fly PCR using the y2039F and y2256R primers.
Real-time PCR:
Real-time PCR was performed on single female pupae to determine the steady-state transcript levels for each of the yellow alleles analyzed. To generate pupae, 12 mating pairs of 2- to 4-day-old adults were placed in a single vial and brooded approximately every 12 hr. Five days after the egg lay, female wandering third instar larvae were identified by the absence of testes and allowed to undergo pupation. Single pupae were harvested at 48, 72, and 96 hr postlarval collection and frozen using liquid nitrogen. At least three pupae were collected at each time point for each allele analyzed. Total RNA was isolated from single pupae using the High Pure RNA isolation kit (Roche). Isolated RNA was then used to make cDNA, using the First Strand Synthesis kit (Invitrogen). Reactions were performed using ABI Syber Green Supermix and 50 ng total cDNA per reaction. The CKIIBF (GTTTTAAATGAGCAGGTACCCAACTA) and CKIIBR (TCGTCCTCCGGTTCCAGT) primers were used to amplify the CKIIβ reference transcript (Karandikar et al. 2003), and the yRT10F (GCCGTGGATATAGGCAAAAA) and yRT10R (CGCCCCATTGGAAGTTAATA) primers were used to amplify yellow transcripts using the ABI 7300 real-time PCR machine.
RESULTS
Mutations of heterologous promoters can, but do not necessarily, release enhancers to act in trans:
We addressed the generality of the mechanism of cis-preference by determining whether disruption of the core elements of promoters other than yellow can also release enhancers to trans action. To this end, we used gene replacement technology to make four new alleles, each carrying a mutation in a heterologous promoter within the context of the yellow gene, and determined whether the resulting alleles released enhancers to act in trans. Two alleles, yeve-tata and yhps70-tata, are identical to yeve and yhsp70 except that the 6-bp TATA boxes in the eve and hsp70 promoters have been replaced with CCCGGG (Figure 3). Another two are identical to yw except that each replaces one of two 4-bp sequences that had been previously recognized as potential DPE motifs (Figure 4; Kutach and Kadonaga 2000). One sequence, GACG, beginning at +26 relative to the start site of transcription, is a perfect match to the 3′-end of the consensus A/G-G-A/T-C/T-G/A/C sequence for DPE motifs (Butler and Kadonaga 2002). The other, GAAG, beginning at +29, is less of a match, but is located at the canonical position for DPE motifs (Kutach and Kadonaga 2000). Importantly, in vitro analysis has demonstrated that replacement of the sequence at +29 with TCAT is more effective in reducing transcription than is replacement of the sequence at +26, consistent with the requirement of DPE motifs to be precisely positioned relative to the start site of transcription (Kutach and Kadonaga 2000). In our studies, we mutated the sequences at +26 and at +29 by replacing them with TCAT to generate yw-DPE26 and yw-DPE29, respectively.
Figure 3.—
Mutation of the TATA box in the eve promoter, but not the hsp70 promoter, releases enhancers to act in trans. Promoter structures of (A) y+ and y1, (B) yeve and yeve-tata-1, and (C) yhsp70 and yhsp70-tata-1 (drawn to scale). Wild-type and mutant sequences are indicated above the core promoter elements, with wild-type bases in uppercase letters and mutant bases in lowercase letters. Thick lines represent regions of yellow substituted with analogous regions from the heterologous promoters. (A) The y1 allele has a mutation of the translation initiation codon, resulting in a lack of protein production and a yellow null phenotype. The yeve-tata-1 (B) and yhsp70-tata-1 (C) alleles have 6-bp replacements of the TATA box as well as a mutation in the translation initiation codon. (D–F) Females heterozygous for y2 and the alleles in A–C. (D) y1/y2 females are phenotypically yellow in the wing and body. (E) yeve-tata-1/y2 females are pigmented in the wing and body, indicating release of enhancers from the yeve-tata-1 allele and activation of the y2 promoter. (F) yhsp70-tata-1/y2 females are phenotypically yellow, indicating that there is no significant release of enhancers.
Figure 4.—
Mutation of the DPE in the white promoter can release enhancers to act in trans. Promoter structure of (A) y+ and y1 and (B and C) yw, yw-DPE26-1, and yw-DPE29-1 (lettering and symbols are as in Figure 3). (A) The DPE of y+ and y1 is that of the natural yellow gene. (B) yw-DPE26-1 has a 4-bp replacement of sequence at +26 as well as a mutation in the translation initiation codon. (C) yw-DPE29-1 has a 4-bp replacement of the DPE at +29 as well as a mutation in the translation initiation codon. (D–F) Females heterozygous for y2 and the alleles in A–C. (D) y1/y2 is phenotypically yellow in the wing and body. (E) yw-DPE26-1/y2 females are phenotypically yellow, indicating that there is no significant release of enhancers. (F) yw-DPE29-1/y2 females are pigmented in the wing and body, indicating release of enhancers from the yw-DPE29-1 allele and activation of the y2 promoter.
The impact of the mutations was assessed by visual inspection of wing and body pigmentation in females homozygous or hemizygous for the mutations using a scale of 1–5, where 1 represents the null or nearly null phenotype and 5 represents the wild-type or nearly wild-type phenotype (Table 1; materials and methods). We found that, compared to yeve, which gives a wild-type score of (5, 5) for pigmentation in wing and body, respectively, in both the homozygous and the hemizygous state, yeve-tata produces a significant reduction in pigmentation, with scores of (3–3+, 4) and (2, 2–3) in the homozygous and hemizygous states, respectively. In contrast, mutation of the hsp70 TATA box produces only a mild reduction in pigmentation; whereas hemizygous yhsp70 females give a score of (5, 5), hemizygous yhsp70-tata females give a score of (4+–5, 4+–5). This reduction in pigmentation is also visible with homozygous yhsp70-tata females, although here the reduction manifests itself as a wild-type (5, 5) level of pigmentation rather than the abnormally dark pigmentation of homozygous yhsp70 females, which show hyperpigmentation in certain abdominal structures. Regarding mutations of the white promoter, yw-DPE29 significantly reduces pigmentation, with scores of (2+–3, 3) and (2, 1+–2) when homozygous and hemizygous, respectively, as compared to yw, which gives a score of (4, 4) in both the homozygous and the hemizygous state. In contrast, yw-DPE26 does not produce a significant reduction, giving scores of (4+, 4–4+) and (4, 4) for the homozygous and hemizygous states, respectively. These data provide in vivo confirmation that the DPE motif at +29 is more critical for transcription than is the sequence at +26.
TABLE 1.
Mutations in heterologous promoters can release enhancers
| Homozygous | Df(1) | 1 | 2 | 82f29 | |
|---|---|---|---|---|---|
| Df(1) | 1, 1 | — | 1, 1 | 1, 1 | 1, 1 |
| Homozygous | — | — | 1, 1 | 1, 1–2 | 1, 1 |
| eve | 5, 5 | 5, 5 | — | — | — |
| hsp70 | 5, 5a | 5, 5 | — | — | — |
| w | 4, 4 | 4, 4 | — | — | — |
| eve-tata | 3–3+, 4 | 2, 2–3 | 2, 2–3 | 4, 4–4+ | 2+–3, 2+–3 |
| hsp70-tata | 5, 5 | 4+–5, 4+–5 | 4–5, 4+–5 | 5, 5 | 4–5, 4+–5 |
| w-DPE26 | 4+, 4–4+ | 4, 4 | 4, 4 | 4–5, 4+–5 | 4, 4–4+ |
| w-DPE29 | 2+–3, 3 | 2, 1+–2 | 2, 1+–2 | 4, 4 | 3, 2–3 |
| eve-tata-1 | 1, 1 | 1, 1 | 1, 1 | 4, 3+–4 | 3, 2–2+ |
| hsp70-tata-1 | 1, 1 | 1, 1 | 1, 1 | 1, 1+–2 | 1, 1+–2 |
| w-DPE26-1 | 1, 1 | 1, 1 | 1, 1 | 1, 1+–2 | 1, 1+–2 |
| w-DPE29-1 | 1, 1 | 1, 1 | 1, 1 | 3+–4, 3+–4 | 3, 3 |
Pigmentation scores (wing, body) when alleles in the first column are homozygous or heterozygous with Df(1) or another allele of yellow listed in the column headings. A score of 1 represents the null or nearly null phenotype, whereas a score of 5 represents wild-type or nearly wild-type pigmentation. Pigmentation scores in the first five rows were previously described by Morris et al. (2004), with only the relevant scores shown for comparison. Note that Morris et al. (2004) used the pigmentation of wild-type Canton S females to define the score of (5, 5), whereas this study used y+ females generated by gene conversion (materials and methods). As such, phenotypes corresponding to a score of 5 in the first five rows may differ slightly from those corresponding to a score of 5 elsewhere in the table. Scores in italics indicate complementation.
yhsp70/yhsp70 females have a dusky appearance.
We next investigated whether the mutations introduced into the heterologous promoters had compromised the capacity of the promoters to maintain cis-preference, allowing the alleles to support interallelic complementation. We did this by determining whether the wing and body enhancers lying upstream of the mutated heterologous promoters could trans-activate either y2, whose wing and body enhancers are blocked by the gypsy insulator, or y82f29, whose wing and body enhancers have been entirely deleted (Morris et al. 1998, 1999a). These two alleles provide sensitive tests for trans-activation because they generate minimal pigmentation on their own. The y2 allele gives scores of (1, 1–2) and (1, 1) and y82f29 gives scores of (1, 1) and (1, 1) when homozygous and hemizygous, respectively. So that this in vivo assay would detect only the pigmentation arising from y2 or y82f29, we eliminated all pigmentation arising from the heterologous promoters through the use of alleles that are identical to the four new yellow alleles described above except that they, like y1, also contain an A-to-C change in their AUG translation initiation codon. These companion alleles, designated as such by the addition of a “-1” in the superscripted part of their name (e.g., yeve-tata-1), are predicted to be protein null and, in fact, give the lowest possible pigmentation score of (1, 1) in the homozygous and hemizygous states as well as when heterozygous with y1.
Excitingly, yeve-tata-1/y2 and yeve-tata-1/y82f29 females produce pigmentation strong enough to indicate complementation, with scores of (4, 3+–4) and (3, 2–2+), respectively (Table 1). These observations demonstrate that a promoter other than yellow can release enhancers to act in trans when harboring a discrete mutation in a core promoter element, providing support for a general importance of promoter competency in the entrainment of enhancers. Interestingly, yhsp70-tata-1/y2 and yhsp70-tata-1/y82f29 females display no increase in pigmentation compared to the minimal pigmentation associated with y2 and y82f29 (Table 1). Therefore, it further appears that while disruption of a single core promoter element can release enhancers from some promoters, it is not always sufficient.
The yellow alleles bearing the white promoter gave additional insight. In particular, the yw-DPE26-1/y2 and yw-DPE26-1/y82f29 heterozygotes support minimal, if any, complementation, while yw-DPE29-1/y2 and yw-DPE29-1/y82f29 females give complementing scores of (3+–4, 3+–4) and (3, 3) respectively (Table 1). These data indicate that disruption of the DPE motif at +29, but not the sequence at +26, is sufficient to release enhancers to act in trans. In fact, complementation arising from disruption of the motif at +29 is strong enough to be observed even with yw-DPE29, which is competent to produce yellow protein on its own. These results are significant at three levels. First, they demonstrate that, of the two potential DPE motifs, the one that is critical for cis-preference is also the one that is important for transcription. Second, they identify a third core promoter element, in addition to the TATA box and Inr, which can participate in cis-preference. Finally, the minimal consequence of mutating the sequence at +26 indicates that not all sequences of the promoter region are key for cis-preference (see also Morris et al. 2004), underscoring the significance of true core promoter elements.
Release of enhancers is not always correlated with disruption of transcription:
As described above, mutation of the eve TATA box and the white DPE motif at +29 resulted in both a reduction in pigmentation and a loss of cis-preference, while mutation of the hsp70 TATA box and white sequence at +26 affected neither pigmentation nor cis-preference. These findings suggest that loss of cis-preference may be a direct consequence of the reduced capacity of a promoter to support transcription. We had investigated this hypothesis through Northern analyses in an earlier study to determine whether promoter competence with regard to cis-preference reflected transcriptional strength or promoter integrity, but were unable to distinguish between the interpretations (Morris et al. 2004). Here, we report our use of quantitative real-time PCR (qRT–PCR) to assess transcriptional strength. These studies were carried out on individual pupae, the developmental stage at which yellow expression is required for pigmentation of the adult wing and body. As yellow transcripts accumulate, reach maximum levels and decrease all within the 4 days of pupation (Nash 1976; Geyer et al. 1990; Walter et al. 1991), we collected populations of wild-type or homozygous mutant female larvae just prior to pupation and then cultured them for another 48, 72, and 96 hr before harvesting them.
Regardless of the allele assayed or the maximum transcript level achieved, all alleles were associated with very little detectable transcript at 48 hr, an apparent peak at 72 hr, followed by lower transcript levels at 96 hr (Figure 5). For example, combining data from the independent assays of 12 wild-type (y+) female pupae, transcript levels started at 2 ± 3%, rose to 100 ± 32%, and fell to 16 ± 18% at the three respective time points, where all data are normalized to the transcript level achieved by wild-type female pupae at 72 hr. As transcript levels at 72 hr invariably represented the maximum obtained for any allele, all levels mentioned below correspond to this time point. Note, also, the considerable variation among assays of some genotypes at a single time point. This variation may reflect the difficulty of synchronizing larval and pupal cultures and, especially in the case of yhsp70-related genotypes, the nature of the promoters. Importantly, this variation did not preclude our ability to detect differences in the transcript levels produced by the different genotypes.
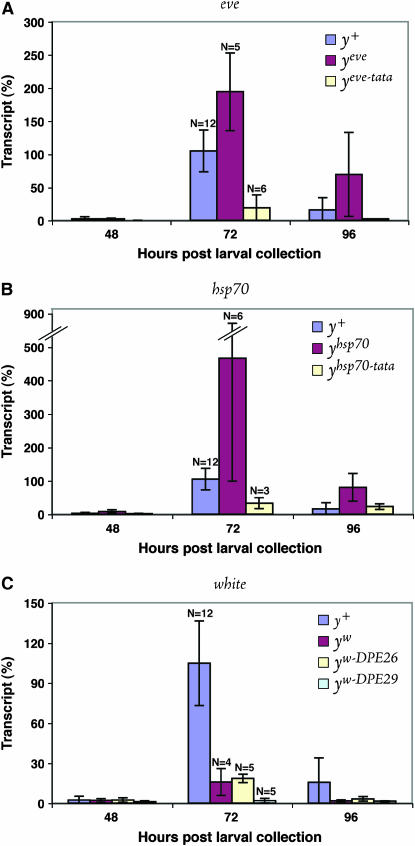
Figure 5.—
Quantitation of transcript levels from yellow alleles using qRT–PCR analysis. Female larvae were collected just prior to pupation and allowed to develop 48, 72, or 96 hr, and then RNA was collected from individual pupae. Transcript levels from 12 y+ control pupae at 72 hr were averaged, set to 100%, and then used as the standard in all panels. Each bar represents the average transcript level from at least three pupae. N, number of pupae sampled at 72 hr. Error bars indicate one standard deviation above and below the mean. (A) yeve compared to yeve-tata. Mutation of the TATA box reduces transcript levels by ∼10-fold at 72 hr. (B) yhsp70 compared to yhsp70-tata. Mutation of the TATA box reduces transcript levels by ∼14-fold at 72 hr. Diagonal double-slash (//), discontinuity in the scale. (C) yw compared to yw-DPE26 and yw-DPE29. Mutation of the DPE at +29 results in reduction of transcript levels by ∼8-fold at 72 hr, while mutation of the sequence at +26 does not significantly alter transcript levels. All reductions are significant (P < 0.05) by Student's t-test (equal or unequal variance).
Our analyses showed, first, that the three heterologous promoters drive yellow expression to vastly different degrees, with yeve, yhsp70, and yw producing transcript levels of 194 ± 59%, 467 ± 367%, and 16 ± 10%, respectively (Figure 5). These transcript levels correlate well with the wild-type and darker-than-wild-type phenotypes associated with yeve and yhsp70, respectively, and the slightly mutant pigmentation associated with yw (Table 1). Compared to our earlier Northern analyses (Morris et al. 2004), these data indicate that the eve and hsp70 promoters are more effective than previously thought. Second, we found that mutation of core elements in any of the three heterologous promoters significantly (P ≤ 0.05) reduced transcript levels to only ∼7–13% of that seen with the unmodified promoters (Figure 5). Specifically, yeve-tata reduced transcript levels from 194 ± 59% to 19 ± 20%, yhsp70-tata reduced levels from 467 ± 367% to 33 ± 16%, and yw-DPE29 reduced levels from 16 ± 10% to 2 ± 2%. This is especially remarkable in the case of yhsp70-tata, which produces wild-type pigmentation. In contrast, yw-DPE26, whose mutated sequence at +26 does not affect pigmentation, did not alter transcript levels. These findings provide molecular evidence that the release of enhancers by the compromised promoters of yeve-tata and yw-DPE29 is accompanied by a severe reduction in transcriptional capacity. Intriguingly, yhsp70-tata is also accompanied by a dramatic reduction in transcriptional strength, although it maintains cis-preference. This outcome may reflect the capacity of yhsp70-tata to generate a significant level of transcripts.
Heterologous promoters can be activated by yellow enhancers in trans:
Thus far, our studies have focused solely on the release of enhancers by the cis promoter. Below, we address the mechanism of trans action from the perspective of the receiving promoter by determining whether the eve, hsp70, and white promoters can receive input in trans from the enhancers of a paired yellow gene. Our strategy entailed the use of targeted gene replacement to replace the yellow promoter with heterologous promoters while simultaneously inserting a gypsy insulator 700 bp upstream of the transcription start site. This location of the insulator separates the wing and body enhancers from the heterologous promoter, minimizing cis-activation of the promoter and, therefore, facilitating the detection of activation in trans (Figure 6A). The gypsy insulator is ∼350 bp in length and contains 12 binding sites, which are bound by the suppressor of Hairy-wing [su(Hw)] protein and are necessary for insulator activity (Geyer et al. 1988; Parkhurst et al. 1988; Peifer and Bender 1988; Spana et al. 1988; Smith and Corces 1992). In our experiments, we used a 593-bp fragment, which contains the entire gypsy insulator (gin) and whose efficacy in blocking interactions between yellow enhancers and the yellow promoter had been previously demonstrated (Geyer and Corces 1992; Kuhn et al. 2003; Parnell et al. 2003). Following the nomenclature of Parnell et al. (2003), we call the new alleles yeve-700gin, yhsp70-700gin, and yw-700gin. Note that while a clean deletion of the enhancers from yellow alleles containing the heterologous promoters would have been ideal, the technological challenge of replacing large segments of genomic DNA by P-element-mediated targeted gene replacement argued, instead, for the use of insulators.
Figure 6.—
Heterologous promoters can participate in two modes of transvection. (A) yeve-700gin and yw-700gin (noted as yhet-700gin) are able to receive input in trans from the yellow enhancers of y1#8 (drawn to scale). The gypsy insulator (solid circle; gin) in a 593-bp fragment of DNA (inverted large triangle), inserted at −700, blocks the cis-action of wing and body enhancers (dashed arrow blocked by boldface T). (B) Insulator bypass is observed when y3c3, which lacks the promoter as well as body enhancer, is heterozygous with yeve-700gin and yw-700gin (noted as yhet-700gin) or y2 (analogous to yhet-700gin except that it contains the entire gypsy retrotransposon at −700 and a wild-type yellow promoter). Pairing with y3c3 may lead to an unpaired loop, resulting in the y2 body enhancer bypassing the gypsy insulator and activating the y2 promoter (solid arrow).
We found that the gypsy insulator effectively blocks cis-activation of the white and eve promoters by the yellow wing and body enhancers. Compared to yeve and yhsp70 (Table 1), both yeve-700gin and yw-700gin produce lower levels of pigmentation, giving scores of (3, 2+–3) or lower in hemizygous females or in females heterozygous with y1 (Table 2). Importantly, both yeve-700gin and yw-700gin nevertheless produce wild-type pigmentation in the bristles, confirming that the heterologous promoters have not been compromised by the insulator and are reasonable targets for trans-activation. Insulator activity was also observed with yhsp70-700gin; the pigmentation produced by yhsp70-700gin in hemizygous females and females heterozygous for y1 gave a score of (5, 4+–5), which is reduced relative to that produced by yhsp70 (Tables 1 and 2). However, since this reduction is too subtle to permit accurate assessment of any potential trans-activation of yhsp70-700gin, the subsequent studies of this allele are not discussed.
TABLE 2.
Heterologous promoters can be activated in trans
| Df(1) | 1 | 1#8 | 59b | 3c3 | |
|---|---|---|---|---|---|
| Df(1) | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 |
| eve-700gin | 3, 2+–3 | 2+–3, 2+a | 4–4+, 4–4+ | 4–4+, 4–4+ | 4–4+, 4–4+ |
| hsp70-700gin | 5, 4+–5 | 5, 4+–5 | 5, 5+ | 5, 5 | 5, 5+ |
| w-700 gin | 3, 2+–3 | 2+–3, 2+a | 4, 4 | 4, 4 | 4, 3+–4 |
Pigmentation scores are as described in Table 1. Pigmentation scores in the first row were previously described (Morris et al. 1999a) and listed here for comparison. Scores in italics indicate complementation.
Pigmentation of these females was slightly less than that of their hemizygous equivalents [yeve-700gin/Df(1) and yw-700gin/Df(1)], but the difference was not considered significant.
We carried out two tests to confirm that the observed decrease in pigmentation is due to the gypsy insulator. First, we demonstrated that excision of the insulator from yeve-700gin and yw-700gin results in restoration of pigmentation. For each allele, exceptionally dark progeny appearing in crosses that excised the insulator (materials and methods) were put into stock, and then PCR analyses of two such lines confirmed that the insulator was no longer present (data not shown). In the second test, we demonstrated that pigmentation can also be restored by disrupting Su(Hw) function. Here, pigmentation of yeve-700gin and yw-700gin females was found to resemble that of yeve and yw, respectively, when placed in a mutant su(Hw)v/su(Hw)f background (data not shown). In short, these studies confirmed that the reduced pigmentation observed with yw-700gin and yeve-700gin is the direct result of the gypsy insulator.
To test the ability of the heterologous promoters to be activated in trans, yw-700gin and yeve-700gin were made heterozygous with y1#8 as well as with another allele that lacks the promoter y59b. Due to their promoter-less natures, both y1#8 and y59b can release their enhancers to activate a receptive promoter in trans (Geyer et al. 1990; Morris et al. 1998). Furthermore, as neither allele produces transcripts, they facilitate the assessment of trans-activation. We find that yeve-700gin/y1#8 and yeve-700gin/y59b heterozygotes give a pigmentation score of (4–4+, 4–4+), representing significant increases in pigmentation compared to the score of (2+–3, 2+) obtained with yeve-700gin/y1. The yw-700gin allele shows similar increases in pigmentation when heterozygous with the promoter-less alleles, giving a score of (4, 4) in both cases (Table 2). These observations demonstrate that both the eve and white promoters are able to receive input in trans, suggesting that the capacity for trans interactions may be a common property of promoters (Figure 6A). That the constellation of core promoter elements of yellow differs significantly from that of white further emphasizes this point.
Heterologous promoters can participate in insulator bypass:
The yeve-700gin and yw-700gin alleles provided an opportunity to investigate a putative second mechanism of transvection, in which homolog pairing permits enhancers to bypass an insulator and activate a cis-linked promoter (Morris et al. 1998). Insulator bypass was first observed with y3c3, a null allele of yellow, which carries a deletion of the body enhancer and the promoter. Surprisingly, females carrying y3c3 in trans to y2, whose wing and body enhancers are blocked by the gypsy insulator, display a significant degree of intragenic complementation in the body. Whereas y3c3 gives a pigmentation score of (1, 1) in the homozygous or hemizygous state and y2 gives a score no greater than (1, 1–2) in either state, y2/y3c3 heterozygotes give a score of (4, 4) (Morris et al. 1998). The unexpected body pigmentation in these heterozygotes is believed to arise from the y2 body enhancer bypassing the insulator to activate its own promoter when topological constraints arising from the pairing of y2 and y3c3 compromise the insulator and/or cause the y2 body enhancer and promoter to interact productively (Figure 6B).
Since the yeve-700gin and yw-700gin alleles are similar to y2 with respect to the position of the insulator, but differ from it in terms of core promoter elements and overall identity, they allowed us to determine whether pairing-mediated insulator bypass can accommodate promoters other than that of yellow. To address this issue, yeve-700gin and yw-700gin were made heterozygous with y3c3and scored for intragenic complementation (Figure 6B). Excitingly, yeve-700gin/y3c3 and yw-700gin/y3c3 heterozygotes displayed complementation in the wing as well as the body, giving a score of (4, 4), comparable to that obtained with y2/y3c3 (Table 2). These results indicate that pairing-mediated insulator bypass is not restricted to the yellow promoter (Figure 6B). Furthermore, as the length of the gypsy retrotransposon (7469 bp) in y2 differs significantly from that of the insulator-bearing fragment (593 bp) in yeve-700gin and yw-700gin, these observations also demonstrate that insulator bypass is not exquisitely sensitive to the precise length of sequences present in one allele but not the other.
DISCUSSION
The capacity of enhancers to act over long distances raises the question of how enhancers specifically activate their own promoter rather than the promoter of another gene that has been brought into proximity. This issue has acquired heightened importance in light of the many phenomena whose mechanistic underpinnings involve the ability of genetic elements to interact in trans (for example, see reviews by Wu and Morris 1999; Duncan 2002; Chandler and Stam 2004; Grant-Downton and Dickinson 2004; Zickler 2006). In this study, we focused on Drosophila, where widespread homolog pairing theoretically places most enhancers in the position of having to make cis–trans decisions.
Previous analyses suggested that the cis–trans decision of an enhancer can be controlled by some feature of promoter competency (Geyer et al. 1990; Martinez-Laborda et al. 1992; Hendrickson and Sakonju 1995; Casares et al. 1997; Sipos et al. 1998; Morris et al. 1999b, 2004). Here, we tested the generality of this model while simultaneously addressing the relative importance of two aspects of promoter competency, transcriptional strength and promoter integrity. Although mechanistically related, these two aspects can be considered separately, in that core promoter elements may serve as a platform for recruitment of the transcription machinery without directly participating in the act of transcription. Our strategy entailed the analysis of wild-type and mutant versions of the eve, hsp70, and white promoters in the context of the yellow gene.
We found that the capacities of the eve and white promoters to maintain cis-preference were compromised when these promoters harbored mutations in the TATA box and DPE, respectively (yeve-tata and yw-DPE29). This observation indicates that the involvement of promoter competency in cis-preference applies to promoters other than that of yellow and may be a general mechanism for control of the cis–trans choice of enhancers. In contrast, mutation of the TATA box of the hsp70 promoter (yhsp70-tata) did not result in a notable loss of cis-preference, revealing that preservation of promoter integrity may not be strictly required for the maintenance of cis-preference.
To determine whether the difference in ability to maintain cis-preference between the promoters might be due to the degree to which mutation of their core elements affects transcriptional strength, we used qRT–PCR to assess transcript levels in the various lines. While disruption of the hsp70, eve, and white promoters resulted in approximately the same severity of reduction in transcript levels, the absolute transcript level obtained for the mutated hsp70 promoter was considerably higher than that obtained for the mutated eve and white promoters (33 ± 16% vs. 19 ± 20% and 2 ± 2% of wild type, respectively). These data strongly suggest that the decision of an enhancer to act in cis or trans may depend at least to some extent on the absolute strength of a promoter. They do not, however, permit us to ascertain whether there exists a threshold of transcript level below which cis-preference is necessarily lost. For example, while the mutated eve promoter releases enhancers to act in trans and produces a transcript level that is 19 ± 20% of wild type, the wild-type white promoter maintains cis-preference, yet yields a similar transcript level of 16 ± 10%.
One complicating factor for the interpretation of these results derives from the considerable variability in transcript levels between individuals of identical genotype. This variability likely reflects the technical limitations of our protocols rather than biological variability, as visualization of wing and body pigmentation shows that pigmentation is generally uniform among flies of the same genotype. Visual assessment of pigmentation further complements our transcript analyses by providing a tissue-specific estimate of gene activity. In general, promoters that maintain cis-preference (yeve, yhsp70, yw, yhsp70-tata, yw-DPE26) have pigmentation scores of (4, 4) or greater, while those that release enhancers (yeve-tata, yw-DPE29) have pigmentation scores lower than (4, 4). In conjunction with our findings regarding transcript levels, these scores are consistent with the cis–trans choice of an enhancer reflecting the absolute strength of a promoter. Note, however, that an allele of yellow where the Inr alone is mutant (yinr) (Morris et al. 1999b) confounds these categorizations. While yinr releases enhancers to act in trans, it gives pigmentation scores of (5, 5) and (5, 4) when homozygous and hemizygous, respectively. Taking current and previous data into consideration, we conclude that neither transcriptional strength nor promoter integrity alone can fully predict the cis–trans choice of an enhancer. In fact, it appears that both these aspects of promoter competency may be influential.
Our studies also addressed whether the capacity to be activated in trans may be an intrinsic property of promoters in general. Here, we replaced the yellow promoter with the eve, hsp70, and white promoters, separating them from the cis-linked wing and body enhancers by the insertion of a gypsy insulator (yeve-700gin, yhsp70-700gin, yw-700gin), and then asked whether the resulting alleles could be activated in trans. The involvement of white in pairing-sensitive phenomena (reviewed by Pirrotta 1999; Duncan 2002; Kassis 2002; Kennison and Southworth 2002) as well as intragenic complementation (Green 1959; Babu and Bhat 1980; Pirrotta et al. 1985; Zachar et al. 1985; Muller et al. 1999) suggested that its promoter would respond to yellow enhancers in trans, but similar indications have not been reported for the eve or hsp70 promoters. While technical limitations precluded analysis of the hsp70 promoter, we found that both the white and the eve promoters can be trans-activated by the yellow wing and body enhancers. This finding, which has been confirmed and extended by an independent study (L. Melnikova, M. Kostuchenko and P. Georgiev, personal communication), suggests that the capacity to be trans-activated may be a common property of promoters.
The effectiveness of the insulator in blocking cis-input to the eve and white promoters (in yeve-700gin and yw-700gin) further allowed us to investigate whether promoter identity can affect pairing-mediated insulator bypass at the yellow gene (Morris et al. 1998). Insulator bypass refers to the ability of enhancers to circumvent an insulator and activate a cis-linked promoter. While several forms of insulator bypass have been described (reviewed by Geyer and Clark 2002; Kuhn and Geyer 2003; Brasset and Vaury 2005), the pairing-mediated form observed at yellow is unique in that it can be induced without alteration to the insulator-bearing allele, disruption of factors that bind the insulator element, or change in the developmental status of the cells in which bypass occurs. Rather, it is believed to result from pairing of the insulator-bearing allele with a structurally dissimilar allele. Our studies have addressed whether this mechanism of bypass is specific for the yellow promoter. For example, the less-than-wild-type pigmentation (4, 4) resulting from bypass in y3c3/y2 females suggests that bypass is not complete in this genotype and, therefore, that promoters of a different composition or weaker transcriptional strength may prove less effective in bypass. In contrast, we find that both yeve-700gin and yw-700gin support bypass when heterozygous with y3c3. This finding indicates that pairing-mediated insulator bypass can accommodate promoters of different types and strengths.
Although our studies have focused primarily on the promoter, enhancers are at least equally important partners in trans interactions. In fact, it may be that enhancers actively seek and draw in functional promoters lying in trans, providing a driving force for the coupling of genes. Furthermore, if the cis–trans choice of enhancers is influenced by the transcriptional status of their cis-linked promoters, it is conceivable that even the enhancers of naturally silenced promoters can be released to participate in trans interactions. As such, trans action of enhancers and its modulation by gene regulatory processes may be a powerful means by which both homologous and nonhomologous interactions are modulated. This interpretation is in line with models in which homolog pairing and transcription are mechanistically intertwined (Jorgensen 1990; Cook 1997; Morris et al. 1998; McKee 2004), as well as with observations suggesting that some pairing sites are associated with transcription or regions containing enhancers and/or promoters (reviewed by McKee 2004; see also Ronshaugen and Levine 2004). Most recently, studies in yeast have suggested that transcription of rDNA genes may link sister chromatids through transcription-mediated catenation or RNA- or RNA-processing-mediated entanglements (Machin et al. 2006). As such, transcription of somatically paired genes may also lead to catenation or entanglements of homologs, perhaps via enhancer–promoter loops.
What, if anything, controls the action of enhancers once they are released? Thus far, yellow enhancers appear to be as promiscuous in trans (this study; L. Melnikova, M. Kostuchenko and P. Georgiev, personal communication) as they are in cis (Morris et al. 2004), although this promiscuity may be explained by the fact that the yellow promoter shares core promoter element motifs with each of the heterologous promoters. One approach for revealing the parameters that govern enhancers when they act in trans would be to test enhancers whose cis activities are known to be restricted to a subset of promoter types. For example, some enhancers preferentially activate either a TATA or a DPE-containing promoter, but not both (Butler and Kadonaga 2001), and retention of such specificity in trans would suggest that the mechanisms governing promoter specificity for these enhancers are the same for both cis and trans interactions. Alternatively, some enhancers may behave indiscriminately when released to act in trans, contacting other promoters or chromosomal regions without preference.
Recent work has suggested that the relevance of trans interactions to gene expression in general may be substantial. Of special interest are observations in mammals, where the abundance of long-range interactions between cis-linked genetic elements indicates that trans interactions may readily occur. In fact, the mammalian β-globin locus supports both long-range cis interactions between the locus control region (LCR) and target genes, as well as trans-activation by plasmids bearing sequences from the locus (Ashe et al. 1997; Carter et al. 2002; Tolhuis et al. 2002; Palstra et al. 2003; reviewed by Chakalova et al. 2005; Dean 2006). Similarly, in T cells, the LCR of the TH2 cytokine locus interacts with its cis-linked promoters (Spilianakis and Flavell 2004) as well as with the Ifng gene located on a separate chromosome (Spilianakis et al. 2005). Communication in trans has also been reported for imprinted loci, olfactory genes, and the X-inactivation center (Xic), where interactions involving the Prader-Willi/Angelman region (Thatcher et al. 2005) and the Xic (Bacher et al. 2006; Xu et al. 2006) are between loci on homologous chromosomes while those occurring at olfactory genes (Lomvardas et al. 2006) and between Igf2/H19 and Wsb1/Nf1 (Ling et al. 2006) are between loci on nonhomologous chromosomes. These observations represent only a few of the many phenomena in which trans interactions participate, but they serve to highlight the potential involvement of such interactions in diverse pathways. Here, we suggest that promoter competency may be one mechanism by which interactions in cis influence those in trans.
Acknowledgments
The authors especially thank P. Georgiev, L. Melnikova, and M. Kostuchenko for their generosity and sharing of information; J. Morris and P. Geyer for their expertise and advice; O. St. Lawrence and M. Hernandez of Applied Biosystems for use of the ABI 7300 real-time PCR system as well as technical assistance; the Bloomington Drosophila Stock Center and P. Geyer for reagents and Drosophila stocks; J. Bateman, R. Emmons, S. Ou, and B. Williams for discussions; and A. Moran and L. Stadelmann for technical support. This work was supported by National Institutes of Health grant GM61936 and by the Harvard Medical School.
References
- Amin, J., R. Mestril, P. Schiller, M. Dreano and R. Voellmy, 1987. Organization of the Drosophila melanogaster hsp70 heat shock regulation unit. Mol. Cell. Biol. 7: 1055–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe, H. L., J. Monks, M. Wijgerde, P. Fraser and N. J. Proudfoot, 1997. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 11: 2494–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baarends, W. M., E. Wassenaar, R. van der Laan, J. Hoogerbrugge, E. Sleddens-Linkels et al., 2005. Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis. Mol. Cell. Biol. 25: 1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu, P., and S. Bhat, 1980. Effect of zeste on white complementation. Basic Life Sci. 16: 35–40. [DOI] [PubMed] [Google Scholar]
- Bacher, C. P., M. Guggiari, B. Brors, S. Augui, P. Clerc et al., 2006. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat. Cell Biol. 8: 293–299. [DOI] [PubMed] [Google Scholar]
- Bean, C. J., C. E. Schaner and W. G. Kelly, 2004. Meiotic pairing and imprinted X chromatin assembly in Caenorhabditis elegans. Nat. Genet. 36: 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen, H. J., C. J. O'Kane, C. Wilson, U. Grossniklaus, R. K. Pearson et al., 1989. P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev. 3: 1288–1300. [DOI] [PubMed] [Google Scholar]
- Biessmann, H., and J. M. Mason, 1988. Progressive loss of DNA sequences from terminal chromosome deficiencies in Drosophila melanogaster. EMBO J. 7: 1081–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasset, E., and C. Vaury, 2005. Insulators are fundamental components of the eukaryotic genomes. Heredity 94: 571–576. [DOI] [PubMed] [Google Scholar]
- Burke, T. W., and J. T. Kadonaga, 1996. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 10: 711–724. [DOI] [PubMed] [Google Scholar]
- Burke, T. W., and J. T. Kadonaga, 1997. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 11: 3020–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, J. E., and J. T. Kadonaga, 2001. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 15: 2515–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, J. E., and J. T. Kadonaga, 2002. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 16: 2583–2592. [DOI] [PubMed] [Google Scholar]
- Cai, H., and M. Levine, 1995. Modulation of enhancer-promoter interactions by insulators in the Drosophila embryo. Nature 376: 533–536. [DOI] [PubMed] [Google Scholar]
- Calhoun, V. C., and M. Levine, 2003. Long-range enhancer-promoter interactions in the Scr-Antp interval of the Drosophila Antennapedia complex. Proc. Natl. Acad. Sci. USA 100: 9878–9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun, V. C., A. Stathopoulos and M. Levine, 2002. Promoter-proximal tethering elements regulate enhancer-promoter specificity in the Drosophila Antennapedia complex. Proc. Natl. Acad. Sci. USA 99: 9243–9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson, M., and V. G. Corces, 2004. Boundary elements and nuclear organization. Biol. Cell 96: 617–629. [DOI] [PubMed] [Google Scholar]
- Carter, D., L. Chakalova, C. S. Osborne, Y. F. Dai and P. Fraser, 2002. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32: 623–626. [DOI] [PubMed] [Google Scholar]
- Casares, F., W. Bender, J. Merriam and E. Sanchez-Herrero, 1997. Interactions of Drosophila Ultrabithorax regulatory regions with native and foreign promoters. Genetics 145: 123–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernilogar, F. M., and V. Orlando, 2005. Epigenome programming by Polycomb and Trithorax proteins. Biochem. Cell Biol. 83: 322–331. [DOI] [PubMed] [Google Scholar]
- Chakalova, L., E. Debrand, J. A. Mitchell, C. S. Osborne and P. Fraser, 2005. Replication and transcription: shaping the landscape of the genome. Nat. Rev. Genet. 6: 669–677. [DOI] [PubMed] [Google Scholar]
- Chandler, V. L., and M. Stam, 2004. Chromatin conversations: mechanisms and implications of paramutation. Nat. Rev. Genet. 5: 532–544. [DOI] [PubMed] [Google Scholar]
- Chen, Q., L. Lin, S. Smith, Q. Lin and J. Zhou, 2005. Multiple promoter targeting sequences exist in abdominal-B to regulate long-range gene activation. Dev. Biol. 286: 629–636. [DOI] [PubMed] [Google Scholar]
- Conte, C., B. Dastugue and C. Vaury, 2002. Promoter competition as a mechanism of transcriptional interference mediated by retrotransposons. EMBO J. 21: 3908–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, P. R., 1997. The transcriptional basis of chromosome pairing. J. Cell Sci. 110(Pt. 9): 1033–1040. [DOI] [PubMed] [Google Scholar]
- Coulthard, A. B., N. Nolan, J. B. Bell and A. J. Hilliker, 2005. Transvection at the vestigial locus of Drosophila melanogaster. Genetics 170: 1711–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, A., 2006. On a chromosome far, far away: LCRs and gene expression. Trends Genet. 22: 38–45. [DOI] [PubMed] [Google Scholar]
- Dillon, N., T. Trimborn, J. Strouboulis, P. Fraser and F. Grosveld, 1997. The effect of distance on long-range chromatin interactions. Mol. Cell 1: 131–139. [DOI] [PubMed] [Google Scholar]
- Dorsett, D., 1999. Distant liaisons: long-range enhancer-promoter interactions in Drosophila. Curr. Opin. Genet. Dev. 9: 505–514. [DOI] [PubMed] [Google Scholar]
- Duncan, I. W., 2002. Transvection effects in Drosophila. Annu. Rev. Genet. 36: 521–556. [DOI] [PubMed] [Google Scholar]
- Galagan, J. E., and E. U. Selker, 2004. RIP: the evolutionary cost of genome defense. Trends Genet. 20: 417–423. [DOI] [PubMed] [Google Scholar]
- Gause, M., H. Hovhannisyan, T. Kan, S. Kuhfittig, V. Mogila et al., 1998. hobo Induced rearrangements in the yellow locus influence the insulation effect of the gypsy su(Hw)-binding region in Drosophila melanogaster. Genetics 149: 1393–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer, P. K., and I. Clark, 2002. Protecting against promiscuity: the regulatory role of insulators. Cell. Mol. Life Sci. 59: 2112–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer, P. K., and V. G. Corces, 1987. Separate regulatory elements are responsible for the complex pattern of tissue-specific and developmental transcription of the yellow locus in Drosophila melanogaster. Genes Dev. 1: 996–1004. [DOI] [PubMed] [Google Scholar]
- Geyer, P. K., and V. G. Corces, 1992. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 6: 1865–1873. [DOI] [PubMed] [Google Scholar]
- Geyer, P. K., M. M. Green and V. G. Corces, 1988. Mutant gene phenotypes mediated by a Drosophila melanogaster retrotransposon require sequences homologous to mammalian enhancers. Proc. Natl. Acad. Sci. USA 85: 8593–8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer, P. K., M. M. Green and V. G. Corces, 1990. Tissue-specific transcriptional enhancers may act in trans on the gene located in the homologous chromosome: the molecular basis of transvection in Drosophila. EMBO J. 9: 2247–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor, G. B., N. A. Nassif, D. M. Johnson-Schlitz, C. R. Preston and W. R. Engels, 1991. Targeted gene replacement in Drosophila via P element-induced gap repair. Science 253: 1110–1117. [DOI] [PubMed] [Google Scholar]
- Goldsborough, A. S., and T. B. Kornberg, 1996. Reduction of transcription by homologue asynapsis in Drosophila imaginal discs. Nature 381: 807–810. [DOI] [PubMed] [Google Scholar]
- Grant-Downton, R. T., and H. G. Dickinson, 2004. Plants, pairing and phenotypes–Two's company? Trends Genet. 20: 188–195. [DOI] [PubMed] [Google Scholar]
- Green, M. M., 1959. The discrimination of wild-type isoalleles at the white locus of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 45: 549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson, J. E., and S. Sakonju, 1995. Cis and trans interactions between the iab regulatory regions and abdominal-A and abdominal-B in Drosophila melanogaster. Genetics 139: 835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, H., M. Lu, M. Anggraini, A. Sikora, Y. Chang et al., 2003. Trans allele methylation and paramutation-like effects in mice. Nat. Genet. 34: 199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdridge, C., and D. Dorsett, 1991. Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol. Cell. Biol. 11: 1894–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack, J., D. Dorsett, Y. Delotto and S. Liu, 1991. Expression of the cut locus in the Drosophila wing margin is required for cell type specification and is regulated by a distant enhancer. Development 113: 735–747. [DOI] [PubMed] [Google Scholar]
- Jorgensen, R., 1990. Altered gene expression in plants due to trans interactions between homologous genes. Trends Biotechnol. 8: 340–344. [DOI] [PubMed] [Google Scholar]
- Kadonaga, J. T., 2004. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell 116: 247–257. [DOI] [PubMed] [Google Scholar]
- Karandikar, U., S. Anderson, N. Mason, R. L. Trott, C. P. Bishop et al., 2003. The Drosophila SSL gene is expressed in larvae, pupae, and adults, exhibits sexual dimorphism, and mimics properties of the beta subunit of casein kinase II. Biochem. Biophys. Res. Commun. 301: 941–947. [DOI] [PubMed] [Google Scholar]
- Kassis, J. A., 2002. Pairing-sensitive silencing, polycomb group response elements, and transposon homing in Drosophila. Adv. Genet. 46: 421–438. [DOI] [PubMed] [Google Scholar]
- Keeler, K. J., T. Dray, J. E. Penney and G. B. Gloor, 1996. Gene targeting of a plasmid-borne sequence to a double-strand DNA break in Drosophila melanogaster. Mol. Cell. Biol. 16: 522–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison, J. A., and J. W. Southworth, 2002. Transvection in Drosophila. Adv. Genet. 46: 399–420. [DOI] [PubMed] [Google Scholar]
- Kermekchiev, M., M. Pettersson, P. Matthias and W. Schaffner, 1991. Every enhancer works with every promoter for all the combinations tested: Could new regulatory pathways evolve by enhancer shuffling? Gene Expr. 1: 71–81. [PMC free article] [PubMed] [Google Scholar]
- Kmita, M., N. Fraudeau, Y. Herault and D. Duboule, 2002. Serial deletions and duplications suggest a mechanism for the collinearity of Hoxd genes in limbs. Nature 420: 145–150. [DOI] [PubMed] [Google Scholar]
- Krebs, J. E., and M. Dunaway, 1998. The scs and scs′ insulator elements impart a cis requirement on enhancer-promoter interactions. Mol. Cell 1: 301–308. [DOI] [PubMed] [Google Scholar]
- Kuhn, E. J., and P. K. Geyer, 2003. Genomic insulators: connecting properties to mechanism. Curr. Opin. Cell Biol. 15: 259–265. [DOI] [PubMed] [Google Scholar]
- Kuhn, E. J., M. M. Viering, K. M. Rhodes and P. K. Geyer, 2003. A test of insulator interactions in Drosophila. EMBO J. 22: 2463–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutach, A. K., and J. T. Kadonaga, 2000. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol. Cell. Biol. 20: 4754–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrange, T., A. N. Kapanidis, H. Tang, D. Reinberg and R. H. Ebright, 1998. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 12: 34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., and M. Noll, 1994. Compatibility between enhancers and promoters determines the transcriptional specificity of gooseberry and gooseberry neuro in the Drosophila embryo. EMBO J. 13: 400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Q., D. Wu and J. Zhou, 2003. The promoter targeting sequence facilitates and restricts a distant enhancer to a single promoter in the Drosophila embryo. Development 130: 519–526. [DOI] [PubMed] [Google Scholar]
- Ling, J. Q., T. Li, J. F. Hu, T. H. Vu, H. L. Chen et al., 2006. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312: 269–272. [DOI] [PubMed] [Google Scholar]
- Loden, M., and B. van Steensel, 2005. Whole-genome views of chromatin structure. Chromosome Res. 13: 289–298. [DOI] [PubMed] [Google Scholar]
- Lomvardas, S., G. Barnea, D. J. Pisapia, M. Mendelsohn, J. Kirkland et al., 2006. Interchromosomal interactions and olfactory receptor choice. Cell 126: 403–413. [DOI] [PubMed] [Google Scholar]
- Macdonald, P. M., P. Ingham and G. Struhl, 1986. Isolation, structure, and expression of even-skipped: a second pair-rule gene of Drosophila containing a homeo box. Cell 47: 721–734. [DOI] [PubMed] [Google Scholar]
- Machin, F., J. Torres-Rosell, G. De Piccoli, J. A. Carballo, R. S. Cha et al., 2006. Transcription of ribosomal genes can cause nondisjunction. J. Cell Biol. 173: 893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M., Y. B. Meng and W. Chia, 1989. Regulatory elements involved in the tissue-specific expression of the yellow gene of Drosophila. Mol. Gen. Genet. 218: 118–126. [DOI] [PubMed] [Google Scholar]
- Martinez-Laborda, A., A. Gonzalez-Reyes and G. Morata, 1992. Trans regulation in the Ultrabithorax gene of Drosophila: alterations in the promoter enhance transvection. EMBO J. 11: 3645–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBryant, S. J., V. H. Adams and J. C. Hansen, 2006. Chromatin architectural proteins. Chromosome Res. 14: 39–51. [DOI] [PubMed] [Google Scholar]
- McKee, B. D., 2004. Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim. Biophys. Acta 1677: 165–180. [DOI] [PubMed] [Google Scholar]
- Merli, C., D. E. Bergstrom, J. A. Cygan and R. K. Blackman, 1996. Promoter specificity mediates the independent regulation of neighboring genes. Genes Dev. 10: 1260–1270. [DOI] [PubMed] [Google Scholar]
- Morris, J. R., J. L. Chen, P. K. Geyer and C. T. Wu, 1998. Two modes of transvection: enhancer action in trans and bypass of a chromatin insulator in cis. Proc. Natl. Acad. Sci. USA 95: 10740–10745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, J. R., J. Chen, S. T. Filandrinos, R. C. Dunn, R. Fisk et al., 1999. a An analysis of transvection at the yellow locus of Drosophila melanogaster. Genetics 151: 633–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, J. R., P. K. Geyer and C. T. Wu, 1999. b Core promoter elements can regulate transcription on a separate chromosome in trans. Genes Dev. 13: 253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, J. R., D. A. Petrov, A. M. Lee and C. T. Wu, 2004. Enhancer choice in cis and in trans in Drosophila melanogaster: role of the promoter. Genetics 167: 1739–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, M., K. Hagstrom, H. Gyurkovics, V. Pirrotta and P. Schedl, 1999. The mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Genetics 153: 1333–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash, W. G., 1976. Patterns of pigmentation color states regulated by the y locus in Drosophila melanogaster. Dev. Biol. 48: 336–343. [DOI] [PubMed] [Google Scholar]
- Ohtsuki, S., M. Levine and H. N. Cai, 1998. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 12: 547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Kane, C. J., and W. J. Gehring, 1987. Detection in situ of genomic regulatory elements in Drosophila. Proc. Natl. Acad. Sci. USA 84: 9123–9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palstra, R. J., B. Tolhuis, E. Splinter, R. Nijmeijer, F. Grosveld et al., 2003. The beta-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 35: 190–194. [DOI] [PubMed] [Google Scholar]
- Parkhurst, S. M., D. A. Harrison, M. P. Remington, C. Spana, R. L. Kelley et al., 1988. The Drosophila su(Hw) gene, which controls the phenotypic effect of the gypsy transposable element, encodes a putative DNA-binding protein. Genes Dev. 2: 1205–1215. [DOI] [PubMed] [Google Scholar]
- Parnell, T. J., M. M. Viering, A. Skjesol, C. Helou, E. J. Kuhn et al., 2003. An endogenous suppressor of hairy-wing insulator separates regulatory domains in Drosophila. Proc. Natl. Acad. Sci. USA 100: 13436–13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer, M., and W. Bender, 1988. Sequences of the gypsy transposon of Drosophila necessary for its effects on adjacent genes. Proc. Natl. Acad. Sci. USA 85: 9650–9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta, V., 1999. Transvection and chromosomal trans-interaction effects. Biochim. Biophys. Acta 1424: M1–M8. [DOI] [PubMed] [Google Scholar]
- Pirrotta, V., H. Steller and M. P. Bozzetti, 1985. Multiple upstream regulatory elements control the expression of the Drosophila white gene. EMBO J. 4: 3501–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt, R. J., D. W. Lee and R. Aramayo, 2004. DNA methylation affects meiotic trans-sensing, not meiotic silencing, in Neurospora. Genetics 168: 1925–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassoulzadegan, M., M. Magliano and F. Cuzin, 2002. Transvection effects involving DNA methylation during meiosis in the mouse. EMBO J. 21: 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronshaugen, M., and M. Levine, 2004. Visualization of trans-homolog enhancer-promoter interactions at the Abd-B Hox locus in the Drosophila embryo. Dev. Cell 7: 925–932. [DOI] [PubMed] [Google Scholar]
- Scott, K. S., and P. K. Geyer, 1995. Effects of the su(Hw) insulator protein on the expression of the divergently transcribed Drosophila yolk protein genes. EMBO J. 14: 6258–6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe, J., S. Nonchev, A. Gould, J. Whiting and R. Krumlauf, 1998. Selectivity, sharing and competitive interactions in the regulation of Hoxb genes. EMBO J. 17: 1788–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearwin, K. E., B. P. Callen and J. B. Egan, 2005. Transcriptional interference—a crash course. Trends Gene 21: 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, P. K., D. Zickler, N. B. Raju, G. Ruprich-Robert and R. L. Metzenberg, 2006. SAD-2 is required for meiotic silencing by unpaired DNA and perinuclear localization of SAD-1 RNA-directed RNA polymerase. Proc. Natl. Acad. Sci. USA 103: 2243–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipos, L., and H. Gyurkovics, 2005. Long-distance interactions between enhancers and promoters. FEBS J. 272: 3253–3259. [DOI] [PubMed] [Google Scholar]
- Sipos, L., J. Mihaly, F. Karch, P. Schedl, J. Gausz et al., 1998. Transvection in the Drosophila Abd-B domain: extensive upstream sequences are involved in anchoring distant cis-regulatory regions to the promoter. Genetics 149: 1031–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale, S. T., 2001. Core promoters: active contributors to combinatorial gene regulation. Genes Dev. 15: 2503–2508. [DOI] [PubMed] [Google Scholar]
- Smale, S. T., and D. Baltimore, 1989. The “initiator” as a transcription control element. Cell 57: 103–113. [DOI] [PubMed] [Google Scholar]
- Smale, S. T., and J. T. Kadonaga, 2003. The RNA polymerase II core promoter. Annu. Rev. Biochem. 72: 449–479. [DOI] [PubMed] [Google Scholar]
- Smith, P. A., and V. G. Corces, 1992. The suppressor of Hairy-wing binding region is required for gypsy mutagenesis. Mol. Gen. Genet. 233: 65–70. [DOI] [PubMed] [Google Scholar]
- Spana, C., D. A. Harrison and V. G. Corces, 1988. The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev. 2: 1414–1423. [DOI] [PubMed] [Google Scholar]
- Spilianakis, C. G., and R. A. Flavell, 2004. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat. Immunol. 5: 1017–1027. [DOI] [PubMed] [Google Scholar]
- Spilianakis, C. G., M. D. Lalioti, T. Town, G. R. Lee and R. A. Flavell, 2005. Interchromosomal associations between alternatively expressed loci. Nature 435: 637–645. [DOI] [PubMed] [Google Scholar]
- Thatcher, K. N., S. Peddada, D. H. Yasui and J. M. Lasalle, 2005. Homologous pairing of 15q11–13 imprinted domains in brain is developmentally regulated but deficient in Rett and autism samples. Hum. Mol. Genet. 14: 785–797. [DOI] [PubMed] [Google Scholar]
- Tolhuis, B., R. J. Palstra, E. Splinter, F. Grosveld and W. de Laat, 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10: 1453–1465. [DOI] [PubMed] [Google Scholar]
- Turner, J. M., S. K. Mahadevaiah, O. Fernandez-Capetillo, A. Nussenzweig, X. Xu et al., 2005. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat. Genet. 37: 41–47. [DOI] [PubMed] [Google Scholar]
- Vyas, M., C. Ravindran and D. P. Kasbekar, 2006. Chromosome segment duplications in Neurospora crassa and their effects on repeat-induced point mutation and meiotic silencing by unpaired DNA. Genetics 172: 1511–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, M. F., B. C. Black, G. Afshar, A. Y. Kermabon, T. R. Wright et al., 1991. Temporal and spatial expression of the yellow gene in correlation with cuticle formation and dopa decarboxylase activity in Drosophila development. Dev. Biol. 147: 32–45. [DOI] [PubMed] [Google Scholar]
- Wittkopp, P. J., K. Vaccaro and S. B. Carroll, 2002. Evolution of yellow gene regulation and pigmentation in Drosophila. Curr. Biol. 12: 1547–1556. [DOI] [PubMed] [Google Scholar]
- Wu, C. T., and J. R. Morris, 1999. Transvection and other homology effects. Curr. Opin. Genet. Dev. 9: 237–246. [DOI] [PubMed] [Google Scholar]
- Xu, N., C. L. Tsai and J. T. Lee, 2006. Transient homologous chromosome pairing marks the onset of X inactivation. Science 311: 1149–1152. [DOI] [PubMed] [Google Scholar]
- Zachar, Z., C. H. Chapman and P. M. Bingham, 1985. On the molecular basis of transvection effects and the regulation of transcription. Cold Spring Harbor Symp. Quant. Biol. 50: 337–346. [DOI] [PubMed] [Google Scholar]
- Zhou, J., and M. Levine, 1999. A novel cis-regulatory element, the PTS, mediates an anti-insulator activity in the Drosophila embryo. Cell 99: 567–575. [DOI] [PubMed] [Google Scholar]
- Zickler, D., 2006. From early homologue recognition to synaptonemal complex formation. Chromosoma 115: 158–174. [DOI] [PubMed] [Google Scholar]