Abstract
The male-specific lethal (MSL) ribonucleoprotein complex is necessary for equalization of X:A expression levels in Drosophila males, which have a single X chromosome. It binds selectively to the male X chromosome and directs acetylation of histone H4 at lysine 16 (H4Ac16), a modification linked to elevated transcription. roX1 and roX2 noncoding RNAs are essential but redundant components of this complex. Simultaneous removal of both roX RNAs reduces X localization of the MSL proteins and permits their ectopic binding to autosomal sites and the chromocenter. However, the MSL proteins still colocalize, and low levels of H4Ac16 are detected at ectopic sites of MSL binding and residual sites on the X chromosome of roX1− roX2− males. Microarray analysis was performed to reveal the effect of roX1 and roX2 elimination on X-linked and autosomal gene expression. Expression of the X chromosome is decreased by 26% in roX1− roX2−male larvae. Enhanced expression could not be detected at autosomal sites of MSL binding in roX1− roX2− males. These results implicate failure to compensate X-linked genes, rather than inappropriate upregulation of autosomal genes at ectopic sites of MSL binding, as the primary cause of male lethality upon loss of roX RNAs.
DROSOPHILA males have one X chromosome and two sets of autosomes (A). The imbalance in X:A gene dosage is addressed by the male-specific lethal (MSL) complex that binds to hundreds of sites along the male X chromosome. While there is general agreement that this complex is responsible for dosage compensation, disagreement about how compensation is accomplished remains (Birchler et al. 2003; Straub et al. 2005a). Two recent studies provide strong evidence that binding of the MSL complex to the X chromosome of Drosophila males increases transcription from almost all X-linked genes (Hamada et al. 2005; Straub et al. 2005b). The MSL complex is composed of five proteins, encoded by the genes maleless (mle), male-specific lethal 1, 2, and 3 (msl1, -2, and -3), and males absent on first (mof) (Fukunaga et al. 1975; Belote and Lucchesi 1980; Uchida et al. 1981; Hilfiker et al. 1997). A sixth protein, JIL-1, is required in both sexes but is enriched on the male X chromosome (Jin et al. 2000). One of the primary functions of the MSL complex is thought to be acetylation of histone H4 at lysine 16 (H4Ac16), a modification associated with increased expression and attributable to MOF, a histone acetyltransferase (Akhtar and Becker 2000; Smith et al. 2000). Two noncoding RNAs, roX1 and roX2, are integral components of the MSL complex and necessary for exclusive binding to the X chromosome (Meller and Rattner 2002). Simultaneous mutation of both roX genes causes male lethality, but mutation of a single roX gene is without phenotype.
Removal of MSL1 or MSL2 prevents any chromatin binding by the remaining subunits. Mutants of mle, msl3, or mof retain a partial MSL complex at ∼35 sites on the X chromosome (Gorman et al. 1995; Lyman et al. 1997; Gu et al. 1998). By contrast, simultaneous mutation of both roX genes prevents the exclusive binding of MSL proteins to the X chromosome. Reduced levels of MSL proteins are retained at some sites on the X chromosome, but they are now detected at a number of ectopic autosomal sites (Meller and Rattner 2002; Deng et al. 2005). Some of these sites are puffed, a chromatin state usually associated with active transcription. Failure to increase expression of X-linked genes could cause male lethality. However, it is possible that elevated transcription at a small number of autosomal sites contributes to male lethality.
To address global changes in gene expression in roX1− roX2− males, we turned to microarray analysis. Previous studies examining the expression of a limited number of genes in msl mutant larvae produced complex, and sometimes conflicting, data (Bhadra et al. 1999, 2000, 2005; Chiang and Kurnit 2003). Two recent studies report a global decrease in X-linked gene expression in Drosophila tissue culture cells following RNA interference (RNAi)-mediated msl2 knockdown (Hamada et al. 2005; Straub et al. 2005b). We now report that the level of X-linked gene expression is also reduced in roX1− roX2− male larvae. Although enrichment of H4Ac16 can be detected at autosomal sites of ectopic MSL binding in roX1− roX2− males, increased expression was not observed at these sites by either microarray analysis or reverse Northern blotting. Thus, failure to compensate X-linked genes, but not inappropriate overexpression of some autosomal genes, appears to be the source for male lethality upon loss of both roX RNAs.
MATERIALS AND METHODS
Fly culture and strains:
Flies were maintained at 25° on standard cornmeal–agar fly food in a humidified incubator. The roX1ex6, roX1SMC17A, and roX2 mutations have been previously described (Meller et al. 1997; Meller and Rattner 2002; Deng et al. 2005).
Microarray expression analysis:
Total RNA was prepared from groups of 50 male third instar larvae by TRIzol (Invitrogen, San Diego), extracted, and purified using the RNeasy kit (QIAGEN, Chatsworth, CA). Three independent RNA preparations for each genotype served as templates for cDNA synthesis. Biotin-labeled probes were produced by in vitro transcription of cDNA (protocol at http://www.Affymetrix.com). Probes were hybridized to Affymetrix Drosophila Genome 2.0 chips (Santa Clara, CA). The Affymetrix Drosophila annotation of December 2004 was used to map genes to their cytological locations. Genes were filtered for present/absent calls by a perfect match–mismatch comparison. Autosomal signals were normalized on a chip-by-chip basis to bring their median values to 100. The identical degree of adjustment was used to normalize X-linked transcripts. Changes in expression were determined by comparing the mean signal intensities of genes on arrays hybridized with roX1SMC17A roX2− probes to those hybridized with roX1+ roX2− probes. Statistical significance was assessed by performing an unpaired two-tail t-test. Complete data for individual genes are supplied in supplemental Table S1 at http://www.genetics.org/supplemental/. The raw data can be downloaded from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; GSE3990).
Quantitative real-time PCR:
One microgram of total RNA was reverse transcribed using random hexamers and ImProm-II reverse transcriptase (Promega, Madison, WI). Real-time PCR was performed as described (Deng et al. 2005). Bigmax, an autosomal gene involved in transcription regulation, was selected as a reliable transcript for normalization of expression (data not shown). The primers used in this study are presented in supplemental Table S2 at http://www.genetics.org/supplemental/.
Reverse Northern analysis of gene expression:
The following genomic bacterial artificial chromosomes (BAC) clones carrying inserts mapped to autosomal sites of ectopic MSL binding in roX1− roX2− males were restriction digested, separated on gels, and blotted to nylon membranes: BAC from 21B, RPC-98 9.J.20; BAC from 49B, RPC-98 24.H.9; BACs from 50 C/D, RPC-98 6.M.19 and RPC-98 13.P.15; BACs from 53E, RPC-98 32.P.8 and RPC-98 48.A.11; BACs from 57B/C, RPC-98 8.P.5, RPC-98 10.P.16, and RPC-98 33.D.17; BACs from 89B, RPC-98 2.G.19, RPC-98 4.C.7, and RPC-98 6.B.8. cDNA probes were generated from 1 μg of poly(A)+ RNA isolated from roX1ex6 roX2− or roX1+ roX2− adult males by reverse transcription in the presence of [32P]CTP.
Histology:
Immunhistochemical detection of MSL3, MOF, and H4Ac16 on polytene chromosomes was done as previously described (Kelley et al. 1999). Rabbit anti-H4Ac16 was purchased from Serotec (Raleigh, NC). Goat anti-MSL3 and rabbit anti-MOF were a gift from M. Kuroda.
Photography:
Visualization and photography were performed with a Zeiss Axioscope 2 fitted with a Qimaging Retiga 2000R digital camera.
RESULTS
MOF colocalizes with other MSL proteins in roX1− roX2− males:
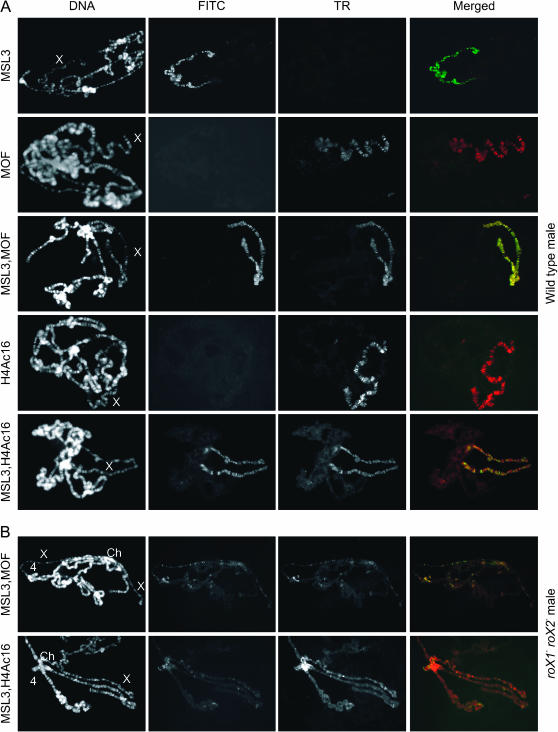
A primary function of the MSL complex is modification of chromatin on the X chromosome. The most dramatic manifestation of this is accumulation of H4Ac16, a modification linked to increased transcription, only on the male X chromosome (Turner et al. 1992; Bone et al. 1994). In an alternative model for the process of dosage compensation, the MSL complex sequesters MOF, thus preventing its uniform distribution. This is proposed to reduce autosomal expression relative to X-linked gene expression (Birchler et al. 2003; Straub et al. 2005a). Although we do not favor this model for dosage compensation, it is possible that release of the MSL proteins from the X chromosome by mutation of both roX genes leads to misregulation of autosomal genes. To estimate the possible extent of autosomal misregulation, we determined the localization and activity of MOF in roX1− roX2− males. Immunostaining of polytene chromosomes from roX1− roX2− mutant larvae reveals disruption of the exclusive X localization of MSL1 and MSL2 (Meller and Rattner 2002; Deng et al. 2005). These proteins are not uniformly redistributed, but are attracted to a small number of autosomal sites. MOF and MSL3 localization on polytene chromosomes from wild-type and roX1− roX2− mutant male larvae was determined. MOF and MSL3 colocalize on the X chromosome in wild-type males (Figure 1A; also see Gu et al. 1998). roX1− roX2− males reveal overlapping MOF and MSL3 localization at residual sites along the X chromosome and also at ectopic sites of MSL binding to the chromocenter and autosomes (Figure 1B). This indicates that MOF is not uniformly distributed in a roX1− roX2− genetic background. Colocalization and mapping studies indicate that MSL1, MSL2, MSL3, and MOF continue to associate at autosomal sites of ectopic MSL binding in roX1− roX2− males (supplemental Table S3 at http://www.genetics.org/supplemental/).
Figure 1.—
H4Ac16 and MSL colocalize on polytene chromosomes from wild-type and roX1− roX2− males. Chromosome preparations from wild-type and roX1ex6 roX2− male larvae were probed with antibodies against MSL3, MOF, and H4Ac16. Patterns of ectopic MSL localization are similar in roX1ex6 roX2− and roX1SMC17A roX2− male larvae, but the salivary glands of roX1ex6 roX2− produce better polytene chromosomes for immunostaining. (A) Wild-type males. (B) roX1ex6 roX2− males. DNA is detected by Hoechst 33258. MSL3 is detected by FITC. MOF and H4AcK16 are detected by Texas Red (TR). An exposure of 40 msec was used for DNA; a 1-sec exposure was used for TR and FITC in wild-type males. Exposure times were lengthened to 4 sec (TR) and 3 sec (FITC) in roX mutants. Exposure for the Texas Red channel of row 1 and the FITC channel of rows 2 and 4 in A was longer to demonstrate the absence of cross-reactivity by the secondary antibodies used. X chromosome (X); fourth chromosome (4); chromocenter (Ch).
Accumulation of H4Ac16 overlaps that of the MSL proteins in roX1− roX2− males:
It is possible that ectopic sites of MSL binding contain genes that are upregulated by the MSL proteins. Acetylation of histones by MOF might contribute to increased transcription or chromatin puffing. In wild-type males, H4Ac16 is highly enriched on the X chromosome (Figure 1A). In roX1− roX2− males, the overall level of acetylation on the X chromosome is sharply reduced, but a number of X-linked sites retaining some MSL proteins show weak acetylation (Figure 1B). The strongest accumulation of H4Ac16 is at the chromocenter, but weak H4Ac16 immunoreactivity is also detected at many autosomal sites of MSL binding in roX1− roX2− mutants. This indicates that removal of roX RNA does not fully eliminate the ability of the MSL proteins to associate and modify chromatin, potentially leading to increased transcription. The nonuniform distribution of MOF and H4Ac16 suggests that a limited number of autosomal genes are candidates for misregulation in roX1− roX2− males. A few autosomal sites are puffed, attract high levels of the MSL proteins, and display modest enrichment of H4Ac16 in roX1− roX2− males. It is possible that toxic overexpression at these autosomal sites contributes to male lethality. However, the vast majority of autosomal genes do not appear to be targets of MOF activity and are thus likely to remain unchanged in roX1− roX2− males.
Mutation of both roX genes leads to a global reduction in X chromosome expression:
To determine the effect of the loss of roX1 and roX2 on gene expression, we performed microarray analysis on roX1− roX2− male larvae. As the changes in expression of most X-linked genes are anticipated to be maximally twofold, efforts were taken to minimize variation in genetic background and to compare X chromosomes that are as similar as possible. roX1+ roX2− male larvae were used as the control. These males have full survival without developmental delay and localize the MSL proteins to the X chromosome in a pattern identical to that of wild-type males (Meller and Rattner 2002). Use of roX1+ roX2− as the control enables a comparison of X chromosomes that are largely identical in sequence. roX1SMC17A is the most severe roX1 allele identified to date and appears to be a genetic null (Deng et al. 2005). While roX1SMC17A roX2− adults are rarely observed, ∼37% of roX1SMC17A roX2− males can be recovered as delayed third instar larvae. Because of its phenotypic severity, the roX1SMC17A roX2− chromosome was used to assess the effect of loss of roX RNA on gene expression.
A failure of dosage compensation in flies causes misregulation of a large portion of the genome. This makes normalization of microarrays problematic. After observing the nonuniform H4Ac16 redistribution in roX1− roX2− males, we decided to normalize microarrays to the median autosomal signals (see materials and methods). This strategy was also based on recent studies documenting reduced X-linked gene expression, rather than increased autosomal expression, following RNAi knockdown of MSL2 in tissue culture cells (Hamada et al. 2005; Straub et al. 2005b). After normalization and filtering for absent genes, roughly equivalent numbers of X-linked genes were detected in all samples (Table 1). Quantitative PCR (QPCR) on selected genes validated the microarray findings. A total of 18 genes (2 autosomal, 16 X-linked) were tested in three independent measurements. Transcript ratios (roX1SMC17A roX2−:roX1+ roX2−) from QPCR closely matched the average ratios derived from microarrays (supplemental Table S4 at http://www.genetics.org/supplemental/). While most changes in expression were modest, two X-linked genes consistently showed dramatic changes. opt1 is immediately proximal to roX1. Expression of opt1 is reduced fivefold in roX1SMC17A roX2− male larvae, but is similarly decreased in roX1SMC17A male larvae (data not shown). This is consistent with disruption of an opt1 regulatory element by the roX1SMC17A mutation, but not with reduced expression due to the failure of dosage compensation. Expression of rala is >10-fold higher in roX1SMC17A roX2− male larvae. This appears due to a low-expressing rala allele on the roX1+ roX2− (control) chromosome. Examination of rala expression in several genotypes is consistent with this interpretation (data not shown). Overall, QPCR analysis strongly supports the changes in gene expression detected by microarray analysis, even when these changes are subtle.
TABLE 1.
Overall expression of X-linked genes decreases in roX1− roX2− males
| Array | No. of X-linked genes with present calls | Median intensity | Mean intensity genes |
|---|---|---|---|
| Mutant 1a | 2060 | 131 | 336 |
| Mutant 2 | 1997 | 133 | 374 |
| Mutant 3 | 2055 | 124 | 347 |
| Control 1b | 2055 | 155 | 456 |
| Control 2 | 2011 | 153 | 471 |
| Control 3 | 2036 | 158 | 466 |
Mutant: roX1SMC17A roX2− male larvae.
Control: roX1+ roX2− male larvae.
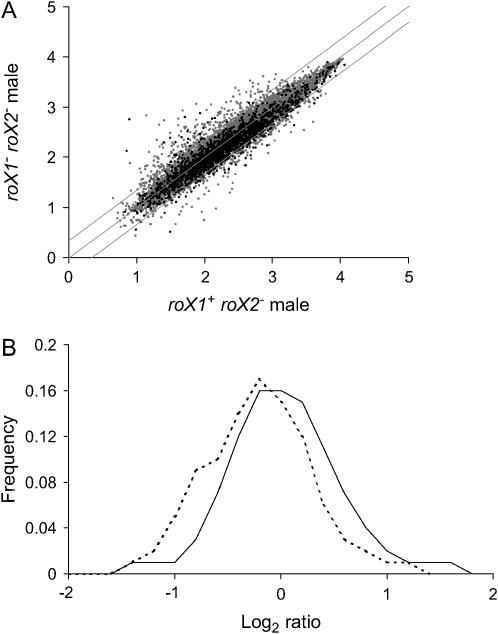
Expression of a substantial number of X-linked genes is decreased as much as twofold in roX1SMC17A roX2− males (Figure 2A). The overall expression of X-linked genes drops by an average of 26% in roX1SMC17A roX2− males (t-test, P < 0.005; Table 1), with a median decrease of 17% (t-test, P < 0.005). A plot of the distribution of log2 ratios (mutant: control) for X-linked and second chromosome genes reveals a peak for the second chromosome close to zero (x = −0.054), while the peak for X-linked genes is shifted left (x = −0.291; Figure 2B). The distribution of third chromosome genes is similar to that of the second chromosome genes (data not shown). ANOVA for multiple comparisons rejects the null hypothesis that the distributions of the X chromosome and the second chromosome are the same (P-value <10−20). These observations indicate a widespread decrease in expression of X-linked genes in roX1SMC17A roX2− males.
Figure 2.—
Expression of the male X chromosome is decreased in roX1− roX2− males. (A) Expression of X-linked genes (solid dots) and autosomal genes (shaded dots) in roX1SMC17A roX2− and roX1+ roX2− male larvae. Log intensity values for each genotype are indicated on the axes. Diagonal lines indicate ratios of 0.5, 1, and 2. (B) Distribution of the log2 ratios for genes detected on roX1SMC17A roX2− and roX1+ roX2− arrays. X chromosome (dotted line) and second chromosome (solid line). Genes with present calls in at least two of three mutant and control arrays were plotted.
A distinct bias toward modestly decreased expression of X-linked genes is detected in roX1SMC17A roX2− males (Table 2). Selection of genes with changes of 1.5- to 1.99-fold or 2- to 2.99-fold reveals a similar proportion of autosomal genes showing slight decrease or increase in expression. However, X-linked genes that decrease slightly are dramatically overrepresented at the expense of those with modest increases. In contrast, similar percentages of X-linked and autosomal genes increase or decrease by ≥3-fold. These more dramatic changes in expression are likely attributable to the indirect effects of dosage compensation failure, such as developmental delay, reduced size, and the overall poor health of roX1SMC17A roX2− male larvae.
TABLE 2.
X-linked genes are biased toward slightly decreased expression in roX1− roX2− males
| Fold increase
|
Fold decrease
|
|||||
|---|---|---|---|---|---|---|
| 1.5–1.99 | 2.0–2.99 | ≥3 | 1.5–1.99 | 2.0–2.99 | ≥3 | |
| X-linked genes | 38a (2.0) | 19 (1.0) | 13 (0.7) | 335 (17.8) | 158 (8.4) | 21 (1.1) |
| Autosomal genes | 505 (4.9) | 162 (1.6) | 68 (0.7) | 967 (9.5) | 332 (2.7) | 62 (0.6) |
The percentage of X-linked or autosomal genes is indicated in parentheses. Only genes present on at least two of three arrays from each genotype were considered (1884 of 3090 X-linked genes and 10,185 of 15,892 autosomal genes).
Genes displaying expression changes (t-test P-value <0.05) in the indicated ranges were determined.
roX1SMC17A roX2− males display gene misregulation similar to that in male cells and larvae lacking MSL proteins:
This analysis of roX1− roX2− males is the first genomewide study of expression after the disruption of dosage compensation in Drosophila larvae. However, several studies have previously looked at the regulation of a small number of genes in larvae carrying msl mutations, and two recent studies have examined gene expression in male S2 cells following msl2 RNAi knockdown. This provides the opportunity to compare the effect of disrupting different components of the dosage compensation system on the expression of X-linked genes. Knockdown of msl2 in S2 cells is particularly valuable as males carrying mutations in msl2 rarely survive into the third instar, the stage at which roX1− roX2− males are collected.
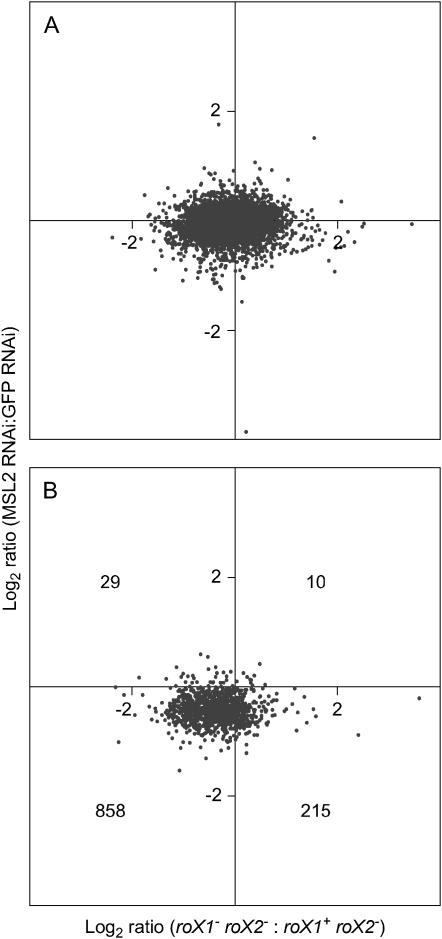
Direct targets of MSL binding were identified, and absolute expression changes for these genes were documented in msl2 RNAi knockdown S2 cells (Straub et al. 2005b). Three individual X-linked genes that are bound by the MSL proteins CG14804, mRpL16, and Arm were reduced in expression by ∼40% upon the depletion of msl2 in S2 cells. These same genes are reduced by ∼30–40% in our analysis. A global comparison of gene expression between S2 cells lacking MSL2 (Hamada et al. 2005) and roX1− roX2− male larvae (this report) was also done. About twice as many genes are detected in male larvae as in S2 cells, presumably because larvae have differentiated tissues expressing genes that are silent in tissue culture cells. However, transcripts detected in both data sets will be enriched for housekeeping and essential genes that are steadily expressed throughout fly development. Most autosomal genes were unchanged in both data sets (Figure 3A). In contrast, of a total of 1112 X-linked genes present in both data sets, 858 decreased in both (Figure 3B). While the overall level of decrease of X-linked gene expression was similar in S2 cells treated with double-stranded RNA to msl2 and in roX1− roX2− male larvae, the response in larvae was more heterogeneous than in S2 cells (Figure 3B). This heterogeneity is most apparent in the weakly expressed genes (supplemental Figure S1, A and B, at http://www.genetics.org/supplemental/). X-linked genes with high expression tend to be uniformly affected in both studies (supplemental Figure S1C at http://www.genetics.org/supplemental/). This may indicate that heavily transcribed genes are more dependent upon the activity of the MSL complex, but it could also reflect more accurate measurement of abundant transcripts.
Figure 3.—
X-linked genes display similar changes in roX1− roX2− males and MSL2-depleted S2 cells. (A) Log2 ratio of change in expression for autosomal genes in response to loss of both roX genes (x-axis; this study) plotted against log2 ratio of change in response to MSL2 depletion in S2 cells (y-axis; Hamada et al. 2005). (B) Log2 ratio of change in expression of X-linked genes in response to loss of both roX genes plotted against log2 ratio of change in S2 cells in response to MSL2 depletion. The number of X-linked genes in each quadrant is indicated. Only genes detected on all 12 arrays contributed to this analysis. Raw microarray data from Hamada et al. (2005) have been renormalized using the methodology of this study (see materials and methods).
Previous studies of msl or Sxl mutant larvae have documented the response of a small number of genes to dosage compensation failure (Bhadra et al. 2000; Chiang and Kurnit 2003). We examined expression changes of these genes upon loss of roX RNAs. Of 20 genes examined in mlepml8 and SxlM male larvae by Northern blot analysis, 8 display similar trends in roX1− roX2−, mlepml8, and SxlM males (supplemental Table S5 at http://www.genetics.org/supplemental/; Bhadra et al. 2000). Considerably more agreement is found between our study and a previous analysis of msl mutants that used a sensitive RT–QPCR assay (Chiang and Kurnit 2003). Of the 12 examined genes, 9 showed the same trend in roX1− roX2−, mle1, and mof1 male larvae (supplemental Table S5 at http://www.genetics.org/supplemental/). We also observed coordinated expression changes of BR-C and Sgs in roX1− roX2− males, consistent with studies showing that BR-C mutations blocked induction of the Sgs genes (supplemental Table S5 at http://www.genetics.org/supplemental/; Guay and Guild 1991; Karim et al. 1993; Chiang and Kurnit 2003). While the agreement between these studies is suggestive, it is worth noting that both msl and roX1− roX2− mutants are developmentally delayed. A broad spectrum of genes normally expressed in late larval development, such as the BR-C and Sgs genes, may be affected by nonspecific mechanisms.
Elevated expression is not detected at ectopic sites of MSL binding:
Strong MSL binding and enrichment for H4Ac16 at ectopic autosomal sites might lead to overexpression of a few autosomal genes in roX1− roX2− males. Changes in gene expression were plotted against chromosomal position for four of the most striking autosomal sites of MSL accumulation (21B, 50D, 53E, and 89B). While there is some variation in expression across each of these regions, none reveals a dramatic increase in expression (supplemental Figure S2 at http://www.genetics.org/supplemental/). A few genes within these sites display approximately twofold increases in roX1− roX2− males, but similar or more pronounced changes occur in surrounding regions that do not attract MSL proteins. To further explore this question, BACs covering six sites that consistently attract high levels of MSL proteins in roX1− roX2−males were examined by reverse Northern blotting. mRNA isolated from roX1+ roX2−and escaping roX1ex6 roX2− adult males was reverse transcribed to generate radiolabeled probes. These probes were hybridized to blots of BAC restriction fragments. No BAC reveals a band with notably stronger hybridization to cDNA probes from roX1ex6 roX2− adult males (supplemental Figure S3 at http://www.genetics.org/supplemental/). While microarray and reverse Northern blotting experiments do not rule out the possibility that a single gene is modestly increased in expression by MSL binding, they do indicate that massive overexpression of a few autosomal genes does not occur at puffed sites that recruit the MSL proteins in roX1− roX2− males.
DISCUSSION
Numerous studies have implicated the roX RNAs in dosage compensation of the male X chromosome of Drosophila. However, this study is the first to address the effect of the elimination of the roX transcripts on gene expression. We find a chromosome-wide decrease in X-linked gene expression, consistent with sharply reduced localization of the MSL proteins on the X chromosome in roX1− roX2− males (Deng et al. 2005). This reduction is not as dramatic as would be anticipated from a complete failure of dosage compensation. It is possible that the minor enrichment of the MSL protein and H4Ac16 that continue to be detectable on the X chromosome of all roX1−roX2− males provides some compensation. However, the magnitude of decrease (∼26%) is comparable to that achieved by knockdown of MSL2 in cells using RNAi (∼30%; Hamada et al. 2005; Straub et al. 2005b). As removal of MSL2 prevents any chromatin binding by the remaining MSL proteins, it appears likely that this level of decrease represents the full effect of compensation failure. The modesty of this effect suggests that, when the roX and MSL mechanism of dosage compensation is disabled, a partial equalization of X-linked gene expression is achieved through the activity of a backup mechanism. The nature of a potential backup system is unknown, but the response to aneuploidy, which partially compensates for large chromosomal deletions, is a likely candidate (Birchler et al. 2001). There is no reason to suspect that the response to reduced expression of the entire X chromosome, which would mimic chromosomal aneuploidy, would be different than the response to aneuploidy for an autosome. Interestingly, the aneuploidy response has been suggested to play a role in normal dosage compensation (Birchler et al. 2003).
While most X-linked genes do appear to be compensated by upregulation in males, it has been proposed that a few are compensated in females through a reduction in translation. This is based on the observation that Sex lethal (SXL) directs compensation of the runt (run) gene, but not through the action of the MSL proteins (Gergen 1987; Bernstein and Cline 1994). The discovery that transcripts with multiple SXL-binding sites were almost exclusively X-linked strengthened the idea of a female component of compensation acting by SXL repression of translation (Kelley et al. 1995). Intriguingly, run is one of the X-linked genes with SXL-binding sites. The run gene does not accumulate H4Ac16 in males, and run expression is not decreased in roX1− roX2− or msl males (supplemental Tables S4 and S5 at http://www.genetics.org/supplemental/; Smith et al. 2001). While this suggests that run is likely to be compensated in females by SXL, about half of the X-linked genes that display multiple SXL-binding sites do decrease in roX1− roX2− males, indicating at least a partial regulation by the MSL complex (data not shown).
No dramatic increase in expression was observed at ectopic sites of MSL binding on autosomes, in spite of puffing and accumulation of H4Ac16 at some of these sites in roX1− roX2− males. A few of these autosomal sites display weak staining of the MSL proteins and enrichment of H4Ac16 even in wild-type males (Gorman et al. 1995; Meller and Rattner 2002). It is possible that this represents genes already under the control of the MSL proteins. If this is indeed the case, further MSL binding and H4Ac16 enrichment might be superfluous. Alternatively, a modest increase might occur, but this increase is so slight that it is indistinguishable from variations in flanking regions of the chromosome. This is somewhat at odds with the dramatic puffing at many of these sites, a condition typically associated with strong transcription (Bonner and Pardue 1977). It is possible that genes not represented on microarrays, such as heterochromatic genes or those producing noncoding RNA, are increased in expression. Nonpolyadenylated transcripts would also escape detection in reverse Northern blots with probes generated from poly(A)+ RNAs. The idea that the MSL proteins might be attracted to the sites of synthesis of noncoding transcripts, many of which have been identified in Drosophila, is plausible (Inagaki et al. 2005). However, the sites of ectopic MSL binding in roX1− roX2− males do not correspond to known noncoding RNAs.
The site of strongest MSL binding and H4Ac16 accumulation on chromosome preparations from roX1− roX2− males is the chromocenter. It is possible that ectopic MSL binding at the chromocenter is due to interaction of the MSL proteins with other proteins. It has been shown that heterochromatin protein 1 (HP1), a major component of heterochromatin, is modestly enriched on the male X chromosome (De Wit et al. 2005). Overexpression of MSL1 and MSL2 leads to ectopic binding of the MSL proteins at the chromocenter (Demakova et al. 2003). These observations suggest an affinity between heterochromatin proteins, possibly HP1 itself, and the MSL proteins. In this study, transcription from the chromocenter, which is primarily heterochromatic, was not specifically addressed. Annotation of heterochromatic genes has lagged behind euchromatic regions of the genome, and these are underrepresented on microarrays (Drosophila Heterochromatin Genome Project, http://www.dhgp.org/; Hoskins et al. 2002).
The failure to identify autosomal transcripts that increase dramatically upon ectopic localization of the MSL complex reinforces the idea that the ultimate cause of male lethality is failure to express the entire X chromosome at appropriate levels. Although many genes are toxic upon massive overexpression, there is only a single locus, Triplolethal (Tpl), for which appropriate dosage is so critical that duplication results in death (Lindsley et al. 1972). We have not detected enhanced MSL binding or increased expression at the Tpl locus in roX1− roX2− larvae (data not shown). This suggests that minor disruptions of autosomal expression are unlikely to contribute to the lethality of roX1− roX2− males.
Acknowledgments
We thank the Applied Genomics and Technology Center at Wayne State University for expert assistance in microarray studies and Bethany Strunk and L. Pile for advice on quantitative PCR. We also thank Ying Huang and C. Freeman for contributing their time and expertise to data analysis. Antibodies were a gift from M. I. Kuroda. This study was supported by start-up funds from Wayne State University and National Institutes of Health grant GM5 8427.
References
- Akhtar, A., and P. Becker, 2000. Activation of transcription through histone H4 acetylation by MOF, an acetyl transferase essential for dosage compensation in Drosophila. Mol. Cell 5: 367–375. [DOI] [PubMed] [Google Scholar]
- Belote, J. M., and J. C. Lucchesi, 1980. Male-specific lethal mutations of Drosophila melanogaster. Genetics 96: 165–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, M., and T. W. Cline, 1994. Differential effects of Sex-lethal mutations on dosage compensation early in Drosophila development. Genetics 136: 1051–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra, U., M. Pal-Bhadra and J. A. Birchler, 1999. Role of the male specific lethal (msl) genes in modifying the effects of sex chromosomal dosage in Drosophila. Genetics 152: 249–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra, U., M. Pal-Bhadra and J. A. Birchler, 2000. Histone acetylation and gene expression analysis of sex lethal mutants in Drosophila. Genetics 155: 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra, M. P., U. Bhadra, J. Kundu and J. A. Birchler, 2005. Gene expression analysis of the function of the male-specific lethal complex in Drosophila. Genetics 169: 2061–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler, J. A., U. Bhadra, M. P. Bhadra and D. L. Auger, 2001. Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev. Biol. 234: 275–288. [DOI] [PubMed] [Google Scholar]
- Birchler, J. A., M. Pal-Bhadra and U. Bhadra, 2003. Dosage dependent gene regulation and the compensation of the X chromosome in Drosophila males. Genetica 117: 179–190. [DOI] [PubMed] [Google Scholar]
- Bone, J. R., J. Lavender, R. Richman, M. J. Palmer, B. M. Turner et al., 1994. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes Dev. 8: 96–104. [DOI] [PubMed] [Google Scholar]
- Bonner, J. J., and M. L. Pardue, 1977. Polytene chromosome puffing and in situ hybridization measure different aspects of RNA metabolism. Cell 12: 227–234. [DOI] [PubMed] [Google Scholar]
- Chiang, P. W., and D. M. Kurnit, 2003. Study of dosage compensation in Drosophila. Genetics 165: 1167–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demakova, O. V., I. V. Kotlikova, P. R. Gordadze, A. A. Alekseyenko, M. I. Kuroda et al., 2003. The MSL complex levels are critical for its correct targeting to the chromosomes in Drosophila melanogaster. Chromosoma 112: 103–115. [DOI] [PubMed] [Google Scholar]
- Deng, X., B. P. Rattner, S. Souter and V. H. Meller, 2005. The severity of roX1 mutations is predicted by MSL localization on the X chromosome. Mech. Dev. 122: 1094–1105. [DOI] [PubMed] [Google Scholar]
- De Wit, E., F. Greil and B. Van Steensel, 2005. Genome-wide HP1 binding in Drosophila: developmental plasticity and genomic targeting signals. Genome Res. 15: 1265–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga, A., A. Tanaka and K. Oishi, 1975. Maleless, a recessive autosomal mutant of Drosophila melanogaster that specifically kills male zygotes. Genetics 81: 135–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergen, J. P., 1987. Dosage compensation in Drosophila: evidence that daughterless and Sex-lethal control X chromosome activity at the blastoderm stage of embryogenesis. Genetics 117: 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman, M., A. Franke and B. S. Baker, 1995. Molecular characterization of the male-specific lethal-3 gene and investigations of the regulation of dosage compensation in Drosophila. Development 121: 463–475. [DOI] [PubMed] [Google Scholar]
- Gu, W., P. Szauter and J. C. Lucchesi, 1998. Targeting of MOF, a putative histone acetyl transferase, to the X chromosome of Drosophila melanogaster. Dev. Genet. 22: 56–64. [DOI] [PubMed] [Google Scholar]
- Guay, P. S., and G. M. Guild, 1991. The ecdysone-induced puffing cascade in Drosophila salivary glands: a Broad-Complex early gene regulates intermolt and late gene transcription. Genetics 129: 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada, F. N., P. J. Park, P. R. Gordadze and M. I. Kuroda, 2005. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev. 19: 2289–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilfiker, A., D. Hilfiker-Kleiner, A. Pannuti and J. C. Lucchesi, 1997. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 16: 2054–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins, R. A., C. D. Smith, J. W. Carlson, A. B. Carvalho, A. Halpern et al., 2002. Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol. 3: RESEARCH0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki, S., K. Numata, T. Kondo, M. Tomita, K. Yasuda et al., 2005. Identification and expression analysis of putative mRNA-like non-coding RNA in Drosophila. Genes Cells 10: 1163–1173. [DOI] [PubMed] [Google Scholar]
- Jin, Y., Y. Wang, J. Johansen and K. M. Johansen, 2000. JIL-1, a chromosomal kinase implicated in regulation of chromatin structure, associates with the male specific lethal (MSL) dosage compensation complex. J. Cell Biol. 149: 1005–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim, F. D., G. M. Guild and C. S. Thummel, 1993. The Drosophila Broad-Complex plays a key role in controlling ecdysone-regulated gene expression at the onset of metamorphosis. Development 118: 977–988. [DOI] [PubMed] [Google Scholar]
- Kelley, R. L., I. Solovyeva, L. M. Lyman, R. Richman, V. Solovyev et al., 1995. Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell 81: 867–877. [DOI] [PubMed] [Google Scholar]
- Kelley, R. L., V. H. Meller, P. R. Gordadze, G. Roman, R. L. Davis et al., 1999. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell 98: 513–522. [DOI] [PubMed]
- Lindsley, D. L., L. Sandler, B. S. Baker, A. T. Carpenter, R. E. Denell et al., 1972. Segmental aneuploidy and the genetic gross structure of the Drosophila genome. Genetics 71: 157–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman, L. M., K. Copps, L. Rastelli, R. L. Kelley and M. I. Kuroda, 1997. Drosophila male-specific lethal-2 protein: structure/function analysis and dependence on MSL-1 for chromosome association. Genetics 147: 1743–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller, V. H., and B. P. Rattner, 2002. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J. 21: 1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller, V. H., K. H. Wu, G. Roman, M. I. Kuroda and R. L. Davis, 1997. roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell 88: 445–457. [DOI] [PubMed] [Google Scholar]
- Smith, E. R., A. Pannuti, W. Gu, A. Steurnagel, R. G. Cook et al., 2000. The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol. Cell. Biol. 20: 312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, E. R., C. D. Allis and J. C. Lucchesi, 2001. Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J. Biol. Chem. 276: 31483–31486. [DOI] [PubMed] [Google Scholar]
- Straub, T., I. K. Dahlsveen and P. B. Becker, 2005. a Dosage compensation in flies: mechanism, models, mystery. FEBS Lett. 579: 3258–3263. [DOI] [PubMed] [Google Scholar]
- Straub, T., G. D. Gilfillan, V. K. Maier and P. B. Becker, 2005. b The Drosophila MSL complex activates the transcription of target genes. Genes Dev. 19: 2284–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, B. M., A. J. Birley and J. Lavender, 1992. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell 69: 375–384. [DOI] [PubMed] [Google Scholar]
- Uchida, S., T. Uenoyana and K. Oishi, 1981. Studies on the sex-specific lethals of Drosophila melanogaster. Jpn. J. Genet. 56: 523–527. [Google Scholar]