THE purpose of this essay is to retrace some of the early steps that I and a few (then) young geneticists took in the late 1960s and early 1970s to define the Escherichia coli functions used by phage to properly execute their developmental cycle. Eventually, this led to the discovery and functional understanding of the so-called DnaK (Hsp70) and GroEL (Hsp60) molecular chaperone machines, universally conserved among the biological kingdoms (Lindquist and Craig 1988; Georgopoulos and Welch 1993). Now we know that these and other molecular chaperone machines are involved in a multitude of biological processes, including protection of nascent polypeptide chains from premature aggregation, disaggregation of protein aggregates, polypeptide transport across biological membranes, and proteolysis (Bukau and Horwich 1998; Hartl and Hayer-Hartl 2002; Craig et al. 2006). Because protein denaturation and aggregation are enhanced by various environmental stresses, e.g., an increase in temperature, it is not surprising that the DnaK (Hsp70) and GroEL (Hsp60) molecular chaperone machines were also discovered independently as “heat-shock” or “stress” proteins. Because of their very short growth cycles, phage have evolved a variety of strategies to subvert and customize host functions for their own use. Phage likely have a greater need than their hosts for quick and abundant chaperone power to carry out their developmental cycle in a timely fashion. If they fail to complete their cycle before the host lyses, infectious phage progeny will not be released into the medium, risking their extinction. This differential need for chaperone power likely explains why many of the bacterial mutations found to block phage development were eventually shown to be in genes encoding the GroEL and DnaK chaperone machines.
THE (THEN) MYSTICAL WORLD OF PHAGE λ
Although my major at Amherst College was physics, I became interested in microbial genetics as a profession in 1961. As an undergraduate, I had taken a part-time job as a laboratory assistant working on phage P22 transduction in the laboratory of Harold H. Plough, a former student of T. H. Morgan. This experience persuaded me to pursue a Ph.D. with Salva Luria at the Massachusetts Institute of Technology (MIT). Under the steadying influence and guidance of Helen Revel (then a senior research associate in Luria's laboratory and a mother hen to me), I finished my Ph.D. thesis on the “sweet and sour” restriction of phage T4 (Revel and Luria 1970). From T4, I moved on to phage λ in Dale Kaiser's laboratory at Stanford. The reason for this leap from the T4 world to that of λ was largely due to the presence of Ethan Signer's laboratory down the hall at MIT. It seemed to me that at that time the smartest workers in biology were those working on phage λ. The developmental cycle and exquisite regulation of this organism are very complicated, and in those early days were way over my head. Thus, I decided to become part of the almost mystical and magical world of phage λ. I simply wanted to be as smart as those guys. It was going to be a great challenge.
Because of a temporary lack of bench space at Stanford, I decided to get a head start on my postdoctoral studies by finding a phage λ genetics project during my waiting period at MIT. Luria supported my decision and generously arranged for me to remain as an Instructor in Microbiology for a few extra months. I have always loved genetics because of its strong reliance on logic. The isolation of mutants and the deciphering of their phenotypes allow the elucidation of concrete biological pathways and, in many instances, the identification of specific interactions among the various participants. Although genetic systems are elegant and powerful, they are also very exacting, occasionally yielding “unexpected” results that can be explained only when one fully understands the applied selection/screen.
IRA HERSKOWITZ AND THE DISCOVERY OF THE FIRST E. coli groP MUTANT
A key person during my initial ventures into lambdology was Ira Herskowitz, at that time a second-year graduate student in Signer's laboratory. Although he was a few years my junior, he was one of my phage λ intellectual fathers simply because he knew so much about it and was always friendly and available. In the spring of 1969, most λ workers were isolating and characterizing phage mutants (mostly of the nonsense/amber, or temperature-sensitive varieties) that were unable to propagate lytically, lysogenize, or recombine (Campbell 1993; Edgar 2004). During strategy-plotting sessions, Ira and I decided to take a completely backward approach, isolating and studying E. coli mutants that block the propagation of phage λ at a step subsequent to injection of its DNA. We reasoned that since the coding capacity of phage λ is rather limited, there must be many interesting host functions that directly or indirectly participate in its developmental cycle. Our hope was to uncover such protein–protein interactions between these two well-studied organisms, shedding additional light on the physiology of important functions in E. coli. Signifying our youthful independence was the fact that Ira and I never discussed our plans or specific experiments with our respective superiors. We simply forged ahead with our collaboration.
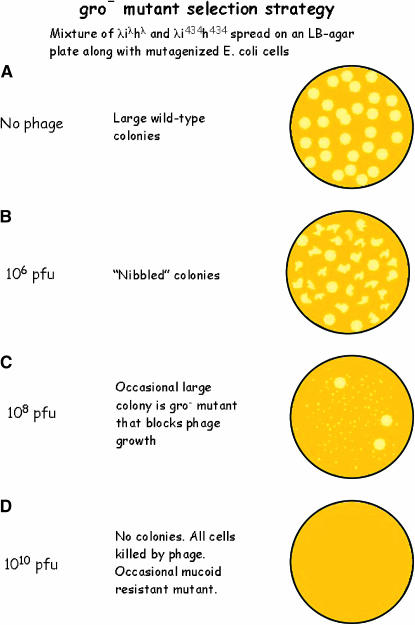
By serendipity, we chose to carry out our selection with the E. coli strain C600, carrying the supE amber (am) nonsense suppressor gene, an important detail whose significance will become obvious shortly. C600 bacteria were mutagenized and spread on LB agar plates along with an appropriate amount of phage λcI− and phage 434cI− (Figure 1). These lambdoid phage variants were chosen because, although they grow well, neither of them can lysogenize, resulting in certain host death following infection. In addition, the fact that each attaches to a different host–surface receptor reduces the frequency of “trivial” bacterial mutants unable to adsorb phage, since both phage receptor-encoding genes would have to mutate to acquire resistance.
Figure 1.—
The genetic strategy used to isolate the E. coli Gro mutants. The diagram shows the effect of increasing phage concentrations on bacterial growth and colony morphology. See text and Georgopoulos (1971) for details.
Normally, when a wild-type bacterium is infected, the released progeny phage will then infect neighboring bacteria in the growing colony, thus limiting the size of the colony. For our experiment, the amount of phage mixed with the bacteria on the plate was critical, since too much phage kills all members of a growing colony (Figure 1D), whereas too little phage results in both “nibbled” colonies and many false positives (Figure 1B). If the infected bacteria are defective in a host function essential for phage development, no viable phage progeny will be released. Hence, although an individual bacterium may be killed as a result of phage infection, the infecting phage are eliminated in the process, thus sparing the neighboring sibling bacteria from being killed, and allowing colony formation (Figure 1C). The procedure was first carried out at 30° in the hope that these mutations would also block bacterial growth at higher temperatures. Under the chosen conditions, wild-type E. coli bacteria formed tiny colonies (Figure 1C) due to killing of the outer cells in a colony by the phage on the plate. However, at a frequency of ∼10−4, large-colony formers appeared on such plates (Figure 1C). The functions defined by these gro bacterial mutations (named for their inability to grow phage; this nomenclature was suggested to me later by Doug Berg at Stanford) did indeed turn out to be important for host growth, as exemplified by the E. coli Gro15 and GroC3 mutants (see below), originally isolated and studied at MIT, which are unable to form colonies at 43°.
PHAGE λ MUTANTS THAT COMPENSATE FOR THE gro15 BLOCK
While at MIT, Ira and I concentrated mostly on the E. coli mutant Gro15. Although the vast majority of λcl− phage are unable to form plaques on this strain, I noted the appearance, at an approximate frequency of 10−7, of phage mutants that were able to somehow compensate for the gro15 block. Interestingly, none of the phage mutants isolated on Gro15 formed plaques on our other mutant, GroC3. Searching for clues as to their nature, I purified many of these gro15 compensatory phage mutants and tested them for growth on standard E. coli laboratory strains, including supD, supE, supF (am-suppressing), and various sup0 (nonsuppressing) wild-type bacterial hosts. To my great surprise and delight, ∼20% of these λ compensatory mutants did not form plaques on any of the wild-type sup0 E. coli tested, but did grow on all of the various am-suppressing wild-type strains. Thus, these phage compensatory mutants carried a suppressible am mutation in some essential, yet unknown, phage gene. This finding highlighted the importance of starting with a strain carrying an am nonsense suppressor allele. Had we started with an E. coli sup0 strain, we would not have isolated such λam compensatory mutants, consequently making it much more tedious to identify the corresponding λ suppressor gene.
I vividly recall our great anticipation and excitement as Ira tested Signer's laboratory collection of essential amber mutants of λ for complementation with our λam compensatory mutants. The very simple, yet elegant technique of “spot” complementation enabled us, in <6 hr, to establish that all of our λam mutants isolated on E. coli gro15 carried mutations in gene P, whose product was already known to be essential for λDNA replication. Because of my Greek origin, I baptized the λ compensatory mutants as “π” to indicate that they map in the P gene. We quickly showed that the bona fide λPam3 or λPam80 mutants of Allan Campbell also grew on the C600 supE gro15 mutant bacteria. Furthermore, recombination experiments showed that most of the λam compensatory mutations were distinct from one another, as well as from Pam3 and Pam80.
Ira and I were perplexed as to how at least nine different am mutations in the λP gene allowed the phage to grow on our C600 supE gro15 mutant host. Since C600 supE gro15 mutants do not grow at high temperature (Table 1), we knew that the corresponding gro15 gene function is essential for bacterial growth, at least at 43°. Our preliminary conclusion was that, for λDNA replication, the λP protein must interact with an essential E. coli protein. Furthermore, we reasoned that our mutant host protein must be at least partially functional, since it carries out its essential bacterial function at all temperatures <43°. Perhaps by simply lowering the λP protein level (because supE is known to suppress am mutations only to ∼5–30% of the wild-type levels), the partially disabled gro15 gene product manages to carry out its function more effectively, thus resulting in some viable phage progeny and hence in plaque formation.
TABLE 1.
Plating properties of groP mutant bacteria
| Growth at 43° | Plaque formation by phage
|
|||
|---|---|---|---|---|
| Bacteria | λ | λπA | λπB | |
| groP+ wild type | + | + | + | + |
| groPA15 (dnaB15) | − | − | + | + |
| groPB558 (dnaB558) | − | − | − | + |
| groPAB756 (groPC756; dnaK756) | − | − | + or − | + |
The original classification scheme devised for the groP class of bacterial mutants is depicted and is taken essentially from Table 1 of Georgopoulos and Herskowitz (1971), except that the later designations of the alleles are indicated in parentheses. The λπA or λπB phage compensatory mutants were isolated as plaque formers on the indicated groP bacterial mutants at an approximate frequency of 10−7 at 37° (see text for details). The subsequent genetic designation(s) of the bacterial alleles is given in parentheses.
THE PHAGE λP PROTEIN INTERACTS WITH E. coli's DnaB PROTEIN
The phage P1-mediated transduction experiments of Ira at MIT (with help from Urs Kühnlein and Madeleine Jolit) showed that gro15 and the vast majority of the groP mutations isolated by me at Stanford mapped in or very near the dnaB locus of E. coli. This finding was rewarding because Fangman and Feiss (1969) had previously shown that λDNA replication was completely blocked in certain dnaB mutants. Furthermore, we showed that previously known bona fide E. coli dnaB mutants behave like our groP mutants, inasmuch as they preferentially support the growth of λπ mutants compared to wild-type λ. Thus, Ira and I specifically proposed at the 1970 Cold Spring Harbor phage λ meeting that, for successful λDNA replication, the host DnaB and phage λP proteins must interact (Figure 2). Later, Wickner (1979) proved and extended this interpretation by showing that the purified E. coli DnaB and phage λP proteins indeed form a complex and that all of DnaB's known biological activities are inhibited in this complex. While the preliminary results on the groP mutants that culminated from my collaboration with Ira were published only in the refereed Cold Spring Harbor Lambda I book edited by Al Hershey (Stahl 1998), I nevertheless consider this publication to be one of my very best (Georgopoulos and Herskowitz 1971).
Figure 2.—
The original diagram used by Ira and me during the 1970 Cold Spring Harbor meeting on phage λ to rationalize the apparent allele specificity observed between some of the E. coli groP mutants and the corresponding phage λP compensatory mutations.
THE DISCOVERY OF THE DnaK/DnaJ/GrpE CHAPERONE MACHINE
It turned out that the only mutation of my Stanford groP collection that did not map at or near the dnaB locus was groPAB756. Interestingly, E. coli groPAB756 exhibited the unique property of plating members of both the λπA and λπB compensatory mutant groups (Table 1). The groPAB756 mutation was eventually mapped during my subsequent stay in Harvey Eisen's laboratory in Geneva, Switzerland, at ∼0.3 min of the E. coli chromosome, near the thr locus, and renamed groPC756 (Georgopoulos 1977). By using an E. coli DNA library cloned into a phage λ vector supplied by Barbara Hohn (Murray and Murray 1975), I was able to easily select for the λgroPC+ recombinant through its ability to form normal-size plaques on the mutant groPC756 host (Georgopoulos 1977). The fact that the groPC756 (λgroPC+) lysogen supports hetero-immune lambdoid phage growth and forms colonies at high temperature proved that the groPC756 mutation is recessive to the wild-type allele and solely responsible for both bacterial temperature sensitivity and the block to lambdoid phage growth. I also concluded that most likely the GroPC protein forms a complex with the λP protein and somehow this interaction is vastly weakened by the groPC756 mutation.
At about this time, Mike Feiss's group had independently isolated an E. coli mutant, GroPC259, on the basis of its inability to propagate both phage λ and phage P2. In their study, published in the same issue as Georgopoulos (1977), Sunshine et al. (1977) showed that their groPC259 mutation is indeed very closely linked to groPC756 and, like groPC756, also affects bacterial growth at high temperature.
Saito and Uchida (1977), selecting for growth of an appropriate λ-defective lysogen at 42°, also isolated bacterial mutations that interfere with λDNA replication and named these mutations grp (groP-like). One of their classes, grpA, was located in the dnaB gene, while another, grpC, mapped near the groPC756 and groPC259 alleles. Ira played a catalytic role in the ensuing collaboration with the Feiss and Uchida laboratories, which quickly established that all of the groPC or grpC alleles indeed fell into two adjacent but distinct complementation groups (Yochem et al. 1978) defined by groPC756 and groPC259. Saito and Uchida (1977, 1978) renamed the genes dnaK and dnaJ, respectively, because they affect host DNA synthesis at high temperatures. Two members of one chaperone machine had been identified.
The third member of the DnaK/DnaJ/GrpE chaperone machine was identified on the basis of genetic properties of the grpE280 mutation of Saito and Uchida (1977), which maps at 56 min (Saito et al. 1978). The GrpE280 mutant was isolated as a colony former at 42°. However, subsequent transductional analysis showed that E. coli GrpE280 mutant cells do not grow at >43° and that the grpE gene is essential even for bacterial growth at all temperatures (Ang et al. 1986; Ang and Georgopoulos 1989). Although the grpE280 mutation also blocks host DNA synthesis at 43°, its gene designation was not changed (Ang et al. 1986).
THE ROLE OF THE DnaK/DnaJ/GrpE CHAPERONE MACHINE IN λDNA REPLICATION AND PROTEIN DISAGGREGATION
The exact role of the DnaK/DnaJ/GrpE chaperone machine in λDNA replication was deciphered over the next several years in various laboratories, including that of Maciej Zylicz (in collaboration with our laboratory), Roger McMacken, the late Hatch Echols, and Sue Wickner (reviewed in Ang et al. 1991). The key step in both E. coli and λDNA replication is the correct positioning of the essential E. coli DNA helicase DnaB at a unique chromosomal site, called oriC in E. coli and oriλ in phage λ. To assist DnaB positioning on λDNA, the λO replication protein binds specifically to oriλ, located within the O structural gene itself (Furth et al. 1978). The phage λP protein, behaving as Dr. Jekyll, then binds DnaB helicase and specifically delivers it to the oriλ site by simultaneously interacting with the λO protein (Furth et al. 1978; Zylicz et al. 1984). However, the Mr. Hyde side of λP's character manifests itself by suppressing all known activities of DnaB in this complex (Wickner 1979). Thus, although the DnaB helicase has been correctly positioned at oriλ, it cannot unwind the DNA until λP is removed from the complex. This is accomplished through disaggregation of the λO–λP–DnaB complex at oriλ by the DnaK/DnaJ/GrpE chaperone machine, resulting in the release of λP and the initiation of DNA replication (reviewed in Ang et al. 1991).
Shortly after, it was shown that the DnaK chaperone machine also disaggregates heat-inactivated E. coli RNA polymerase (Skowyra et al. 1990), as well as aggregated DnaA protein, whose monomerization is essential for bacterial DNA replication (Hwang et al. 1990). This finding provides the rationale for why dnaK, dnaJ, or grpE mutations interfere with bacterial DNA replication at high temperature.
One of the ways in which the DnaJ and GrpE proteins assist the DnaK chaperone to carry out its biological function is by accelerating its very slow ATPase activity. DnaJ specifically accelerates the hydrolysis of DnaK-bound ATP, while GrpE accelerates DnaK's ATPase cycle by acting at the level of nucleotide release (Liberek et al. 1991; Bukau and Horwich 1998; Hartl and Hayer-Hartl 2002). DnaJ also facilitates DnaK's chaperone chores by binding and “presenting” specific substrates to DnaK (reviewed in Craig et al. 2006). In this respect, it is interesting to note that the original dnaJ259 and grpE280 mutations specifically interfere with the interaction of their corresponding gene products with DnaK (Johnson et al. 1989; Liberek et al. 1991; Keppel et al. 2002; Kerner et al. 2005), whereas the original dnaK756 mutation interferes with the interaction of the DnaK756 protein and GrpE (Georgopoulos et al. 1972, 1973; Johnson et al. 1989; Buchberger et al. 1996).
THE DISCOVERY OF THE GroEL/GroES CHAPERONE MACHINE
The discovery of the E. coli genes encoding the GroEL chaperone machine was made in the early 1970s, again through the efforts of bacteriophage geneticists isolating gro-type bacterial mutants that blocked either phage λ (Georgopoulos et al. 1972, 1973; Sternberg 1973) or phage T4 (Takano and Kakefuda 1972; Coppo et al. 1973; Revel et al. 1980). My studies on groE at Stanford were carried out in collaboration with a fellow postdoc, Roger Hendrix. Roger played an important role in both the early stages and the subsequent development of the GroEL chaperone machine story (see below). Again, serendipity played key roles in the GroE story. For example, 30% of the λ compensatory mutants isolated at Stanford as plaque formers on GroC3, isolated at the same time as Gro15, had either am or temperature-sensitive mutations in the capsid-encoding gene E of λ (originally referred to as λɛ). Thus, the GroC3 mutant was renamed GroEAC3 to designate this fact.
The various groE-type mutants in my Stanford collection were arbitrarily classified as GroEA or GroEB on the basis of their ability to propagate various λɛ compensatory mutants (Table 2). I observed that members of both the groEA and the groEB classes did not propagate other lambdoid phage (nor did the virulent phage T5) and interfered with bacterial growth at high temperature. A subsequent detailed genetic analysis of λɛ missense mutants revealed that whereas the λɛA mutations always mapped in the E gene, the λɛB mutations mapped in either the E or the B gene, required for correct morphogenesis of the phage capsid. This result was gratifying because Kaiser's electron micrographs of groE bacteria infected by wild-type λ clearly showed that the λE capsid protein was assembled, but in an aberrant manner (Georgopoulos et al. 1973). In this respect, infection of groE bacteria by wild-type λ resembled infection of wild-type E. coli sup0 bacteria by either λBam or λCam phage. Thus, it appeared from those early studies that the block exerted by our groE mutations on phage λ assembly was very likely at the level of λB action. In this respect, the groE designation is a misnomer, and the name groB would have been more appropriate! Kochan and Murialdo (1983) later showed that the GroEL machine proteins indeed play a primary role in the proper assembly of the λB dodecameric structure, an early step in λ prohead assembly. All of the above observations left the long-standing impression that the GroEL machine's primary role is the correct assembly of macromolecular structures. It would have been almost impossible to realize in the early 1970s that the GroE proteins act uniquely at the level of folding single nascent polypeptide chains, and that their subsequent assembly into macromolecular structures follows spontaneously (see Figure 3 and below).
TABLE 2.
Plating properties of groE mutant bacteria
| Growth at 43° | Plaque formation by phage
|
|||||
|---|---|---|---|---|---|---|
| Bacteria | λ | λɛA | λɛB | T4 | T4ɛ1 | |
| groE+ wild type | + | + | + | + | + | + |
| groEASC3 (groESC3) | − | − | + | − | + | + |
| groEA44 (groEL44) | − | − | − | − | − | + |
| groEB515 (groEL515) | + | − | − | + | + | − |
The original E. coli groE mutant isolates were arbitrarily divided into classes A or B on the basis of the ability of the various λɛ compensatory mutations to form plaques on them. It turned out later that all groES mutations belonged to the groEA group, while groEL mutations fell into both the groEA and the groEB groups (new designation given in parentheses).
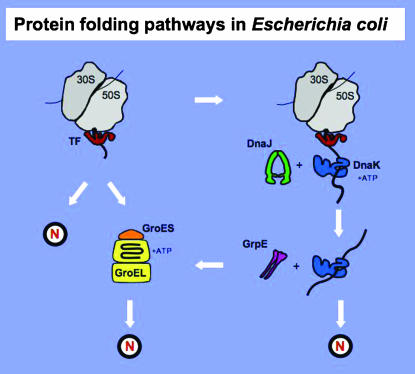
Figure 3.—
Role of DnaK and GroE chaperone machines in nascent protein folding. The diagram depicts the ribosome, the ribosome-bound TF chaperone, and the DnaK/DnaJ/GrpE and GroES/GroEL chaperone machines. The diagram depicts the various pathways that nascent polypeptides may follow during the intracellular folding process to arrive at their native state (N). It is essentially adapted from Hartl and Hayer-Hartl (2002), except all structures are drawn to scale. See text for details.
The second lucky finding was the observation that one of the E. coli mutant hosts, GroEA44, in addition to not propagating λ, was unique among my isolates in not propagating my old friend phage T4 as well, although I had exerted no selection for such a phenotype in the original isolation procedure (Table 2). However, again, at a frequency of ∼10−7, T4 compensatory plaque-forming mutants (originally referred to as T4ɛ) could be isolated. The third stroke of luck was that one of these phage T4 mutants, ɛ1, simultaneously lost its ability to propagate on some of the other groE mutant hosts (e.g., groEB515), which otherwise allow normal growth of wild-type T4 phage (Table 2). The inability of T4ɛ1 to grow on the GroEB515 mutant host led to the quick assignment of its mutation to the morphogenetic gene 31, again by spot complementation tests using a battery of known T4 am mutants (in collaboration with William Wood, then at Caltech) (Georgopoulos et al. 1972). In this respect, T4-infected GroEL mutants exhibit a phenotype identical to wild-type sup0 bacteria infected by T4 31 am mutants (Laemmli et al. 1970). The effect of a single mutation on both λ and T4 phage head assembly pointed to a possible direct interaction between the groEA44 gene product and the T4-encoded 31 gene product, Gp31, in proper protein assembly. Essentially similar conclusions about Gp31/GroEL interactions were reached by Takano and Kakefuda (1972), Coppo et al. (1973), and Revel et al. (1980), who named their corresponding groE-like mutations mop, tabB, and hdh, respectively.
HOST- AND PHAGE-ENCODED GroEL CO-CHAPERONES
By using the first phage λ recombinant libraries of E. coli DNA, λgroE+-transducing phage were easily selected as plaque formers on various groE mutant hosts (Georgopoulos and Hohn 1978; Hendrix and Tsui 1978). Barbara Hohn played a catalytic role in the isolation of a λgroE+-transducing phage by obtaining the λ library from Ken Murray, whereas Hendrix independently obtained his library from Ron Davis's laboratory. Both groups easily identified the “GroE” product as an ∼60,000-Da protein following infection of UV-irradiated bacteria. The easy overproduction of “GroE” from λgroE+-infected cells led to its purification by Hendrix (1979) and Hohn et al. (1979), followed by determination of its large, tetradecameric structure and ATPase activity.
Subsequently, a more careful genetic analysis of various λgroE+-deleted derivatives led us to the realization that there are two groE cistrons, named groEL (to signify the large 60,000-Da GroEL polypeptide) and groES (to indicate the small ∼15,000-Da polypeptide) (Tilly et al. 1981). Approximately half of my groE mutant alleles mapped in the groEL gene and the rest mapped in groES. Purification of GroES demonstrated its ATP-dependent interaction with GroEL (Chandrasekhar et al. 1986), and its negative modulation of GroEL's ATPase activity. These biochemical results were anticipated by earlier genetic suppressor studies (Tilly and Georgopoulos 1982), and they helped explain why mutations in either groES or groEL exert the same phenotypes on the growth of E. coli or phage λ (Georgopoulos et al. 1973). The genetic experiments of Olivier Fayet clearly demonstrated that the groES and groEL genes are the only major chaperone-encoding genes absolutely essential for E. coli viability under all conditions tested (Fayet et al. 1989). This essentiality has been traced to its unique ability to fold a small number of essential E. coli proteins (Kerner et al. 2005).
It took a few more years to demonstrate directly that Gp31 is a bona fide GroEL co-chaperone (van der Vies et al. 1994; Richardson et al. 1999). Further studies showed that many large bacteriophage encode Gp31-like proteins capable of substituting for GroES in E. coli growth (reviewed in Ang et al. 2000, 2001; Keppel et al. 2002). However, it is still not clear why Gp31, but not GroES, is uniquely required for correct folding of the Gp23 capsid protein.
It is interesting to point out again that the use of phage λ in our original selection was serendipitous because it led to the isolation of mutations in both groES and groEL, thus revealing the existence of the two genes. If we had used only phage T4, which does not need GroES, only groEL would have been identified under these circumstances.
Because of the GroE chaperone machine's effect on phage morphogenesis and the fact that Hsp60, the eukaryotic GroEL homolog, is also involved in the correct assembly of the large, oligomeric Rubisco protein in plants, the notion that GroE is uniquely involved in macromolecular protein assembly persisted until 1988 (Hemmingsen et al. 1988). This concept began to unravel when Bochkareva et al. (1988) clearly demonstrated that GroEL crosslinks in crude extracts to the nascent chain of β-lactamase, a monomeric enzyme destined for the E. coli periplasm. Shortly after, George Lorimer's group demonstrated that the GroES/GroEL chaperone machine assists the correct folding of prokaryotic Rubisco in a purified system (Goloubinoff et al. 1989). The ensuing years resulted in an explosion in our knowledge of the mechanistic details of the GroES/GroEL machinery (Bukau and Horwich 1998; Hartl and Hayer-Hartl 2002).
THE DnaK AND GroEL CHAPERONE MACHINES AND INTRACELLULAR PROTEIN FOLDING
Protein folding, protein aggregation, and protein–chaperone interactions are highly dynamic processes (Bukau and Horwich 1998; Hartl and Hayer-Hartl 2002; Bukau et al. 2006; Chiti and Dobson 2006; Craig et al. 2006). Figure 3 outlines the role of the DnaK and GroEL chaperone machines in E. coli to prevent premature nascent polypeptide aggregation, thus promoting proper folding. Trigger Factor (TF; the tig gene product) is a highly abundant chaperone that reversibly binds to the ribosome in the vicinity of the polypeptide channel exit and thus interacts first with the emerging nascent chains. Subsequently, the DnaK chaperone machine can step in and bind some of these nascent chains. However, because of their substrate-binding promiscuity, additional chaperones may also interact transiently, and with varying affinities, with a given protein substrate. The key role that the TF and DnaK chaperone machines play in protein folding is highlighted by the resulting synthetic lethality of the tig dnaK double mutant above 30° (Genevaux et al. 2004; Vorderwulbecke et al. 2004). The DnaK (Hsp70) machine, often in collaboration with the ClpB (Hsp104) chaperone, can also disaggregate certain protein aggregates once formed (Bukau et al. 2006). In contrast to TF and the DnaK machine, the GroEL machine is generally thought to act mostly at a post-translational level to help some polypeptides fold properly. The potential in vivo interchangeability and plasticity of the various chaperone machines is exemplified by several recent findings. First, Ying et al. (2006) have shown that GroEL can unexpectedly associate cotranslationally with nascent chains in a well-defined in vitro system. Second, the overproduction of either the GroEL machine or the SecB chaperone enables both growth of the tig dnaK double mutant and a reduction of protein aggregates at otherwise lethal temperatures (Genevaux et al. 2004; Ullers et al. 2004; Vorderwulbecke et al. 2004). Third, both the E. coli DnaK chaperone machine and its various eukaryotic Hsp70 homologs can function interchangeably both in vivo and in vitro with various DnaJ-like and GrpE-like factors (Brodsky and Chiosis 2006; Craig et al. 2006). Thus, it is likely that in vivo some nascent polypeptide chains can be “cradled” by more than one chaperone machine “midwife.”
Recent studies have also highlighted the potential role of the Hsp70 chaperone machine in modulating a variety of important cellular functions, such as apoptosis, tumor growth, and certain human diseases caused by protein aggregation, e.g., Parkinson's and Huntington's diseases. In turn, this has spurred the development of small molecule modulators of Hsp70 activity as therapeutic agents (Brodsky and Chiosis 2006). The Hsp60 human chaperone machine has also been shown to play an important role in the development of a particular form of human spastic paraplegia (Hansen et al. 2002).
EPILOGUE
Ira visited Geneva a few months before his untimely death in the spring of 2003 (Botstein 2004). One evening we took a long walk along the beautiful lake shore and reminisced about the “good old days.” We both felt lucky that we lived through the relatively early stages of the 1953 “big bang” era of molecular biology. In the spring of 1969, armed with sterile toothpicks, we embarked on a long journey of adventure and discovery but, unlike Odysseus, with no clear destination in mind. We were heartened that our genetic studies, with such primitive tools as toothpicks and phage spot tests, had contributed to a deeper understanding of the intracellular protein-folding process. We were certain that plenty of important discoveries were still to be made, hiding at the tips of toothpicks wielded by future generations of microbial geneticists. Finally, we agreed that genetic analyses of simple organisms, such as phage, bacteria, and yeast, is the easiest and fastest way to arrive at “Hershey's Heaven” (Stahl 1998).
Acknowledgments
I thank all of my past and present collaborators who have contributed to progress in the chaperone field, Debbie Ang for Figure 1, and Pierre Genevaux for Figure 3. Finally, I am grateful to Debbie Ang for joining me on this trail of adventure and for her critical suggestions in making the text more concise and palatable. I am also grateful to the National Institutes of Health, the Swiss National Fund, and the Canton of Geneva for financial support.
I dedicate this article to the memory of my friend Ira Herskowitz. Without his seminal contributions and insights, I never would have embarked on this type of research.
References
- Ang, D., and C. Georgopoulos, 1989. The heat-shock-regulated grpE gene of Escherichia coli is required for bacterial growth at all temperatures but is dispensable in certain mutant backgrounds. J. Bacteriol. 171: 2748–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang, D., G. N. Chandrasekhar, M. Zylicz and C. Georgopoulos, 1986. Escherichia coli grpE gene codes for heat shock protein B25.3, essential for both lambda DNA replication at all temperatures and host growth at high temperature. J. Bacteriol. 167: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang, D., K. Liberek, D. Skowyra, M. Zylicz and C. Georgopoulos, 1991. Biological role and regulation of the universally conserved heat shock proteins. J. Biol. Chem. 266: 24233–24236. [PubMed] [Google Scholar]
- Ang, D., F. Keppel, G. Klein, A. Richardson and C. Georgopoulos, 2000. Genetic analysis of bacteriophage-encoded cochaperonins. Annu. Rev. Genet. 34: 439–456. [DOI] [PubMed] [Google Scholar]
- Ang, D., A. Richardson, M. P. Mayer, F. Keppel, H. Krisch et al., 2001. Pseudo-T-even bacteriophage RB49 encodes CocO, a cochaperonin for GroEL, which can substitute for Escherichia coli's GroES and bacteriophage T4's Gp31. J. Biol. Chem. 276: 8720–8726. [DOI] [PubMed] [Google Scholar]
- Bochkareva, E. S., N. M. Lissin and A. S. Girshovich, 1988. Transient association of newly synthesized unfolded proteins with the heat-shock GroEL protein. Nature 336: 254–257. [DOI] [PubMed] [Google Scholar]
- Botstein, D., 2004. Ira Herskowitz: 1946–2003. Genetics 166: 653–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky, J. L., and G. Chiosis, 2006. Hsp70 molecular chaperones: emerging roles in human disease and identification of small molecule modulators. Curr. Top. Med. Chem. 6: 1215–1225. [DOI] [PubMed] [Google Scholar]
- Buchberger, A., H. Schroder, T. Hesterkamp, H. J. Schonfeld and B. Bukau, 1996. Substrate shuttling between the DnaK and GroEL systems indicates a chaperone network promoting protein folding. J. Mol. Biol. 261: 328–333. [DOI] [PubMed] [Google Scholar]
- Bukau, B., and A. L. Horwich, 1998. The Hsp70 and Hsp60 chaperone machines. Cell 92: 351–366. [DOI] [PubMed] [Google Scholar]
- Bukau, B., J. Weissman and A. Horwich, 2006. Molecular chaperones and protein quality control. Cell 125: 443–451. [DOI] [PubMed] [Google Scholar]
- Campbell, A. M., 1993. Thirty years ago in Genetics: prophage insertion into bacterial chromosomes. Genetics 133: 433–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekhar, G. N., K. Tilly, C. Woolford, R. Hendrix and C. Georgopoulos, 1986. Purification and properties of the groES morphogenetic protein of Escherichia coli. J. Biol. Chem. 261: 12414–12419. [PubMed] [Google Scholar]
- Chiti, F., and C. M. Dobson, 2006. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75: 333–366. [DOI] [PubMed] [Google Scholar]
- Coppo, A., A. Manzi, J. F. Pulitzer and H. Takahashi, 1973. Abortive bacteriophage T4 head assembly in mutants of Escherichia coli. J Mol Biol 76: 61–87. [DOI] [PubMed] [Google Scholar]
- Craig, E. A., P. Huang, R. Aron and A. Andrew, 2006. The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev. Physiol. Biochem. Pharmacol. 156: 1–21. [DOI] [PubMed] [Google Scholar]
- Edgar, B., 2004. The genome of bacteriophage T4: an archeological dig. Genetics 168: 575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangman, W. L., and M. Feiss, 1969. Fate of lambda DNA in a bacterial host defective in DNA synthesis. J. Mol. Biol. 44: 103–116. [DOI] [PubMed] [Google Scholar]
- Fayet, O., T. Ziegelhoffer and C. Georgopoulos, 1989. The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J. Bacteriol. 171: 1379–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth, M. E., C. McLeester and W. F. Dove, 1978. Specificity determinants for bacteriophage lambda DNA replication. I. A chain of interactions that controls the initiation of replication. J. Mol. Biol. 126: 195–225. [DOI] [PubMed] [Google Scholar]
- Genevaux, P., F. Keppel, F. Schwager, P. S. Langendijk-Genevaux, F. U. Hartl et al., 2004. In vivo analysis of the overlapping functions of DnaK and trigger factor. EMBO Rep. 5: 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos, C., 1971. Bacterial mutants in which the gene N function of bacteriophage lambda is blocked have an altered RNA polymerase. Proc. Natl. Acad. Sci. USA 68: 2977–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos, C. P., 1977. A new bacterial gene (groPC) which affects lambda DNA replication. Mol. Gen. Genet. 151: 35–39. [DOI] [PubMed] [Google Scholar]
- Georgopoulos, C. P., and I. Herskowitz, 1971. Escherichia coli mutants blocked in lambda DNA synthesis, pp. 553–565 in The Bacteriophage Lambda, edited by A. D. Hershey. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Georgopoulos, C. P., and B. Hohn, 1978. Identification of a host protein necessary for bacteriophage morphogenesis (the groE gene product). Proc. Natl. Acad. Sci. USA 75: 131–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos, C., and W. J. Welch, 1993. Role of the major heat shock proteins as molecular chaperones. Annu. Rev. Cell Biol. 9: 601–634. [DOI] [PubMed] [Google Scholar]
- Georgopoulos, C. P., R. W. Hendrix, A. D. Kaiser and W. B. Wood, 1972. Role of the host cell in bacteriophage morphogenesis: effects of a bacterial mutation on T4 head assembly. Nat. New Biol. 239: 38–41. [DOI] [PubMed] [Google Scholar]
- Georgopoulos, C. P., R. W. Hendrix, S. R. Casjens and A. D. Kaiser, 1973. Host participation in bacteriophage lambda head assembly. J. Mol. Biol. 76: 45–60. [DOI] [PubMed] [Google Scholar]
- Goloubinoff, P., J. T. Christeller, A. A. Gatenby and G. H. Lorimer, 1989. Reconstitution of active dimeric ribulose bisphosphate carboxylase from an unfolded state depends on two chaperonin proteins and Mg-ATP. Nature 342: 884–889. [DOI] [PubMed] [Google Scholar]
- Hansen, J. J., A. Durr, I. Cournu-Rebeix, C. Georgopoulos, D. Ang et al., 2002. Hereditary spastic paraplegia SPG13 is associated with a mutation in the gene encoding the mitochondrial chaperonin Hsp60. Am. J. Hum. Genet. 70: 1328–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl, F. U., and M. Hayer-Hartl, 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295: 1852–1858. [DOI] [PubMed] [Google Scholar]
- Hemmingsen, S. M., C. Woolford, S. M. van der Vies, K. Tilly, D. T. Dennis et al., 1988. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature 333: 330–334. [DOI] [PubMed] [Google Scholar]
- Hendrix, R. W., 1979. Purification and properties of groE, a host protein involved in bacteriophage assembly. J. Mol. Biol. 129: 375–392. [DOI] [PubMed] [Google Scholar]
- Hendrix, R. W., and L. Tsui, 1978. Role of the host in virus assembly: cloning of the Escherichia coli groE gene and identification of its protein product. Proc. Natl. Acad. Sci. USA 75: 136–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey, A. D. (Editor), 1971. The Bacteriophage Lambda. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Hohn, T., B. Hohn, A. Engel, M. Wurtz and P. R. Smith, 1979. Isolation and characterization of the host protein groE involved in bacteriophage lambda assembly. J. Mol. Biol. 129: 359–373. [DOI] [PubMed] [Google Scholar]
- Hwang, D. S., E. Crooke and A. Kornberg, 1990. Aggregated dnaA protein is dissociated and activated for DNA replication by phospholipase or dnaK protein. J. Biol. Chem. 265: 19244–19248. [PubMed] [Google Scholar]
- Johnson, C., G. N. Chandrasekhar and C. Georgopoulos, 1989. Escherichia coli DnaK and GrpE heat shock proteins interact both in vivo and in vitro. J. Bacteriol. 171: 1590–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppel, F., M. Rychner and C. Georgopoulos, 2002. Bacteriophage-encoded cochaperonins can substitute for Escherichia coli's essential GroES protein. EMBO Rep. 3: 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner, M. J., D. J. Naylor, Y. Ishihama, T. Maier, H. C. Chang et al., 2005. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122: 209–220. [DOI] [PubMed] [Google Scholar]
- Kochan, J., and H. Murialdo, 1983. Early intermediates in bacteriophage lambda prohead assembly. II. Identification of biologically active intermediates. Virology 131: 100–115. [DOI] [PubMed] [Google Scholar]
- Laemmli, U. K., F. Beguin and G. Gujer-Kellenberger, 1970. A factor preventing the major head protein of bacteriophage T4 from random aggregation. J. Mol. Biol. 47: 69–85. [DOI] [PubMed] [Google Scholar]
- Liberek, K., J. Marszalek, D. Ang, C. Georgopoulos and M. Zylicz, 1991. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc. Natl. Acad. Sci. USA 88: 2874–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist, S., and E. A. Craig, 1988. The heat-shock proteins. Annu. Rev. Genet. 22: 631–677. [DOI] [PubMed] [Google Scholar]
- Murray, K., and N. E. Murray, 1975. Phage lambda receptor chromosomes for DNA fragments made with restriction endonuclease III of Haemophilus influenzae and restriction endonuclease I of Escherichia coli. J. Mol. Biol. 98: 551–564. [DOI] [PubMed] [Google Scholar]
- Revel, H. R., and S. E. Luria, 1970. DNA-glucosylation in T-even phage: genetic determination and role in phagehost interaction. Annu. Rev. Genet. 4: 177–192. [DOI] [PubMed] [Google Scholar]
- Revel, H. R., B. L. Stitt, I. Lielausis and W. B. Wood, 1980. Role of the host cell in bacteriophage T4 development. I. Characterization of host mutants that block T4 head assembly. J. Virol. 33: 366–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, A., S. M. van der Vies, F. Keppel, A. Taher, S. J. Landry et al., 1999. Compensatory changes in GroEL/Gp31 affinity as a mechanism for allele-specific genetic interaction. J. Biol. Chem. 274: 52–58. [DOI] [PubMed] [Google Scholar]
- Saito, H., and H. Uchida, 1977. Initiation of the DNA replication of bacteriophage lambda in Escherichia coli K12. J. Mol. Biol. 113: 1–25. [DOI] [PubMed] [Google Scholar]
- Saito, H., and H. Uchida, 1978. Organization and expression of the dnaJ and dnaK genes of Escherichia coli K12. Mol. Gen. Genet. 164: 1–8. [DOI] [PubMed] [Google Scholar]
- Saito, H., Y. Nakamura and H. Uchida, 1978. A transducing lambda phage carrying grpE, a bacterial gene necessary for lambda DNA replication, and two ribosomal protein genes, rpsP (S16) and rplS (L19). Mol. Gen. Genet. 165: 247–256. [DOI] [PubMed] [Google Scholar]
- Skowyra, D., C. Georgopoulos and M. Zylicz, 1990. The E. coli dnaK gene product, the hsp70 homolog, can reactivate heat-inactivated RNA polymerase in an ATP hydrolysis-dependent manner. Cell 62: 939–944. [DOI] [PubMed] [Google Scholar]
- Stahl, F. W., 1998. Hershey. Genetics 149: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg, N., 1973. Properties of a mutant of Escherichia coli defective in bacteriophage lambda head formation (groE). II. The propagation of phage lambda. J. Mol. Biol. 76: 25–44. [DOI] [PubMed] [Google Scholar]
- Sunshine, M., M. Feiss, J. Stuart and J. Yochem, 1977. A new host gene (groPC) necessary for lambda DNA replication. Mol. Gen. Genet. 151: 27–34. [DOI] [PubMed] [Google Scholar]
- Takano, T., and T. Kakefuda, 1972. Involvement of a bacterial factor in morphogenesis of bacteriophage capsid. Nat. New Biol. 239: 34–37. [DOI] [PubMed] [Google Scholar]
- Tilly, K., and C. Georgopoulos, 1982. Evidence that the two Escherichia coli groE morphogenetic gene products interact in vivo. J. Bacteriol. 149: 1082–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly, K., H. Murialdo and C. Georgopoulos, 1981. Identification of a second Escherichia coli groE gene whose product is necessary for bacteriophage morphogenesis. Proc. Natl. Acad. Sci. USA 78: 1629–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullers, R. S., J. Luirink, N. Harms, F. Schwager, C. Georgopoulos et al., 2004. SecB is a bona fide generalized chaperone in Escherichia coli. Proc. Natl. Acad. Sci. USA 101: 7583–7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vies, S. M., A. A. Gatenby and C. Georgopoulos, 1994. Bacteriophage T4 encodes a co-chaperonin that can substitute for Escherichia coli GroES in protein folding. Nature 368: 654–656. [DOI] [PubMed] [Google Scholar]
- Vorderwulbecke, S., G. Kramer, F. Merz, T. A. Kurz, T. Rauch et al., 2004. Low temperature or GroEL/ES overproduction permits growth of Escherichia coli cells lacking trigger factor and DnaK. FEBS Lett. 559: 181–187. [DOI] [PubMed] [Google Scholar]
- Wickner, S. H., 1979. DNA replication proteins of Escherichia coli and phage lambda. Cold Spring Harbor Symp. Quant. Biol. 43(Pt. 1): 303–310. [DOI] [PubMed] [Google Scholar]
- Ying, B. W., H. Taguchi and T. Ueda, 2006. Co-translational binding of GroEL to nascent polypeptides is followed by post-translational encapsulation by GroES to mediate protein folding. J. Biol. Chem. 281: 21813–21819. [DOI] [PubMed] [Google Scholar]
- Yochem, J., H. Uchida, M. Sunshine, H. Saito, C. P. Georgopoulos et al., 1978. Genetic analysis of two genes, dnaJ and dnaK, necessary for Escherichia coli and bacteriophage lambda DNA replication. Mol. Gen. Genet. 164: 9–14. [DOI] [PubMed] [Google Scholar]
- Zylicz, M., I. Gorska, K. Taylor and C. Georgopoulos, 1984. Bacteriophage lambda replication proteins: formation of a mixed oligomer and binding to the origin of lambda DNA. Mol. Gen. Genet. 196: 401–406. [DOI] [PubMed] [Google Scholar]