Abstract
Secretagogin is a hexa EF-hand protein, which has been identified as a novel potential tumour marker. In the present study, we show that secretagogin binds four Ca2+ ions (log K1=7.1±0.4, log K2=4.7±0.6, log K3=3.6±0.7 and log K4=4.6±0.6 in physiological salt buffers) with a [Ca2+]0.5 of approx. 25 μM. The tertiary structure of secretagogin changes significantly upon Ca2+ binding, but not upon Mg2+ binding, and the amount of exposed hydrophobic surface in secretagogin increases upon Ca2+ binding, but not upon Mg2+ binding. These properties suggest that secretagogin belongs to the ‘sensor’ family of Ca2+-binding proteins. However, in contrast with the prototypical Ca2+ sensor calmodulin, which interacts with a very large number of proteins, secretagogin is significantly less promiscuous. Only one secretagogin-interacting protein was reproducibly identified from insulinoma cell lysates and from bovine and mouse brain homogenates. This protein was identified as SNAP-25 (25 kDa synaptosome-associated protein), a protein involved in Ca2+-induced exocytosis in neurons and in neuroendocrine cells. Kd was determined to be 1.2×10−7 M in the presence of Ca2+ and 1.5×10−6 M in the absence of Ca2+. The comparatively low Ca2+ affinity for secretagogin and the fact that it undergoes Ca2+-induced conformational changes and interacts with SNAP-25 raise the possibility that secretagogin may link Ca2+ signalling to exocytotic processes.
Keywords: calcium binding, conformational change, EF-hand, 25 kDa synaptosome-associated protein (SNAP-25), secretagogin, sensor
Abbreviations: ANS, 8-anilinonaphthalene-l-sulfonic acid; BAPTA, bis-(o-aminophenoxy)ethane-N,N,N′,N′-tetra-acetic acid; 5,5′-Br2-, 5,5′-dibromo-; DTT, dithiothreitol; GST, glutathione S-transferase; HEF, hexa EF-hand; NHS, N-hydroxysuccinimide; ITC, isothermal titration calorimetry; LC, liquid chromatography; MALDI–TOF, matrix-assisted laser-desorption ionization–time-of-flight; MS/MS, tandem MS; 5N-, 5-nitro-; SNAP-25, 25 kDa synaptosome-associated protein; SNARE, soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor; SPR, surface plasmon resonance
INTRODUCTION
Secretagogin is a recently described protein that is expressed in pancreatic islets [1] and the central nervous system, particularly neuroendocrine cells [2,3]. The secretagogin mRNA was identified as one of the most abundant transcripts in pancreatic islets, highlighting the potential importance of secretagogin in this tissue [1]. Secretagogin is down-regulated in pancreatic islets exposed to high concentrations of glucose [4], in human non-functional pituitary adenomas [5] and in adenocarcinomas [6]. These results suggest that secretagogin may be involved in suppressing cell growth, and it has been proposed as a novel biomarker for diagnosis of different forms of cancer [7].
Secretagogin belongs to the HEF (hexa EF-hand) protein family, a group of Ca2+-binding proteins within the calmodulin superfamily. Well-known members of the HEF group are calbindin D28k and calretinin. A high-resolution structure was recently presented for calbindin D28k [8], showing a single globular domain comprising all six EF-hands in agreement with earlier fragment complementation studies [9,10]. Secretagogin exists in three different forms, two of which are characterized by a single amino acid exchange [glutamine (Q)/arginine (R)] at residue 22 (secretagogin Q-22 and secretagogin R-22) [11]. The third form, setagin, does not bind Ca2+ and consists of only 49 amino acids [11]. Many EF-hand proteins belong to one of two classes: (i) signalling proteins or (ii) buffering proteins. EF-hand proteins that function as Ca2+-buffers or transporters have high Ca2+ affinities (Kd<0.1 μM) and undergo only minor conformational changes upon binding of Ca2+ (for a review, see [12]). An important biological function of Ca2+-buffer proteins is to maintain non-toxic levels of free intracellular Ca2+ and to restore the low resting level after signalling events. The signalling proteins (also called Ca2+ sensors) have lower Ca2+ affinities (Kd>1 μM) and undergo significant Ca2+-induced conformational changes, which result in exposure or modulation of binding surfaces for target proteins. The target proteins are often activated or attenuated upon complex formation. For example, a Ca2+ sensor protein may activate protein kinases that initiate cytoplasmic and nuclear actions. These processes are tightly controlled by the Ca2+ concentration in the cell and the Ca2+-binding affinity of the EF-hand protein. In the present study, we have studied the Ca2+-binding properties of secretagogin, its conformational response upon Ca2+ binding and the binding of secretagogin to other proteins.
EXPERIMENTAL
Proteins
Human recombinant calbindin D28k was expressed in Escherichia coli and purified to homogeneity as described in [13]. Human recombinant secretagogin and SNAP-25 (25 kDa synaptosome-associated protein) were expressed as GST (glutathione S-transferase)-fusion proteins in E. coli. The cells were pelleted and resuspended in ice-cold PBS, and then lysed by passage through a French pressure cell. Insoluble material was pelleted, and the supernatants were loaded on to glutathione–Sepharose columns, which were then washed thoroughly with PBS at 4 °C. Thrombin was added to the columns, which were then incubated on a rocking table at room temperature for 12 h for secretagogin or 2 h for SNAP-25. The thrombin activity was stopped with PMSF, and the cleaved proteins were eluted with PBS. The proteins were further purified on a Pharmacia HiLoad® 26/60 Superdex 75 prep grade gel filtration column. The concentrations of purified SNAP-25 and secretagogin were determined by amino acid analysis after acid hydrolysis. Ca2+-free (apo) protein was prepared in a similar manner as described previously for calbindin D28k [13]. 1H NMR spectroscopy was used to verify that the apo samples were free from EGTA.
Far-UV CD spectroscopy
Secretagogin (9 μM; 0.3 mg/ml) in 1 mM DTT (dithiothreitol), 0.5 mM EGTA and 2 mM Tris/HCl (pH 7.5) was used to record the far-UV CD spectra in the far-UV range (185–250 nm). Then, 5 mM MgCl2 (pH 7.5) was added, and the same measurements were performed again. Finally, 1 mM CaCl2 (pH 7.5) was added, and the same measurements were repeated. CD spectra were obtained using a Jasco J-720 spectropolarimeter at 25 °C (thermostatically maintained).
Near-UV CD spectroscopy and near-UV absorbance spectroscopy
A protein solution (37 μM secretagogin, in 1 mM DTT, 0.5 mM EGTA and 2 mM Tris, pH 7.5) was used to record the CD and UV spectra in the near-UV range (250–300 nm). Then, 5 mM MgCl2 (pH 7.5) was added, and the same measurements were performed again. Finally, 1 mM CaCl2 (pH 7.5) was added, and the same measurements were repeated. UV absorbance measurements were done in a GBC Scientific Equipment (Dandenong, Australia) UV–visible 920 spectrophotometer. CD spectra were obtained using a Jasco J-720 spectropolarimeter at 25 °C (thermostatically maintained). Fluorescence cuvettes made of quartz with path lengths of 10 mm were used.
ANS (8-anilinonaphthalene-l-sulfonic acid) and tryptophan fluorescence spectroscopy
Fluorescence spectra were obtained using a PerkinElmer lumine-scence spectrometer LS 50 B connected to a Julabo F25 water bath. All data were collected at 25.0 °C using a quartz cuvette (10 mm path length). For tryptophan fluorescence spectra, the excitation wavelength was 295 nm and the emission was scanned between 300 and 450 nm. The excitation slit was 3 nm and the emission slit 3 nm. The ANS fluorescence spectra were recorded between 400 and 600 nm with excitation at 385 nm. The excitation slit was 3 nm and the emission slit 8 nm. A protein solution (5 μM secretagogin in 1 mM DTT, 0.5 mM EGTA, 2 mM Tris/HCl and 0.15 mM KCl, pH 7.5) was used to record the intrinsic (tryptophan) fluorescence emission spectrum or the ANS fluorescence spectra, in which case the protein was saturated with 120 μM ANS. Then, 5 mM MgCl2 (pH 7.5) was added and the measurements were performed again. Finally, 1 mM CaCl2 (pH 7.5) was added and the measurements were repeated.
Macroscopic Ca2+-binding constants by the chelator method
The protein (30 μM) was titrated with Ca2+ in the presence of a chromophoric chelator (25 μM) in 2 mM Tris/HCl (pH 7.5) and 0.15 M KCl. Experiments were performed using the chelators 5,5′-Br2-BAPTA [5,5′-dibromobis-(o-aminophenoxy)ethane-N,N,N′,N′-tetra-acetic acid] and 5N-BAPTA (5-nitro-BAPTA). The chelator concentration was determined by absorbance, the Ca2+ concentrations by atomic-absorbtion spectroscopy and the protein concentration by amino acid analysis after acid hydrolysis. The chelator–protein solutions (1.25 ml) were titrated with 4 μl aliquots of a 2.94 mM CaCl2 solution, and a few 4 μl aliquots of a 10.7 mM CaCl2 solution were added at the end of the titration. The titration was monitored as a change in absorbance at 263 nm (5,5′-Br2-BAPTA) or 430 nm (5N-BAPTA). Two to three sets of data were obtained for each chelator–protein combination. Parallel experiments were performed in a buffer without salt (2 mM Tris/HCl, pH 7.5). The data were analysed using the Caligator software [14] using a model with a chelator of known Ca2+ affinity and up to four macroscopic Ca2+ binding constants for the protein.
ITC (isothermal titration calorimetry)
ITC was carried out using a VP-ITC instrument from Microcal at constant temperature (25 °C) in 2 mM Hepes/NaOH buffer (pH 7.5) with 0 or 150 mM NaCl. Deionized water was used in the reference cell. All solutions were thoroughly degassed by stirring under vacuum before use. CaCl2 (1 mM) was titrated into 25 μM secretagogin in the sample cell (1.4 ml). For each titration, a 2 μl injection was followed by 40 injections of 4 μl each into the sample cell. The mixture was allowed to react for 4 min between injections. Heats due to injection and dilution were obtained by titrating CaCl2 into buffer. The data were fitted using the modified Origin software supplied by Microcal. ΔH0 (enthalpy change) and log K values were obtained by fitting a macroscopic binding model and fits allowing for two, three or four binding sites were attempted.
Limited proteolysis of secretagogin
Secretagogin was dissolved in 50 mM Tris/HCl and 150 mM KCl, containing either 1 mM CaCl2 or 1 mM EDTA at a protein concentration of 1 mg/ml. The pH was adjusted to 7.5. Sequencing grade modified trypsin (Promega) was dissolved in 50 mM ammonium bicarbonate, yielding a stock solution with a concentration of 0.5 mg/ml. Proteolysis was initiated by mixing 700 μl of secretagogin (1 mg/ml) with 7 μl of the trypsin stock at room temperature. Aliquots of 50 μl were withdrawn at various time points and the digestion was blocked by the addition of 2 μl of soya-bean trypsin inhibitor (1 mg/ml) (Boehringer Mannheim; Roche). Peptide fragments from the digestions were analysed by MALDI–TOF (matrix-assisted laser-desorption ionization–time-of-flight). Spectra were acquired on a MALDI-Micro MX (Waters) in linear mode. Sinapinic acid was used as the matrix and an external calibration standard was used for calibration of the spectra. The experimentally measured peptide masses were compared with the theoretical peptides calculated from the human secretagogin sequence by the FindPept tool (http://us.expasy.org/tools/find-pept.html). The digested fragments were also separated by SDS/PAGE (16.5% gel) in 100 mM Tris/Tricine buffer, pH 8.3. Some Coomassie Blue-stained protein fragments were excised from the gel and identified by LC (liquid chromatography)-MS/MS (tandem MS) as described previously [15].
Affinity chromatography, SDS/PAGE and identification of proteins
Human recombinant secretagogin or calbindin D28k were immobilized on CNBr-activated Sepharose. A number of columns were prepared with 3 mg of immobilized protein on 1 ml of CNBr-activated Sepharose. Bovine brain tissue (10 g) from two animals or mouse brain tissue (3 g) from ten mice was homogenized using a mixer followed by a Potter–Elvehjem tissue grinder in 20 ml of 50 mM Tris, 150 mM NaCl, 1 mM DTT and 200 μl of protease inhibitor cocktail (Sigma P8340). The homogenate was then centrifuged three times for 6 min at 3000 g and the supernatant was collected and ultracentrifuged at 45000 rev./min for 90 min using a Beckman 50.2 Ti rotor. The pellet was collected and dissolved by homogenization by a Potter–Elvehjem tissue grinder in 20 ml of a buffer containing detergent [1% β-dodecyl maltoside, 50 mM Tris, 150 mM NaCl, 1 mM DTT and 200 μl of protease inhibitor cocktail (Sigma P8340)]. The pellet fraction was then incubated for 1 h and ultracentrifuged at 100000 g for 90 min, and the supernatant was collected and divided into two equal parts. CaCl2 (2 mM) was added to one part and EGTA (2 mM) was added to the other. The samples were ultracentrifuged again at 100000 g for 90 min and the supernatants were collected and filtered through 45 μM filters. Then, 7 ml of the supernatants was incubated with secretagogin–Sepharose or calbindin D28k–Sepharose respectively for 2 h. The affinity columns incubated in the presence of 2 mM CaCl2 were rinsed with 20 column volumes of 50 mM Tris, 0.15 M NaCl, 1 mM DTT and 2 mM CaCl2 (pH 7.5) containing 0.25% β-dodecyl maltoside, followed by a two-step elution. First, the columns were eluted with 50 mM Tris, 0.15 NaCl, 1 mM DTT, 2 mM EGTA and 0.25% β-dodecyl maltoside (pH 7.5), and then by 0.1 M glycine/HCl and 0.25% β-dodecyl maltoside (pH 2.5). The affinity columns incubated in the presence of 2 mM EGTA were rinsed with 20 column volumes of 50 mM Tris, 0.15 M NaCl, 1 mM DTT and 2 mM EGTA (pH 7.5) containing 0.25% β-dodecyl maltoside, followed by a two-step elution. First, the columns were eluted with 50 mM Tris, 0.15 NaCl, 1 mM DTT, 2 mM CaCl2 and 0.25% β-dodecyl maltoside (pH 7.5) and then by 0.1 M glycine/HCl and 0.25% β-dodecyl maltoside (pH 2.5). In all cases we checked that no material appeared in the wash (monitored by UV absorption at 280 nm) before elution. The eluates were concentrated by ultra-filtration to 50 μl. Then, 10 μl of the eluates from the secretagogin or calbindin D28k columns were separated by SDS/PAGE (12% gel). After the separation of proteins by SDS/PAGE, bands observed only on the secretagogin-gel but not on the calbindin D28k-gel, were excised from the gel and identified by LC-MS/MS as described previously [15].
GST pull-down assay
Exponentially growing Rin-5F insulinoma cells were washed in ice-cold PBS and then lysed in cell lysis buffer {TBS [Tris-buffered saline (20 nM Tris/137 nM NaCl)], pH 7.5, containing 1% Triton X-100, 10 μg/ml aprotinin, 1 μg/ml pepstatin, 10 μg/ml leupeptin, 50 mM PMSF and 0.8 mM Pefabloc}. The cell lysate was precleared by a 5 min spin at 12000 g. An aliquot (30 μl) of packed fusion protein-loaded glutathione–Sepharose 4B beads (GST–secretagogin or GST–setagin) was incubated in 500 μl of Rin-5F cell lysate for 4 h at 4 °C under constant rotation. The beads were then washed four times in cell lysis buffer containing protease inhibitors and finally suspended in SDS sample buffer under reducing conditions. Boiled samples were applied on to an SDS/PAGE (12% gel) and after electrophoretic protein separation, gels were either stained or blotted on to a nitrocellulose membrane and incubated using a mouse monoclonal SNAP-25 antibody (Sigma; product number: S5187, clone SP12) followed by a goat anti-mouse horseradish-peroxidase-conjugated antibody (Dako; product number P 0447).
SPR (surface plasmon resonance) studies
The interaction between calmodulin, calbindin D28k secretagogin and SNAP-25 was studied using SPR technology on a BIAcore™ biosensor system. All buffers were sterile filtered using a 0.22 μm filter. The flow buffer consisted of 10 mM Hepes/KOH (pH 7.4) with 0.15 M NaCl, 0.005% Tween 20 and 0.02% NaN3, containing either 2 mM CaCl2 (buffer A) or 3.4 mM EDTA (buffer B).
Immobilization of calmodulin, calbindin D28k and secretagogin to a CM-5 sensor chip surface was performed at a constant flow rate of 5 μl/min, using buffer B as flow buffer. Equal volumes of 0.1 M NHS (N-hydroxysuccinimide) and 0.4 M EDC [1-ethyl-3-(3-dimethylaminopropyl)carbodi-imide hydrochloride] were first mixed, and 40 μl of the mixture was allowed to flow over the sensor chip surface to activate the carboxymethylated dextran. Secretagogin was then injected over the sensor chip (20 μl at 0.020 mg/ml in 10 mM sodium acetate, pH 4.5). Deactivation of unchanged NHS ester was achieved by the flow of 20 μl of 1 M ethanolamine hydrochloride (pH 8.5).
The association and the dissociation of the analyte (SNAP-25) from the chip-immobilized components were studied at several different analyte concentrations. Control experiments were performed using a flow cell, which had been prepared using the immobilization protocol above, but without coupling the protein to the chip dextran surface. The surface was regenerated by injecting 0.1 M HCl for 15 min to remove residual bound analyte.
The association and the dissociation kinetics in the absence or presence of Ca2+ were studied using buffer A or B as the flow buffers. SNAP-25 was diluted from a concentrated stock into buffer A or B. The surface was regenerated by injecting 0.1 M HCl for 5 min to remove residual bound analyte.
The data were evaluated by a non-linear least-square analysis method using the software KaleidaGraph™ as described previously [9,10].
RESULTS
Since secretagogin has six EF-hand motifs, it is reasonable to expect that the protein binds Ca2+. EF-hand Ca2+-binding proteins are often classified as either Ca2+-sensor or Ca2+-buffer proteins. In order to investigate if secretagogin is likely to have a role in Ca2+-induced signal transduction as a Ca2+ sensor protein or if it is likely to function as a high-affinity Ca2+-buffer protein, we used five different methods: limited proteolysis, ITC, titration with Ca2+ in the presence of a chromophoric chelator, and titration with Ca2+ monitored by ANS and tryptophan fluorescence. The initial plan was to also monitor Ca2+ binding by NMR spectroscopy [e.g. by heteronuclear two-dimensional 15N-1H HSQC (heteronuclear single-quantum coherence) NMR spectroscopy]. However, Ca2+ titrations of secretagogin at high protein concentrations monitored by 1H NMR spectroscopy indicated that the spectral resolution gets poorer as more Ca2+ is added. At very high Ca2+ concentrations (3 equiv. or higher), precipitation is seen; hence the protein aggregates at high protein (100 μM) and Ca2+ concentration. Therefore any future NMR experiments should be performed with the apo-protein, at low concentrations of Ca2+, or in the presence of protein targets that stabilize the protein in Ca2+ solutions.
Far-UV CD spectroscopy
Far-UV CD spectroscopy was used to investigate the secondary structure of secretagogin in the apo, Ca2+- and Mg2+-bound states. We observed an intense negative ellipticity 208 and 222 nm in the spectra of apo, Ca2+- and Mg2+-secretagogin (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/401/bj4010353add.htm). This indicates that the protein has significant α-helical character in all forms. Addition of either Ca2+ or Mg2+ causes only minor differences in the intensity of each of the 208 and 222 nm bands, suggesting that the binding of each ion induces only minor changes in the secondary structure of the protein. We conclude that the secondary structure is not significantly altered by the binding of Ca2+ to secretagogin. Similar results have been obtained for the prototypical Ca2+ sensor calmodulin that displays a modest increase in CD signal on Ca2+ binding although the high-resolution structures of the Ca2+ and apo states reveal the same number of residues in helical conformation [16–18].
Near-UV CD spectroscopy
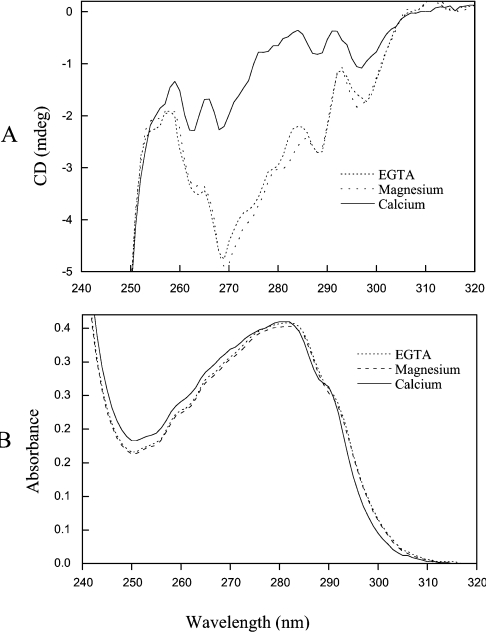
Secretagogin contains 19 phenylalanine, three tryptophan and five tyrosine residues. The tryptophan and tyrosine signals from approx. 270–295 nm and the strong phenylalanine signals below 270 nm indicate that secretagogin has a significant amount of tertiary structure in the absence of any bound metal ions (Figure 1A). No significant change in the near-UV CD spectrum is seen upon addition of 5 mM MgCl2. However, the shape of the near-UV CD spectrum of secretagogin, especially between 270 and 290 nm, is markedly altered by addition of 1 mM Ca2+. As near-UV CD spectroscopy is sensitive to structural changes in the vicinity of aromatic amino acid side chains or to changes in mobility of aromatic side chains, these results suggest that the tertiary structure of secretagogin is rearranged upon Ca2+ binding.
Figure 1. Near-UV CD (A) and absorbance spectroscopy (B).
(A) Near-UV CD spectroscopy. A near UV CD spectrum was first obtained for the apo-protein (37 μM apo-secretagogin in 1 mM DTT, 0.5 mM EGTA and 2 mM Tris/HCl, pH 7.5) (dotted line). MgCl2 was added to a concentration of 5 mM, and a spectrum for the Mg2+ form was recorded (broken line). Finally CaCl2 was added to a concentration of 1 mM, and a spectrum for the Ca2+ form was recorded (continuous line). (B) Near-UV absorbance spectroscopy. The UV spectrum at room temperature was recorded for a solution of 37 μM secretagogin in 1 mM DTT, 0.5 mM EGTA and 2 mM Tris/HCl (pH 7.5) (apo-secretagogin; dotted line). Then, 5 mM MgCl2 was added to the same solution (broken line). Finally, 1 mM CaCl2 was added to the same solution (continuous line).
UV absorbance spectroscopy
The UV absorbance spectra generated in 0.5 mM EGTA and 5 mM MgCl2 are highly similar, whereas the shape of the spectrum changes upon addition of 1 mM CaCl2 (Figure 1B). The UV absorbance spectra of secretagogin in the apo and Mg2+ form display maxima at 282 nm, which is shifted to 280.5 nm when Ca2+ is added. The UV difference spectrum (results not shown) induced by Ca2+ binding shows a positive absorbance at 240–283 nm and a negative absorbance between 283 and 310 nm, indicating that a conformational change has occurred in the local environment around aromatic residues upon Ca2+ binding. A peak seen around 243 nm may reflect increased light scattering in the Ca2+ state or effects on phenylalanine residues.
Intrinsic fluorescence spectra
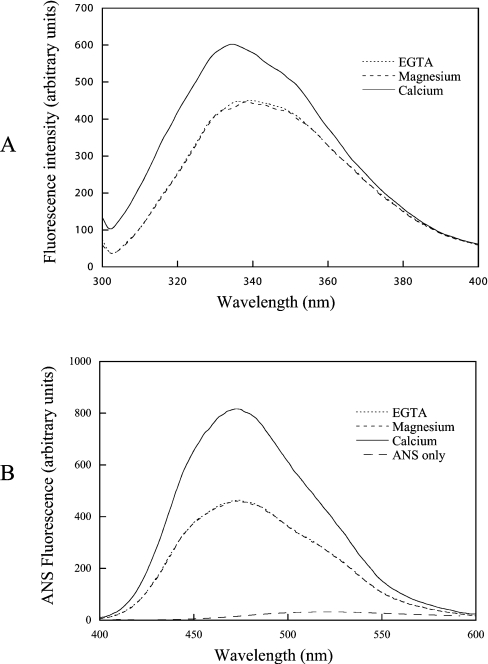
Secretagogin contains three tryptophan residues. Two of these are located in the first helix of EF-hand 1 and one is located in the N-terminal helix of EF-hand 3. The tryptophan fluorescence spectrum of the apo form has an emission maximum at 339.5 nm. Addition of 5 mM MgCl2 does not change the spectrum by any significant degree. Addition of 1 mM CaCl2, however, results in a change in the spectrum (Figure 2A). The Ca2+-loaded form displays a higher fluorescence intensity than the apo and Mg2+ forms (by a factor of ∼1.35) and a blue-shift in the maximum emission from 339.5 to 334.5 nm (Figure 2A). Addition of excess EGTA returns the emission intensity to the initial value (results not shown). The blue-shift in tryptophan fluorescence shows that one or more tryptophan residues become more buried when Ca2+ is bound. The changes in tryptophan fluorescence may reflect global structural changes. However, they could also be a consequence of more localized rearrangements around the Ca2+ sites.
Figure 2. Tryptophan (A) and ANS (B) fluorescence.
(A) Tryptophan fluorescence spectra were recorded for a sample containing 5 μM secretagogin, 1 mM DTT, 0.5 mM EGTA, 0.15 M KCl and 2 mM Tris/HCl (pH 7.5) (dotted line), after which MgCl2 (pH 7.5) was added to a concentration of 5 mM (broken line). Then, 1 mM CaCl2 (pH 7.5) was added to the same solution (continuous line). (B) ANS fluorescence spectra were recorded for a sample containing 120 μM ANS, 5 μM secretagogin, 1 mM DTT, 0.5 mM EGTA, 0.15 M KCl and 2 mM Tris/HCl (pH 7.5) (dotted line). MgCl2 (pH 7.5) was then added to a concentration of 5 mM (broken line). Finally, 1 mM CaCl2 (pH 7.5) was added to the same solution (continuous line).
Interaction of secretagogin with ANS
The degree of exposed hydrophobic surface in the Ca2+ and apo states of human recombinant secretagogin was measured using the hydrophobic probe ANS. In the presence of secretagogin, the fluorescence intensity of ANS increases significantly accompanied by a strong blue-shift both in Ca2+ (60 nm) and EGTA (58 nm) buffers (Figure 2B). This suggests that secretagogin has exposed hydrophobic surfaces both in the presence and absence of Ca2+. The fluorescence intensity is 2.3 times higher in the Ca2+ state than in the apo state, and there is a further blue-shift of 2 nm after Ca2+ addition. The fact that a wavelength shift accompanies the intensity change indicates a true difference in the conformational response between the two forms (i.e. fluorescence quenching cannot explain the observed difference). By monitoring the ANS fluorescence before and after the addition of Ca2+, followed by addition of excess of EGTA, we can conclude that the Ca2+-induced ANS-fluorescence intensity change is reversible. Mg2+ addition to secretagogin does not induce any changes in the ANS fluorescence spectrum compared with the apo form, suggesting that the presence of this ion does not change the amount of exposed hydrophobic surface in secretagogin (Figure 2B).
Limited proteolysis of secretagogin
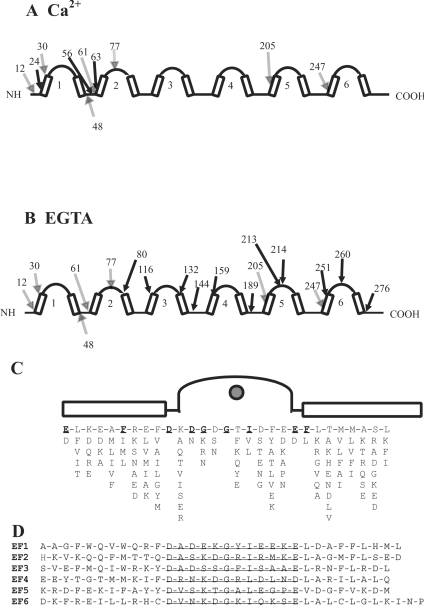
Limited tryptic digestion of human recombinant secretagogin was performed in the presence of EGTA and Ca2+. The reaction was quenched at different time points ranging from 2 min to 24 h and analysed by SDS/PAGE in a Tris/Tricine buffer system (see Supplementary Figure 2 at http://www.BiochemJ.org/bj/401/bj4010353add.htm). MALDI–TOF MS of the digests showed a number of masses corresponding to specific tryptic peptides of secretagogin (see Supplementary Table 1 at http://www.BiochemJ.org/bj/401/bj4010353add.htm). A major fragment with a molecular mass of approx. 23 kDa was seen by electrophoresis both in the presence and absence of Ca2+ (Supplementary Figure 2). This protein band was identified by MALDI–TOF and sub-digestion followed by peptide identification by LC-MS/MS as consisting of two fragments: residues 48–246 and 12–204. These results suggest that major Ca2+-independent cleavage sites are at amino acids 12, 48, 205 and 247. Supplementary Figure 3 (http://www.BiochemJ.org/bj/401/bj4010353add.htm)shows the mass spectra of the digests at 5 and 30 min respectively, and the first and last amino acids are given for all peptides that can be assigned to a specific mass within the secretagogin sequence. Both the rate and pattern of tryptic digestion are Ca2+-dependent, although some of the identified peptides are similar in both forms. As shown in Figure 3(A), peptides that are identified in the presence of Ca2+ derive mainly from the N-terminal part of the protein. Major cleavage sites in this region are at amino acids 24, 56, 61 and 63. These cleavage sites are all located in EF-hand 1 or in the loop between EF-hands 1 and 2. In contrast, peptides identified in the absence of Ca2+ derive from the whole protein with many cleavage sites in EF-hands 4, 5 and 6 (Figure 3B).
Figure 3. Secretogogin primary and secondary structure.
(A) Limited proteolysis of secretagogin. Schematic diagram of secretagogin outlining secondary structure elements, including the helix–loop–helix motifs and major tryptic cleavage sites in (A) 1 mM CaCl2 and (B) 1 mM EGTA. The residue number of cleavage sites identified both in CaCl2 and EGTA are indicated by shaded arrows. Cleavage sites identified only in CaCl2 or EGTA are indicated by solid arrows. EF-hand motifs 1–6 are shown by arabic numbers. (C) The EF-hand consensus sequence. (D) The amino acid sequence of the six EF-hands of secretagogin. Loops are underlined.
Ca2+ binding determined from ITC
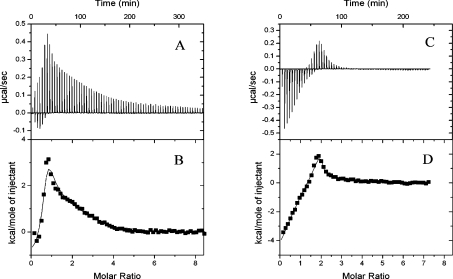
ITC was used to analyse the energetics of Ca2+ binding to secretagogin. For these experiments (and for the titration with Ca2+ in the presence of chromophoric chelators; see below) we used two different batches of recombinant secretagogin to ensure that our results were reproducible. The purity of both batches was checked by agarose gel electrophoresis in the presence of 1 mM EDTA or 2 mM calcium lactate, SDS/PAGE, isoelectric focusing and MS. 1H NMR spectroscopy was used to verify that all samples were free from EGTA after preparation of apo-protein. To quantify the residual Ca2+ concentration in the apo-protein, we performed competition experiments with the high-affinity chelator quin-2. The results suggest that both our batches of recombinant secretagogin are homogenous, intact, free from EGTA and contain 0.3 equiv. of Ca2+ (results not shown). Titration of CaCl2 into apo-secretagogin in physiological salt concentrations (0.15 M KCl) results in a biphasic curve indicating the presence of two classes of binding sites (Figures 4C and 4D). Data were fitted by the Origin software according to a model allowing for co-operative binding. To determine the stoichiometry of binding, we varied the number of binding sites from 1 to 6. The data could not be fitted with one or two binding sites, but was well fitted with three or more binding sites. Binding of Ca2+ to the first binding site proceeds with a negative enthalpy change, indicating an exothermic process with a ΔH of −17 kJ/mol, while Ca2+ binding to the second class of sites is an endothermic process with an average ΔH of +7 kJ/mol. As the data were fitted equally well with three or four Ca2+-binding sites in physiological salt concentrations, we also performed the experiments in low salt (Figures 4A and 4B). Again we observed two classes of binding sites. However, the initial exothermic process is significantly less pronounced at low salt. In low salt solutions, Ca2+ binding to the first site is exothermic with ΔH of −3 kJ/mol, followed by an endothermic process. At low salt concentrations, the data cannot be fitted with less than one high-affinity site and three low-affinity sites, suggesting that the protein binds four Ca2+ ions. The Ca2+ binding affinity (Ka) of the first site is too high to be accurately determined by the present experiments, but could be estimated to approx. 5×106 M−1 or higher. In low salt solutions, the macroscopic Ca2+-binding constants for the three low-affinity sites (K2, K3 and K4) are obtained as log K2=4.3±1.0, log K3=5.5±1.0 and log K4=7.0±1.0 respectively. The average ΔH for these three sites is +20 kJ/mol, and the data are best fitted when log K4>log K3>log K2, suggesting a high degree of co-operativity of Ca2+ binding to the low-affinity sites.
Figure 4. ITC analysis of the Ca2+ binding to secretagogin.
Calorimetric titration of 40×4 μl aliquots of 1 mM CaCl2 into 25 μM secretagogin in 2 mM Hepes/NaOH (pH 7.5) with raw data minus baseline in (A) and fitted data in (B). The same experiment performed in 2 mM Hepes/NaOH (pH 7.5) with 0.15 M NaCl is shown with raw data minus baseline in (C) and fitted data in (D).
Ca2+ binding determined from competition with chromophoric chelators
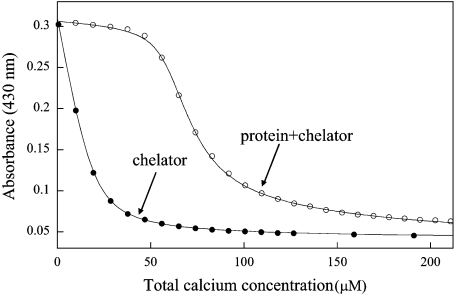
To further characterize the Ca2+-binding properties of secretagogin, we used titration with Ca2+ in the presence of chromophoric chelators to determine the macroscopic Ca2+-binding constants of secretagogin as described in [19,20]. In a previous study we used the chelators quin-2 or 5,5′-Br2-BAPTA [Kd (Ca2+) of 0.12 and 2.3 μM respectively in 0.15 M KCl] for the determination of the Ca2+ affinity of the related HEF protein calbindin D28k [13]. Since the ITC experiments indicated that secretagogin may contain a set of sites with lower affinity, we used both 5,5′-Br2-BAPTA and the chelator 5N-BAPTA, which has lower Ca2+ affinity [Kd (Ca2+)=27 μM in 0.15 M KCl] than that of quin-2 or 5′-Br2-BAPTA. Titrations were also performed in the same buffer with no added salt (Figure 5 and Table 1). The data were analysed by fitting each titration by a model that allows for up to four macroscopic binding constants in the protein using the Caligator software [14]. In accordance with the ITC results, the data suggest that secretagogin binds Ca2+ to one site with comparatively high affinity (log K1=7.1±0.4 in 0.15 M KCl), which is manifested as a rather flat part in the beginning of the titrations in the presence of 5N-BAPTA (Figure 5), and as a slight curvature in the presence of 5,5′-Br2-BAPTA (results not shown). The titrations in the presence of 5N-BAPTA suggest that there are also at least two sites with lower affinity in 0.15 M KCl. Incorporation of a fourth site leads to lower χ2 (for a definition of χ2, see [14]), but the change is not significant given that more parameters are introduced. Thus the data from the chelator experiments are equally well fitted with three or four binding-sites. If we assume that secretagogin binds four Ca2+ ions, as suggested by the ITC experiments, the following affinities are obtained in physiological salt buffers: log K1=7.1±0.4, log K2=4.7±0.6, log K3=3.6±0.7 and log K4=4.6±0.6. All data sets are best fitted if K4 is at least an order of magnitude larger than K3, implying strong positive co-operativity of Ca2+ binding to the low-affinity sites [21]. This is seen as a sigmoidal shape in the titration data after the saturation of the high-affinity site. The Ca2+ concentration required to achieve half-maximal saturation of secretagogin in 0.15 M KCl is approx. 25 μM. At low salt concentrations secretagogin binds Ca2+ to four sites with the following affinities: log K1=8.0±0.3, log K2=5.0±0.1, log K3=3.0±0.1 and log K4=5.5±0.3. Again, the data are better fitted if K4 is at least an order of magnitude larger than K3, implying positive co-operativity among the low-affinity sites. We used two batches of recombinant secretagogin, both of which gave similar results. Altogether nine titrations (four and five titrations each of the two batches of recombinant secretagogin) were performed using 5N-BAPTA.
Figure 5. Ca2+ titration of secretagogin.
Titration of 22 μM 5N-BAPTA with Ca2+ in the presence of 57 μM secretagogin in 2 mM Tris/HCl (pH 7.5). The experimental data are shown as symbols and the curves of optimal fit to the data points as continuous lines. ○, Titration of a mix of protein and chelator. ●, titration of only chelator.
Table 1. Macroscopic Ca2+-binding constants for secretagogin determined by competition with the chromophoric chelator 5N-BAPTA.
The mean values and S.D. are presented.
| Number of titrations | [KCl] | log K1 | log K2 | log K3 | log K4 |
|---|---|---|---|---|---|
| 4 | – | 8.0±0.3 | 4.9±0.1 | 3.0±0.1 | 5.5±0.3 |
| 5 | 0.15M | 7.1±0.4 | 4.7±0.6 | 3.6±0.7 | 4.6±0.6 |
Ca2+ binding to secretagogin as monitored by ANS and tryptophan fluorescence
In order to confirm that the binding constants obtained by the chelator method are in the correct range, we monitored Ca2+ binding by tryptophan and ANS fluorescence. Although the titration monitored by tryptophan and ANS fluorescence does not provide the high precision obtained using the competitive chromophoric chelator method, it does confirm that the binding constants obtained by the chelator method are in the correct range (see supplementary material at http://www.BiochemJ.org/bj/401/bj4010353add.htm).
Identification of secretagogin-binding proteins
Ca2+ sensor proteins such as calmodulin often interact with target proteins. We investigated if secretagogin binds to specific targets by affinity chromatography using immobilized recombinant secretagogin. In order to identify bona fide interacting proteins versus sample contaminants, we conducted negative controls in parallel using Sepharose 4B cross-linked to human recombinant calbindin D28k. Calbindin D28k was selected as control protein because it is very similar to secretagogin based on homology searches performed using the BLAST 2 software (58% sequence similarity). Like secretagogin, calbindin D28k is an HEF protein belonging to the calmodulin protein superfamily [3,22]. We performed the experiments using brain homogenates from two different species (mouse and cow) and by using pull-downs from the rat insulinoma cell line RIN-5F. All eluates were concentrated and separated by SDS/PAGE. Protein bands that appeared to bind specifically to the secretagogin column were excised and digested with trypsin and the resulting peptides were identified by LC-MS/MS. Experiments were performed using both cytosolic fractions (tissue homogenized without detergent) and membrane fractions (tissue homogenized in the presence of detergent in order to solubilize membrane-associated proteins). Although a small number of proteins (e.g. protein kinase C, N-ethylmaleimide-sensitive fusion protein and caskin-1) were identified from secretagogin affinity chromatography experiments using cytosolic fractions, these proteins did not pass the criteria we set up for accepting a protein as a specific secretagogin-binding protein. Thus they were either not reproducibly identified or they were also found in the eluates from the calbindin D28k control columns (results not shown). When using membrane fractions for the affinity chromatography experiments, however, SNAP-25 was identified as a specifically binding protein in all experiments. Below we therefore describe only the affinity chromatography experiments performed using membrane fractions.
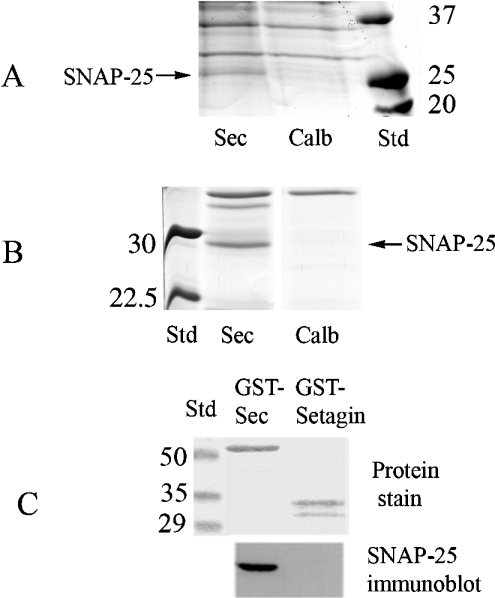
The first experiment was designed to purify proteins binding to Ca2+-secretagogin in mouse brain membrane fractions (Figure 6A). CaCl2 (2 mM) was included when solubilizing the brain extract, which was then incubated with secretagogin–Sepharose or calbindin D28k–Sepharose. Each column was eluted with 5 ml of a buffer containing 2 mM EGTA. A protein band that was found in the eluate from the secretagogin column, but not in the eluate from the calbindin D28k column, was identified by LC-MS/MS as SNAP-25 with a Mascot score of 50 (corresponding to a P value of <0.0005, i.e. the statistical probability of a correct protein identification is 99.95%) and five identified peptides (MLQLVEESK, EQMAISGGFIR, ADQLADESLESTRR, MLQLVEESKDAGIR and TLVMLDEQGEQLER).
Figure 6. Identification of secretagogin-binding proteins.
Mouse brain tissue (A) and bovine brain tissue (B) were homogenized as described in the Experimental section and incubated with secretagogin (Sec)–Sepharose or calbindin D28k (‘Calb’)–Sepharose respectively for 2 h. The affinity columns were then rinsed followed by elution with a buffer containing 2 mM EGTA. The eluates from all experiments were concentrated by ultra-filtration to 50 μl, and 10 μl was separated by SDS/PAGE. Bands indicated with arrows were cut and processed for MS/MS (gel slice named ‘Sec’). (C) GST–secretagogin or GST–setagin was incubated with cell extracts from RIN-5F cells and pulled down using glutathione beads. After washing the beads, they were boiled in SDS/PAGE sample buffer, centrifuged and transferred on to PVDF membranes and stained or probed using anti-SNAP-25 antibodies.
The second experiment was designed to purify proteins binding to Ca2+-secretagogin from bovine brain membrane fractions. The secretagogin and calbindin D28k columns were incubated with bovine brain homogenates in the presence of 2 mM CaCl2 and washed and proteins were eluted with 5 ml of a buffer containing EGTA. The identity of a 30 kDa band present in the eluate from the secretagogin column was determined to be SNAP-25 by LC-MS/MS (Figure 6B). This band was not seen in the eluate from the calbindin D28k column (Figure 6B). Three different peptides (CCGLFICPCNKLK and TLVMLDEQLDR from SNAP-25a and TLVMLDEQLER from SNAP-25b) were identified with a Mascot score of 68 (corresponding to a P value of <0.00005, i.e. the statistical probability of a correct protein identification is 99.995%).
The third experiment was designed to identify proteins binding to apo-secretagogin from mouse brain membrane fractions. Two columns with immobilized secretagogin and two columns with immobilized calbindin D28k were prepared and incubated separately with homogenized mouse brain tissue in the presence of 2 mM EGTA. Proteins binding to these columns were first eluted by a buffer containing 2 mM CaCl2. No protein bands could bee seen in the eluates from the secretagogin columns (results not shown). The columns were then eluted with glycine/HCl (pH 2.5). LC-MS/MS identified SNAP-25 in the eluates from the two secretagogin columns, but not in the eluates from the two calbindin D28k columns (results not shown). SNAP-25 was identified from the first secretagogin column with a Mascot score of 82 (corresponding to a P value of <10−6) and with seven identified tryptic peptides (MLQLVEESK, IEEGMDQINK, EQMAISGGFIR, ADQLADESLESTR, MLQLVEESKDAGIR, TLVMLDEQGEQLER and HMALDMGNEIDTQNR) and from the second secretagogin column with a Mascot score of 193 (corresponding to a P value of <10−6) and with seven identified tryptic peptides (MLQLVEESK, IEEGMDQINK, EQMAISGGFIR, ADQLA-DESLESTR, IEEGMDQINKDMK, RADQLADESLESTR and MLQLVEESKDAGIR). These results suggest that secretagogin binds to SNAP-25 also in the absence of Ca2+.
Secretagogin is not only expressed in the brain, but also abundantly expressed in neuroendocrine cells in the pancreas. The fourth experiment was designed to investigate the binding between secretagogin and SNAP-25 in a pancreatic cell line. For this purpose, immobilized GST–secretagogin fusion protein was used to affinity-purify (‘pull-down’) proteins from a membrane preparation of the β-cell line RIN-5F. GST–secretagogin protein was able to pull down endogenous SNAP-25 from RIN-5F cells. In contrast, control GST or GST–setagin was unable to pull down SNAP-25 (Figure 6C). Setagin is a 5.6 kDa splice variant of secretagogin that contains only a segment of the first EF-hand [11].
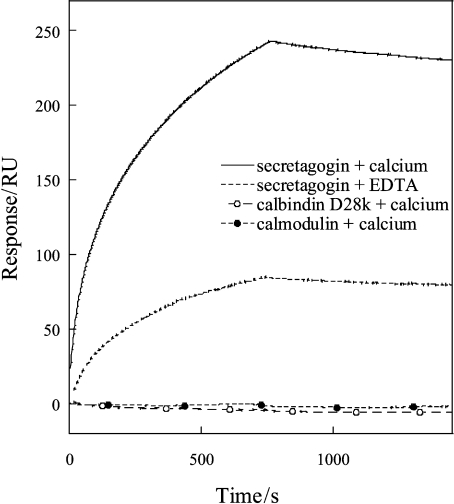
SPR was used to study the binding kinetics and affinity between secretagogin and SNAP-25 in vitro. SNAP-25 was overexpressed in E. coli and purified as described in the Experimental section. The amount of immobilized secretagogin was approx. 2500 resonance units in pg/mm2. The interaction between the two proteins was studied at four different SNAP-25 concentrations ranging from 25 nM to 4.5 μM. The curves representing the three lowest SNAP-25 concentrations (25, 180 and 900 nM) were used for fitting. The sensorgrams indicate that the two proteins interact with high affinity in the presence of Ca2+. Based on the values of kon and koff (see Table 2), the equilibrium dissociation constant Kd in the presence of Ca2+ was determined to be 1.2×10−7 M. The affinity is reduced by a factor of 8 in the absence of Ca2+ (Kd=1.5×10−6 M). The difference between the Ca2+ and EDTA forms seems to be in koff, indicating that secretagogin dissociates from SNAP-25 more rapidly in the absence of Ca2+ (see Table 2). We also performed SPR control experiments with immobilized calbindin D28k and calmodulin under identical conditions as above. Secretagogin binds strongly to SNAP-25 in Ca2+ buffers and somewhat weaker in EDTA buffers (Figure 7). Calbindin D28k and calmodulin, on the other hand, do not bind to SNAP-25 in the presence of Ca2+ (Figure 7) or in the presence of EDTA (results not shown).
Table 2. SPR results.
The immobilized protein was secretagogin and the analyte in the flow phase was SNAP-25.
| Ca2+/EDTA | Analyte concentrations tested (μM) | Kd (M) | kon (s−1·M−1) | koff (s−1) |
|---|---|---|---|---|
| Ca2+ | 0.025–0.9 | 1.2×10−7 | 3.7×103 | 4.4×10−5 |
| EDTA | 0.025–0.9 | 1.5×10−6 | 2.6×103 | 2.6×10−4 |
Figure 7. Binding between SNAP-25 and secretagogin, calbindin D28k or calmodulin investigated by SPR.
Calmodulin, calbindin D28k and secretagogin were immobilized in different flow cells on the same chip. SNAP-25 (1.5 μM) was first passed over calmodulin, then calbindin D28k and finally secretagogin. The flow buffer contained either 2 mM CaCl2 or 3.4 mM EDTA. The signal intensity before injecting SNAP-25 has been subtracted from all curves. RU, resonance units.
DISCUSSION
An important role of Ca2+-buffer proteins is to equip cells with high-affinity Ca2+-binding sites in order to reduce the intracellular Ca2+ concentration and to regulate Ca2+ signals. For this purpose, this group of proteins does not need to undergo any Ca2+-induced conformational changes. Ca2+-sensor proteins, however, undergo conformational changes, which allow them to regulate their activities in a Ca2+-dependent manner. Often a hydrophobic binding surface is exposed, which results in Ca2+-sensor/effector complexes that regulate specific cellular events. We found evidence for Ca2+-induced conformational changes in the tertiary structure of secretagogin by several spectroscopic methods. The results indicate that both the apo form and Ca2+ form of secretagogin bind significant amounts of ANS, suggesting that hydrophobic groups are exposed on the surface of the protein both in the Ca2+-free and Ca2+-loaded states. Secretagogin shares this property with the related HEF proteins calretinin and calbindin D28k [13,23,24]. The ANS-fluorescence studies also indicate that the Ca2+-loaded form of secretagogin is more hydrophobic than the Ca2+-free form. The exposure of hydrophobic surfaces is thermodynamically unfavourable. An obvious way of minimizing the exposed hydrophobic surfaces is by the interaction with other proteins. Thus it is possible that the hydrophobic patch at the surface of secretagogin may be instrumental for interaction with (and functional regulation of) other proteins.
Our results suggest that secretagogin binds four Ca2+ ions. We find it likely that EF-hands 3, 4, 5 and 6 are responsible for the binding of Ca2+ ions to secretagogin. This is because the amino acid sequences of EF-hands 1 and 2 do not conform to the EF-hand consensus sequence. EF-hand 1 has a lysine residue at a position where normally an aspartic acid or asparagine side chain provides an oxygen to co-ordinate Ca2+. The loop of EF-hand 2 has the standard Ca2+-co-ordinating groups, but in one position there is a positively charged arginine where a normally negative or hydrophilic side chain hydrogen-bonds to a Ca2+-co-ordinating water molecule in the same loop. Therefore these two EF-hands are most likely the ones that are non-functional in terms of Ca2+ binding. The sequences of EF-hands 3, 5 and 6 are typical for Ca2+-binding EF-hands (Figures 3C and 3D). EF-hand 4 has an aspartic acid residue in position 12, which is uncommon, but found in a number of active Ca2+-binding sites, for example in the sarcoplasmic Ca2+-binding protein.
The ITC data for secretagogin stand in bright contrast with those of calbindin D28k. Calbindin D28k binds four Ca2+ ions with high affinity and positive co-operativity [13,25]. ITC experiments performed with calbindin D28k show a single exothermic process that requires 4 equiv. of Ca2+ to reach saturation and the affinity is too high to be quantified (S. Linse, unpublished work). However, for secretagogin ITC data are clearly biphasic, and the sites act in two groups; one with high–moderate affinity and negative reaction enthalpy, and one group with lower affinity, positive enthalpy and co-operativity. The apparent Ca2+ affinity of secretagogin (Ka) is over ten times lower than that of the highly related HEF proteins calbindin D28k. At resting potential ([Ca2+]=10−9–10−7 M), 0.5–14% of secretagogin would be saturated by Ca2+. This indicates that the high-affinity site may provide some buffering capacity. However, Ca2+-buffering proteins such as parvalbumin (Kd≈0.05 μM) usually have much higher Ca2+ affinities, suggesting that secretagogin is unlikely to function as a traditional Ca2+-buffer protein. If we assume that secretagogin functions as a Ca2+ sensor protein, the affinity (Kd≈25 μM) is still rather low. For example, calmodulin has a Kd of approx. 5 μM (in the absence of bound targets). There are, however, several examples of Ca2+-sensor proteins with low affinity. The apparent Ca2+ affinity of the Ca2+ sensor for exocytosis in neural cells, synaptotagmin I, is 5–50 μM in the presence of phospholipid membranes [26–28], which is in the same range as measured here for secretagogin. This information is particularly interesting since synaptotagmin I binds to SNAP-25, the same protein that was identified in this study as a potential secretagogin-binding protein. Alternatively, the requirement for higher Ca2+ concentrations may be similar to the situation with the cytoplasmic calpains (for a review, see [29]). Calpains participate in intracellular signal processing via limited proteolysis of their targets, altering the function of these proteins. The Ca2+ concentration giving half-maximal activity, [Ca2+]0.5, ranges from 2 to 75 μM for l-calpain (e.g. [30–32]) and 200–1000 μM for m-calpain [31,33]. Since Ca2+ concentrations in this range do not exist in intact living cells, calpains may function at the initial, ascending limb of their Ca2+ saturation curves, allowing only a small fraction of the calpain pool to become activated [29].
We recently completed a study in which we identified a large number of proteins in EGTA eluates from calmodulin affinity columns [15]. When comparing those results with those from the present study, it is clear that secretagogin does not bind a very large number of proteins as observed for calmodulin. Although we cannot exclude the existence of other secretagogin-interacting proteins, SNAP-25 was the only protein that we could identify from all tissues and species used. Unlike secretagogin, the related proteins calbindin D28k and the promiscuous Ca2+ sensor calmodulin were unable to bind SNAP-25. These results indicate that the interaction between secretagogin and SNAP-25 is highly specific. SNAP-25 is one of the basic components of the SNARE (soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor) complex. The SNARE complex is the core complex for membrane fusion, a process that is regulated by changes in intracellular Ca2+ levels (for reviews, see [34–36]). Secretagogin is expressed in neuroendocrine cells in the brain and in the pancreatic islets [1–3], which are sites of exocytosis of neurotransmitters and insulin respectively. Thus secretagogin and SNAP-25 seem to be expressed in the same type of cells.
SNAP-25 could be eluted from secretagogin columns by buffers containing Ca2+ chelators, suggesting that the interaction is at least partially Ca2+-dependent. The apparent affinity of the interaction increases approx. 10-fold following addition of Ca2+. However, although the interaction is enhanced by Ca2+ binding, it occurs with high affinity both in the presence and absence of Ca2+. Does this mean that secretagogin is unlikely to function as a Ca2+ sensor protein? EF-hand Ca2+-sensor proteins that are prebound to targets in their Ca2+-free states are not uncommon. Prebound calmodulin can for example be activated and modify associated proteins as demonstrated by its regulation of voltage-gated Ca2+ channels [37–39]. SNAP-25 interacts with another Ca2+-binding protein, the Ca2+ sensor for exocytosis in neural cells, synaptotagmin I. Synaptotagmin I binds to SNAP-25 in the absence of Ca2+ [40] with the apparent affinity of the interaction increasing only approx. 6-fold following addition of Ca2+ [41]. Thus the triggering of fast exocytosis by Ca2+ binding to synaptotagmin I is not executed via Ca2+-induced binding of synaptotagmin I to SNAP-25 and other SNAREs [42]. Ca2+-independent binding of synaptotagmin I to SNAREs serves to position synaptotagmin I next to SNARE complexes after these have been assembled but before the fusion pore has opened [34]. Ca2+ then induces a conformational change in the prebound synaptotagmin that results in a series of events that opens the fusion pore. Similarly, formation of a complex between secretagogin and SNAP-25 at resting Ca2+ concentrations could, in theory, allow Ca2+-induced structural changes of secretagogin to be rapidly transmitted to elicit a functional change. An investigation of the possible role of secretagogin in exocytosis is an important question for the future.
Conclusions
Secretagogin binds four Ca2+ ions with a [Ca2+]0.5 of approx. 25 μM in physiological salt buffers, which could be compared with a [Ca2+]0.5 of approx. 1 μM for calbindin D28k [13] and 1.5 μM for calretinin [43]. The differences in Ca2+ affinities probably reflect important biological and evolutionary differences between these proteins. The comparatively low Ca2+ affinity of secretagogin makes it unlikely to function as an efficient Ca2+-buffer protein, although the high-affinity site may provide some buffering capacity. The amount of exposed hydrophobic surface in secretagogin increases upon Ca2+ binding and it undergoes Ca2+-induced conformational changes, suggesting the possibility that it can transduce intracellular Ca2+-concentration changes to other proteins. Secretagogin does not interact with a large number of proteins as observed for e.g. calmodulin. Only SNAP-25, a protein involved in Ca2+ regulated exocytosis, could be reproducibly identified as a secretagogin-binding protein.
Online data
Acknowledgments
This work was supported by grants from Stiftelsen Lars Hiertas Minne (T.B.), Magnus Bergvalls Foundation (T.B.), the SWEGENE Foundation (T.B.) and by grants from the Swedish Research Foundation (S.L. and P.J.)
References
- 1.Cras-Meneur C., Inoue H., Zhou Y., Ohsugi M., Bernal-Mizrachi E., Pape D., Clifton S. W., Permutt M. A. An expression profile of human pancreatic islet mRNAs by serial analysis of gene expression (SAGE) Diabetologia. 2004;47:284–299. doi: 10.1007/s00125-003-1300-8. [DOI] [PubMed] [Google Scholar]
- 2.Gartner W., Lang W., Leutmetzer F., Domanovits H., Waldhausl W., Wagner L. Cerebral expression and serum detectability of secretagogin, a recently cloned EF-hand Ca2+-binding protein. Cereb. Cortex. 2001;11:1161–1169. doi: 10.1093/cercor/11.12.1161. [DOI] [PubMed] [Google Scholar]
- 3.Wagner L., Oliyarnyk O., Gartner W., Nowotny P., Groeger M., Kaserer K., Waldhausl W., Pasternack M. S. Cloning and expression of secretagogin, a novel neuroendocrine- and pancreatic islet of Langerhans-specific Ca2+-binding protein. J. Biol. Chem. 2000;275:24740–24751. doi: 10.1074/jbc.M001974200. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed M., Bergsten P. Glucose-induced changes of multiple mouse islet proteins analysed by two-dimensional gel electrophoresis and mass spectrometry. Diabetologia. 2005;48:477–485. doi: 10.1007/s00125-004-1661-7. [DOI] [PubMed] [Google Scholar]
- 5.Zhan X., Evans C. O., Oyesiku N. M., Desiderio D. M. Proteomics and transcriptomics analyses of secretagogin down-regulation in human non-functional pituitary adenomas. Pituitary. 2003;6:189–202. doi: 10.1023/b:pitu.0000023426.99808.40. [DOI] [PubMed] [Google Scholar]
- 6.Birkenkamp-Demtroder K., Christensen L. L., Olesen S. H., Frederiksen C. M., Laiho P., Aaltonen L. A., Laurberg S., Sorensen F. B., Hagemann R., Orntoft T. F. Gene expression in colorectal cancer. Cancer Res. 2002;62:4352–4363. [PubMed] [Google Scholar]
- 7.Birkenkamp-Demtroder K., Wagner L., Brandt Sorensen F., Bording Astrup L., Gartner W., Scherubl H., Heine B., Christiansen P., Orntoft T. F. Secretagogin is a novel marker for neuroendocrine differentiation. Neuroendocrinology. 2005;82:121–138. doi: 10.1159/000091207. [DOI] [PubMed] [Google Scholar]
- 8.Kojetin D. J., Venters R. A., Kordys D. R., Thompson R. J., Kumar R., Cavanagh J. Structure, binding interface and hydrophobic transitions of Ca2+-loaded calbindin-D28K. Nat. Struct. Mol. Biol. 2006;13:641–667. doi: 10.1038/nsmb1112. [DOI] [PubMed] [Google Scholar]
- 9.Berggard T., Thulin E., Akerfeldt K. S., Linse S. Fragment complementation of calbindin D28k. Protein Sci. 2000;9:2094–2108. doi: 10.1110/ps.9.11.2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linse S., Thulin E., Gifford L. K., Radzewsky D., Hagan J., Wilk R. R., Akerfeldt K. S. Domain organization of calbindin D28k as determined from the association of six synthetic EF-hand fragments. Protein Sci. 1997;6:2385–2396. doi: 10.1002/pro.5560061112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zierhut B., Daneva T., Gartner W., Brunnmaier B., Mineva I., Berggard T., Wagner L. Setagin and secretagogin-R22: posttranscriptional modification products of the secretagogin gene. Biochem. Biophys. Res. Commun. 2005;329:1193–1199. doi: 10.1016/j.bbrc.2005.02.093. [DOI] [PubMed] [Google Scholar]
- 12.Pauls T. L., Cox J. A., Berchtold M. W. The Ca2+-binding proteins parvalbumin and oncomodulin and their genes: new structural and functional findings. Biochim. Biophys. Acta. 1996;1306:39–54. doi: 10.1016/0167-4781(95)00221-9. [DOI] [PubMed] [Google Scholar]
- 13.Berggard T., Miron S., Onnerfjord P., Thulin E., Akerfeldt K. S., Enghild J. J., Akke M., Linse S. Calbindin D28k exhibits properties characteristic of a Ca2+ sensor. J. Biol. Chem. 2002;277:16662–16672. doi: 10.1074/jbc.M200415200. [DOI] [PubMed] [Google Scholar]
- 14.Andre I., Linse S. Measurement of Ca2+-binding constants of proteins and presentation of the CaLigator software. Anal. Biochem. 2002;305:195–205. doi: 10.1006/abio.2002.5661. [DOI] [PubMed] [Google Scholar]
- 15.Berggard T., Arrigoni G., Olsson O., Fex M., Linse S., James P. 140 mouse brain proteins identified by Ca2+-calmodulin affinity chromatography and tandem mass spectrometry. J. Proteome Res. 2006;5:669–687. doi: 10.1021/pr050421l. [DOI] [PubMed] [Google Scholar]
- 16.Finn B. E., Evenas J., Drakenberg T., Waltho J. P., Thulin E., Forsen S. Calcium-induced structural changes and domain autonomy in calmodulin. Nat. Struct. Biol. 1995;2:777–783. doi: 10.1038/nsb0995-777. [DOI] [PubMed] [Google Scholar]
- 17.Kuboniwa H., Tjandra N., Grzesiek S., Ren H., Klee C. B., Bax A. Solution structure of calcium-free calmodulin. Nat. Struct. Biol. 1995;2:768–776. doi: 10.1038/nsb0995-768. [DOI] [PubMed] [Google Scholar]
- 18.Zhang M., Tanaka T., Ikura M. Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nat. Struct. Biol. 1995;2:758–767. doi: 10.1038/nsb0995-758. [DOI] [PubMed] [Google Scholar]
- 19.Linse S., Brodin P., Drakenberg T., Thulin E., Sellers P., Elmden K., Grundstrom T., Forsen S. Structure-function relationships in EF-hand Ca2+-binding proteins. Protein engineering and biophysical studies of calbindin D9k. Biochemistry. 1987;26:6723–6735. doi: 10.1021/bi00395a023. [DOI] [PubMed] [Google Scholar]
- 20.Linse S., Helmersson A., Forsen S. Calcium binding to calmodulin and its globular domains. J. Biol. Chem. 1991;266:8050–8054. [PubMed] [Google Scholar]
- 21.Klotz I. M. Introduction to Biomolecular Energetics. Orlando, FL: Academic Press; 1986. [Google Scholar]
- 22.Hof P. R., Glezer I. I., Conde F., Flagg R. A., Rubin M. B., Nimchinsky E. A., Vogt Weisenhorn D. M. Cellular distribution of the calcium-binding proteins parvalbumin, calbindin, and calretinin in the neocortex of mammals: phylogenetic and developmental patterns. J. Chem. Neuroanat. 1999;16:77–116. doi: 10.1016/s0891-0618(98)00065-9. [DOI] [PubMed] [Google Scholar]
- 23.Berggard T., Silow M., Thulin E., Linse S. Ca2+- and H+-dependent conformational changes of calbindin D28k. Biochemistry. 2000;39:6864–6873. doi: 10.1021/bi992394g. [DOI] [PubMed] [Google Scholar]
- 24.Palczewska M., Groves P., Batta G., Heise B., Kuznicki J. Calretinin and calbindin D28k have different domain organizations. Protein Sci. 2003;12:180–184. doi: 10.1110/ps.0215303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Veenstra T. D., Johnson K. L., Tomlinson A. J., Naylor S., Kumar R. Determination of calcium-binding sites in rat brain calbindin D28K by electrospray ionization mass spectrometry. Biochemistry. 1997;36:3535–3542. doi: 10.1021/bi9628329. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X., Rizo J., Sudhof T. C. Mechanism of phospholipid binding by the C2A-domain of synaptotagmin I. Biochemistry. 1998;37:12395–12403. doi: 10.1021/bi9807512. [DOI] [PubMed] [Google Scholar]
- 27.Fernandez I., Arac D., Ubach J., Gerber S. H., Shin O., Gao Y., Anderson R. G., Sudhof T. C., Rizo J. Three-dimensional structure of the synaptotagmin 1 C2B-domain: synaptotagmin 1 as a phospholipid binding machine. Neuron. 2001;32:1057–1069. doi: 10.1016/s0896-6273(01)00548-7. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez-Chacon R., Konigstorfer A., Gerber S. H., Garcia J., Matos M. F., Stevens C. F., Brose N., Rizo J., Rosenmund C., Sudhof T. C. Synaptotagmin I functions as a calcium regulator of release probability. Nature. 2001;410:41–49. doi: 10.1038/35065004. [DOI] [PubMed] [Google Scholar]
- 29.Friedrich P. The intriguing Ca2+ requirement of calpain activation. Biochem. Biophys. Res. Commun. 2004;323:1131–1133. doi: 10.1016/j.bbrc.2004.08.194. [DOI] [PubMed] [Google Scholar]
- 30.Baki A., Tompa P., Alexa A., Molnar O., Friedrich P. Autolysis parallels activation of μ-calpain. Biochem. J. 1996;318:897–901. doi: 10.1042/bj3180897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cong J., Goll D. E., Peterson A. M., Kapprell H. P. The role of autolysis in activity of the Ca2+-dependent proteinases (μ-calpain and m-calpain) J. Biol. Chem. 1989;264:10096–10103. [PubMed] [Google Scholar]
- 32.Goll D. E., Thompson V. F., Li H., Wei W., Cong J. The calpain system. Physiol. Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 33.Dutt P., Spriggs C. N., Davies P. L., Jia Z., Elce J. S. Origins of the difference in Ca2+ requirement for activation of μ- and m-calpain. Biochem. J. 2002;367:263–269. doi: 10.1042/BJ20020485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sudhof T. C. The synaptic vesicle cycle. Annu. Rev. Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 35.Jahn R., Lang T., Sudhof T. C. Membrane fusion. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 36.Rizo J. SNARE function revisited. Nat. Struct. Biol. 2003;10:417–419. doi: 10.1038/nsb0603-417. [DOI] [PubMed] [Google Scholar]
- 37.Lee A., Wong S. T., Gallagher D., Li B., Storm D. R., Scheuer T., Catterall W. A. Ca2+/calmodulin binds to and modulates P/Q-type calcium channels. Nature. 1999;399:155–159. doi: 10.1038/20194. [DOI] [PubMed] [Google Scholar]
- 38.Peterson B. Z., DeMaria C. D., Adelman J. P., Yue D. T. Calmodulin is the Ca2+ sensor for Ca2+-dependent inactivation of L-type calcium channels. Neuron. 1999;22:549–558. doi: 10.1016/s0896-6273(00)80709-6. [DOI] [PubMed] [Google Scholar]
- 39.Zuhlke R. D., Pitt G. S., Deisseroth K., Tsien R. W., Reuter H. Calmodulin supports both inactivation and facilitation of L-type calcium channels. Nature. 1999;399:159–162. doi: 10.1038/20200. [DOI] [PubMed] [Google Scholar]
- 40.Schiavo G., Stenbeck G., Rothman J. E., Sollner T. H. Binding of the synaptic vesicle v-SNARE, synaptotagmin, to the plasma membrane t-SNARE, SNAP-25, can explain docked vesicles at neurotoxin-treated synapses. Proc. Natl. Acad. Sci. U.S.A. 1997;94:997–1001. doi: 10.1073/pnas.94.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerona R. R., Larsen E. C., Kowalchyk J. A., Martin T. F. The C terminus of SNAP25 is essential for Ca(2+)-dependent binding of synaptotagmin to SNARE complexes. J. Biol. Chem. 2000;275:6328–6336. doi: 10.1074/jbc.275.9.6328. [DOI] [PubMed] [Google Scholar]
- 42.Shin O. H., Rhee J. S., Tang J., Sugita S., Rosenmund C., Sudhof T. C. Sr2+ binding to the Ca2+ binding site of the synaptotagmin 1 C2B domain triggers fast exocytosis without stimulating SNARE interactions. Neuron. 2003;37:99–108. doi: 10.1016/s0896-6273(02)01145-5. [DOI] [PubMed] [Google Scholar]
- 43.Schwaller B., Durussel I., Jermann D., Herrmann B., Cox J. A. Comparison of the Ca2+-binding properties of human recombinant calretinin-22k and calretinin. J. Biol. Chem. 1997;272:29663–29671. doi: 10.1074/jbc.272.47.29663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.