Abstract
The identification of conserved sequence tags (CSTs) through comparative genome analysis may reveal important regulatory elements involved in shaping the spatio-temporal expression of genetic information. It is well known that the most significant fraction of CSTs observed in human–mouse comparisons correspond to protein coding exons, due to their strong evolutionary constraints. As we still do not know the complete gene inventory of the human and mouse genomes it is of the utmost importance to establish if detected conserved sequences are genes or not. We propose here a simple algorithm that, based on the observation of the specific evolutionary dynamics of coding sequences, efficiently discriminates between coding and non-coding CSTs. The application of this method may help the validation of predicted genes, the prediction of alternative splicing patterns in known and unknown genes and the definition of a dictionary of non-coding regulatory elements.
INTRODUCTION
A fundamental task in genome analysis is the annotation of the various sequence features that constitute the genetic program of each organism. In this respect the identification of genes and of the regulatory elements controlling level, location and chronology of their expression represents a major challenge for biologists in the genomic era.
It should be noted that we still have not established, with any degree of confidence, exactly the number of genes encoded by any of the completed (at least at draft level) prokaryotic and eukaryotic genomes. The problem is not trivial even for prokaryotic genomes, where the typical high gene density and the absence of introns makes the task of gene detection and annotation somewhat more tractable. For example, it can be difficult to accurately predict some of the shortest genes that often lack identifiable homologs in other species. The gene-finding problem becomes even more daunting in large eukaryotic genomes, where coding regions are generally scattered in a vast sea of non-coding noise.
The simplest way to predict a coding region is the observation of a statistically significant similarity to a known protein (for example by BlastX analysis). However, in many cases no homolog can be identified in the protein databanks. Furthermore, given that most of the proteins collected in public databases merely represent the conceptual translation of predicted ORFs, the observation of a protein match does not guarantee the identification of a true gene and the correct identification of its exon/intron structure. For this reason it is attractive to use several approaches, including computational methods performing ab initio gene predictions, concurrently. These methods function by integrating the detection of specific signals (e.g. splice sites, start codon context, etc.) with the observation of sequence statistical features peculiar to protein coding regions (e.g. long ORFs, asymmetric composition of the three codon positions, presence of upstream CpG islands, etc.). Gene finding tools integrating both content and signal sensors perform particularly well when adopting hidden Markov models (HMMs) applying probabilistic models to interconnect the sequence and boundary signals considered. Among the most popular programs are Glimmer (1) and GeneMark (2) for bacterial genomes and Genscan (3) and HMMgene (4) for eukaryotic genes, with prediction accuracies >90% (5). However, auxiliary experimental information, such as EST or cDNA matches, are needed to confirm a gene prediction.
The availability of both genome and high throughput transcript collections for several model organisms, such as human and mouse, opens new possibilities for the identification of protein coding genes based on comparative analysis of homologous sequences (6,7). Several methods have been proposed that use a strategy taking into account similarity at the nucleotide and amino acid levels as well as conservation of splice sites, exon length and codon usage. Indeed, a comparison of the mRNA sequences of 1880 orthologous human and mouse gene pairs (8) showed ∼85% identity for coding exons, in contrast to an average 35% identity for introns (close to the expected level of identity for random sequences).
As it is known that sequences regulating gene expression tend to be conserved between species (9), the problem of discriminating between potentially coding and non-coding conserved sequence tags (CSTs) arises. Only these latter may represent potential regulatory elements whose activity deserves further investigation.
Here we present a new heuristic method based on pairwise genome comparison which has been implemented in software called CSTfinder. Following the identification of high scoring segment pairs (HSPs) through a Blast-like sequence comparison, CSTfinder assesses the potential coding capacity of CSTs delimited by each HSP. The measure of coding capacity, expressed by a coding potential score (CPS), is related to the observation of a constrained evolutionary process specific to coding regions (and not observable in non-coding regions) that can be observed through cross-species comparisons. Firstly, substitutions in homologous coding regions tend to be strongly biased toward synonymous changes. Secondly, non-synonymous substitutions (i.e. base changes involving amino acid substitution) tend to be more conservative than those randomly expected, favoring interconversions between structurally similar amino acids. A similar strategy is adopted by some gene finding algorithms (10,11) and in QRNA (12), a program specifically designed for non-coding RNA gene detection. However, this latter requires aligned sequences as input.
The method proposed here has been specifically designed to detect CSTs in large-scale genome comparisons, and also to assess their coding capacity using a novel scoring formula. It is extremely fast and accurate and has proven very valuable in the recognition of known protein coding regions in genomic and cDNA nucleotide sequences and in detecting novel potentially coding exons in large unannotated homologous genome regions.
MATERIALS AND METHODS
CSTfinder algorithm
Given a CST of length L identified by Blast search (13) for a pair of sequences S1 and S2 the analysis is carried out for all the three possible frames in the forward and reverse strand orientation, i.e. f = +1, +2, +3, –1, –2 and –3, respectively. For each frame the pairs of Nf aligned trinucleotides (i.e. hypothetical codons) are considered that show at least one nucleotide substitution. The Nf triplet pairs considered for frame f can be classified as synonymous (S) or non- synonymous (A) so that Nf = Sf + Af + G, where G is the number of gapped or stop codon-containing triplet pairs. A CPS is then calculated for the CST under investigation as:
where Sf and Af are the number of triplet pairs where synonymous and replacement changes are observed, respectively, in frame f, and AA_simi is a measure of the structural similarity between compared amino acids for the ith triplet pair expressed in terms of a PAM or Blosum score. The PAM or Blosum matrix is selected by the user (default Blosum80) and assigns a penalty of –9 for gap lengths not a multiple of three and for stop codon-containing triplet pairs. L is the nucleotide length of the relevant HSP. Given the dependence between the CPS and the CST size, the CPS is normalized to a fixed size of 100 codons.
Alternatively, CSTfinder allows the CPS to be calculated over triplet windows scanning the CST under investigation. This approach can allow the approximate identification of the boundaries, if any, of the protein coding region within the CST.
Parameters used in the Blast-like sequence similarity search carried out by the CSTfinder software included a word size of 10, a minimum CST length of 30 nt and a maximum E-value of 10–5.
Data
The CDS dataset, including 1880 human and rodent nucleotide sequences coding for proteins (CDS), was obtained from the collection of orthologous mRNA sequences analyzed by Makalowski et al. (14). This set has been divided into five parts according to observed identities in encoded proteins: cds1 (399 CDSs, 97–100%), cds2 (421 CDSs, 92–97%), cds3 (413 CDSs, 73–85%), cds4 (383 CDSs, 73–85%), cds5 (263 CDSs, 40–73%).
The GENE dataset (15) included 15 orthologous pairs of single gene sequences from human and mouse collected in the IMOG dataset.
The 5′-UTR mRNA sequences of the human and rodent division from UTRdb (16) were analyzed to reveal wrongly annotated CDSs in the EMBL/GenBank entry.
The GENOME dataset included 199 orthologous paired genome regions from human and mouse, accounting for a total of ∼140 Mb, extracted from the EnsEMBL database (17), including at least one known gene and flanking 5′ and 3′ genomic regions containing other partial or complete genes classified as ‘novel’, ‘EST’ or ‘GenScan/Twinscan’ predicted genes.
RANDOM and SYNTENY datasets were also generated to evaluate CSTfinder performance. For the RANDOM dataset a thousand pairs of nucleotide sequences were generated by Monte Carlo simulation using the nucleotide composition of sequences in the CDS dataset. To make simulated sequences resembling natural sequences the simulation generated sequences made of five ‘exons’ and four ‘introns’, all 100 nt long, with exon sequences diverging by 60–90% and intron sequences made of a poly(A) in the first and a poly(C) in the second sequence. The SYNTENY dataset included eight conserved syntenic regions for human chromosome 22 and the mouse genome (EnsEMBL release 11.31), accounting for ∼31 Mb for human and 28 Mb for mouse and a total of 2215 annotated human genes. We expect that in these syntenic regions virtually all human coding exons have been discovered and annotated (18).
CSTfinder software and trial data are available from the authors upon request.
RESULTS
To test the effectiveness of the algorithm we named CSTfinder, we analyzed a set of 1880 pairs of human and mouse orthologous CDS sequences (14). Results are summarized separately for five groups of orthologous gene pairs showing a decreasing level of protein identity in Table 1. As anticipated, synonymous codons significantly outnumbered non-synonymous codons at all levels of identity. The percentage of synonymous codons was fairly homogeneous among all five classes as well as the AA_sim score (see Materials and Methods), whereas the percentage of non-synonymous codons was higher in more divergent human–mouse pairs.
Table 1. Result of the application of CSTfinder to the CDS dataset (see Materials and Methods).
| Class | N genes | Positives (%) | S | A | AA_sim | CPS |
|---|---|---|---|---|---|---|
| 97–100% | 399/399 | 100 | 30.3 | 6.4 | 234.2 | 3318.7 |
| 92–97% | 421/421 | 100 | 26.9 | 8.5 | 210.8 | 1117.2 |
| 85–92% | 413/413 | 100 | 26.7 | 11.9 | 202.8 | 727.3 |
| 73–85% | 383/383 | 100 | 24.7 | 17.1 | 188.4 | 389.6 |
| 40–73% | 249/263 | 95 | 23.0 | 20.3 | 161.8 | 270.8 |
| RANDOM set | 2.8 | 7.3 | 0.3 | 13.0 |
The number of synonymous (S) and non-synonymous (A) substitutions, the Blosum80 score (AA_sim) and the average CPS are shown for each group of genes, all values normalized to 100 codons.
Virtually all CDSs in the five groups of sequences showed a CPS > 30 (100% positives in Table 1). The average CPS ranged between 270.8 and 3318.7 for the five groups. Only 14 of the total 1880 sequence pairs in the CDS dataset, all in the 40–73% similarity group, did not show CPS > 30. However, in these cases no HSP was detected through the Blast search, thus preventing CPS calculation.
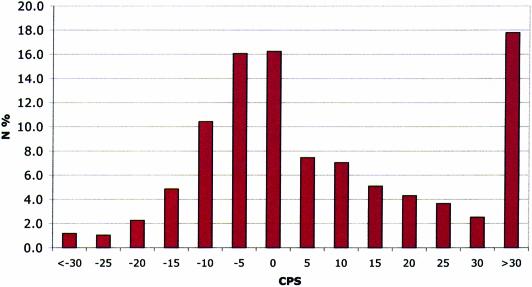
The RANDOM dataset (see Materials and Methods) has been used as a negative training set carrying out the analysis on each detected HSP. Results shown in Table 1 show a clear-cut discrimination between genuine coding and non-coding highly conserved regions, with the latter showing non-synonymous changes greatly outnumbering synonymous changes and an average CPS of 13.0. Figure 1 shows the distribution of the CPSs obtained in the analysis of the RANDOM training set. Using a CPS threshold of 30 the rate of false positives can be estimated at 17.8%. However, only less than 5% of simulated gene pairs in the random dataset showed two or more concurrent HSPs with CPS > 30 and in only one case was a CPS > 500 observed.
Figure 1.
Distribution of CPSs from the CSTfinder analysis of the RANDOM set.
To further assess the reliability of the algorithm in detecting coding exons when comparing homologous genomic regions we analyzed 15 pairs of human–mouse orthologous genes contained in the GENE dataset (see Materials and Methods). Results are summarized in Table 2. For all genes at least one exon was detected with CPS > 500, with an overall rate of 66/73 exons and an average CPS of 864.5. The method performed equally well at various degrees of conservation of the corresponding protein products, although more clear-cut signals were obtained from more conserved proteins.
Table 2. Result of the application of CSTfinder to the GENE database (see Materials and Methods).
| Gene | Hsa ID | Rod ID | ID (%) (aa) | Average CPS | Matching exons |
|---|---|---|---|---|---|
| BAL | M94579 | M69157 | 75.8 | 640.6 | 9/11 (9/11) |
| COXD | U66875 | U34801 | 79.4 | 167.9 | 3/3 (3/3) |
| ENOB | X56832 | X61600 | 97.7 | 817.4 | 10/11 (8/11) |
| H1T | M97755 | L28753 | 53.2 | 975.1 | 1/1 (0/1) |
| H4 | M16707 | V00753 | 100.0 | 5414.1 | 1/1 (1/1) |
| HS71 | M11717 | M32218 | 87.3 | 590.3 | 1/1 (0/1) |
| KCRB | X15334 | M74149 | 96.6 | 686.8 | 7/7 (6/7) |
| MIF | L19686 | U20156 | 89.5 | 193.7 | 3/3 (3/3) |
| MT3 | S72043 | S72046 | 86.8 | 104.3 | 3/3 (2/3) |
| OSTP | U20758 | X51834 | 48.0 | 117.5 | 4/6 (2/6) |
| PAP1 | L15533 | D63360 | 69.1 | 294.8 | 4/5 (4/5) |
| PSPA | M68519 | S48768 | 71.0 | 321.5 | 3/4 (3/4) |
| ROM1 | M96759 | M96760 | 84.6 | 311.8 | 3/3 (3/3) |
| RS7 | Z25749 | AF043285 | 100.0 | 1768.4 | 6/6 (6/6) |
| SPEE | M64231 | Z67748 | 94.7 | 563.7 | 8/8 (6/8) |
The last column reports the fraction of CSTs matching to exons correctly classified as coding (values in parentheses refer to a QRNA application with the same BLAST parameters used in CSTfinder).
The CSTfinder algorithm was also used to compare 5′-UTR sequences of human and rodent mRNAs from the UTRdb database (16) in order to detect potentially coding CSTs (present in the database due to mis-annotation of the coding region or to UTRs that are part of the coding portion in alternatively spliced mRNAs). Table 3 shows the 10 highest CPS matches for both 5′- and 3′-UTR analysis. In all cases the hypothetical peptide encoded by the human sequence of the detected CST is identical or highly similar to an already annotated human protein. These results imply that mis-annotation of CDS regions (or indeed known or undetected alternative splicing patterns) have contributed to the presence of a number of coding regions in UTRdb.
Table 3. Results of CSTfinder application to the comparison of human and mouse 5′- and 3′-UTRs collected in UTRdb (16).
| Rod UTRdb ID | Hsa UTRdb ID | S | A | CPS | Protein ID |
|---|---|---|---|---|---|
| 5′-UTR | |||||
| 5RNO000568 | 5HSA011649 | 26 | 1 | 826.8 | NP_036234.2 |
| 5MMU013664 | 5HSA024541 | 13 | 1 | 497.3 | NP_004796.1 |
| 5RNO000682 | 5HSA003514 | 12 | 1 | 437.2 | AAC61479.1 |
| 5RNO000568 | 5HSA011649 | 18 | 1 | 416.3 | NP_036234.2 |
| 5MMU012957 | 5HSA021834 | 13 | 1 | 402.7 | AAG35479.1 |
| 5MMU012175 | 5HSA033873 | 18 | 1 | 391.8 | AAH32597.1 |
| 5MMU014128 | 5HSA028662 | 53 | 3 | 339.1 | BAB85047.1 |
| 5MMU013456 | 5HSA031624 | 20 | 3 | 259.0 | AF381996.1 |
| 5MMU010706 | 5HSA029926 | 10 | 1 | 256.4 | NP_079011.2 |
| 5RNO005888 | 5HSA011221 | 8 | 1 | 239.6 | P39192 |
| 3′-UTR | |||||
| 3MMU007952 | 3HSA031829 | 33 | 1 | 1020.6 | NP_001010.2 |
| 3MMU007222 | 3HSA026992 | 37 | 2 | 879.4 | NP_004455.1 |
| 3RNO000947 | 3HSA024970 | 46 | 2 | 762.7 | AAB41498.1 |
| 3MMU013840 | 3HSA004501 | 21 | 1 | 565.8 | NP_067642.1 |
| 3MMU010062 | 3HSA034129 | 13 | 1 | 495.2 | XP_172586.1 |
| 3MMU008684 | 3HSA001895 | 15 | 1 | 367.1 | AAC51213.1 |
| 3RNO006623 | 3HSA012556 | 21 | 2 | 296.0 | BAC04618.1 |
| 3MMU010353 | 3HSA020623 | 9 | 1 | 278.6 | NP_038267.1 |
| 3MMU005948 | 3HSA001570 | 10 | 1 | 269.7 | NP_001558.2 |
| 3MMU008634 | 3HSA009559 | 9 | 1 | 246.2 | P78536 |
The 10 highest scoring HSPs are shown for both 5′- and 3′-UTRs with the accession number of the relevant matching protein.
In order to provide a conservative estimate of the false positive rate in real biological data we analyzed the SYNTENY dataset. 1801 of 1845 CTSs with CPS > 500 (∼98%) overlapped exons of EnsEMBL annotated genes, as well as 4015 of 5755 CSTs with CPS > 30 (70%). This implies a false positive rate of ∼2% if a CPS threshold of 500 is adopted, but also indicates that CSTs with CPS > 30 merit further investigation. In contrast, only 414 of 1579 CSTs with CPS < 30 (26%) corresponded to exons of EnsEMBL annotated genes.
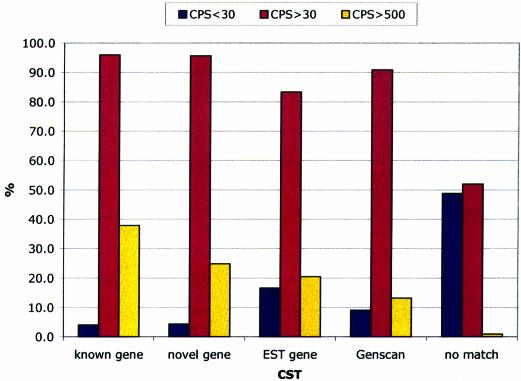
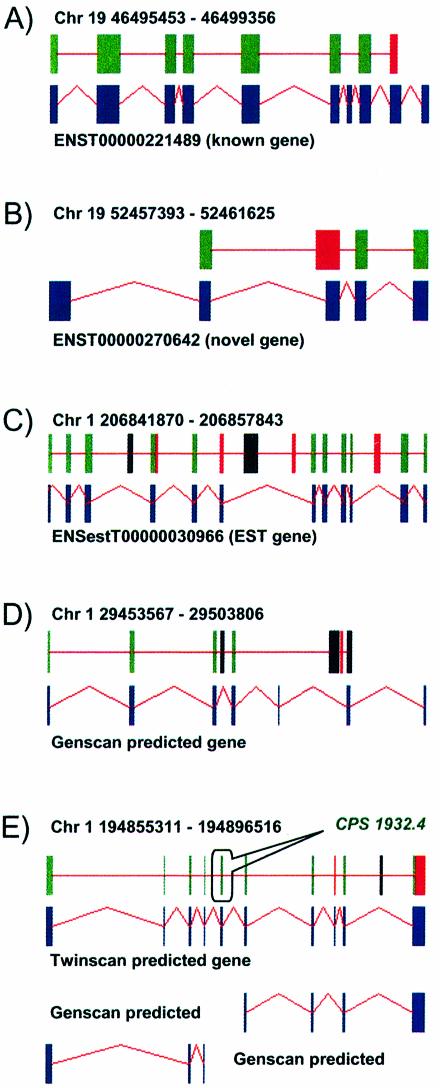
Finally, we carried out an extensive analysis on 199 pairs of human and mouse gene loci accounting for a total of ∼140 Mb. A total of 10 899 CSTs were identified. We classified these CSTs on the basis of the detected overlap between CST coordinates and genome feature coordinates annotated in EnsEMBL as ‘known gene’, ‘novel gene’, ‘EST gene’ or ‘GenScan predicted gene’. Remaining CSTs were classified as ‘no match’. Results are summarized in Figure 2. Interestingly, only 4.1% of CSTs overlapping with exons belonging to ‘known genes’ showed a CPS < 30, with a remarkable fraction of ∼40% with a CPS > 500 denoting a highly significant coding capacity. High CPSs have also been observed for CSTs falling in ‘novel genes’, ‘EST genes’ and ‘GenScan predicted genes’, thus providing significant confirmation of a large number of unknown genes. We have also investigated those few cases were the CSTs overlapping known genes presented low CPSs (< 30). We noted that in most cases the CSTs covered not only a coding portion of the gene but comprised conserved regions falling in introns or 5′- or 3′-UTRs. Some examples are reported in Figure 3, where a graphic representation of CSTs mapping on known genes (Fig. 3A), novel genes (Fig. 3B), EST genes (Fig. 3C) and GenScan (Fig. 3D) and TwinScan (Fig. 3E) predicted genes is shown. It is evident that CSTfinder confirms most known exons (see also data in Table 1) and, more importantly, indicates novel coding exons including those missing the experimental validation of a matching transcript sequence and only computationally predicted by GenScan or TwinScan. In addition, most CSTs not showing a significantly high CPS fall outside the coding portion of gene exons.
Figure 2.
Results obtained from the CSTfinder analysis of the GENOME set showing the percentage of CSTs falling in different gene annotation categories.
Figure 3.
Detailed result of CSTfinder analysis on five human–mouse homologous gene loci in the GENOME dataset belonging to different EnsEMBL gene classes. (A) Known gene; (B) novel gene; (C) EST gene; (D and E) GenScan predicted gene. Upper boxes represent identified CSTs (green, CPS > 500; red, 30 > CPS ≤ 500; black, CPS ≤ 30) with lower boxes corresponding to known or predicted exons. For each gene the EnsEMBL ID, the chromosome position and the coordinates (NCBI 30 release) are reported. The arrow highlights a CSTfinder predicted coding sequence missed by GenScan but coincident with a TwinScan predicted exon.
A large fraction of identified CSTs (∼50%, 4577 of 10 899), designated as ‘no match’, do not correspond to sequence stretches with known function. Although some of these may represent as yet unannotated exons it is likely that most of them actually represent non-genic sequences endowed with structural or regulatory activities. A comparable estimate of the abundance of non-genic conserved sequences has been made from an analysis of the syntenic region of human chromosome 21 (19).
DISCUSSION
The completion of the draft genome sequence of human, mouse, rat and Fugu, as well as the steady progression of other vertebrate sequencing projects, has opened new avenues in the genomic (or post-genomic) era that will give us the extraordinary opportunity of investigating evolutionary changes that have shaped genome structure and content during evolution. The comparison of two or more genome sequences revealing conservative and diverging patterns may highlight common and species-specific molecular mechanisms that control gene expression.
In particular, the comparative sequence analysis of genomes of two or more species gives us the possibility of identifying the entire inventory of conserved genomic elements playing a role in the control of gene expression. It is increasingly clear that to decrypt the genetic information encoded in the genome it is necessary, but not sufficient, to define the structure of all its genes. Indeed, most of the genome space (>95% in mammals) is non-coding and thus potentially involved in the concerted regulation of spatio-temporal expression of genes. The identification of CSTs may be critical in shedding light on this ‘dark side’ of the genome. However, given that we still do not know the complete gene inventory of almost any of the genomes sequenced to date, it is particularly important to assign the genome CSTs identified in cross-species comparisons to the coding or non-coding portion of the genome. Indeed, most annotated coding exons correspond to conserved regions in human–mouse comparisons (20). We have here proposed a novel heuristic approach that can effectively discriminate between coding and non-coding CSTs. It is based on the simple idea that coding and non-coding portions of the genome are subjected to remarkably different evolutionary dynamics. In fact, specific constraints operate on coding regions where synonymous outnumber non-synonymous changes, and these latter more likely result in conservative amino acid replacements.
Although a similar strategy underlies other methods recently proposed, such as QRNA (12), these methods generally require pairwise sequence alignments as input and are computationally unfeasible for large-scale genome comparisons. Furthermore, CSTfinder proved generally more accurate in assessing CST coding capacity than QRNA (see Table 2).
We have shown that the proposed CPS effectively discriminates between these two different dynamics as >99% of CSTs identified in the comparison of human and mouse coding sequences displayed a CPS above the cut-off value of 30, with an average value much higher (see Table 1). Of the few genes that CSTfinder failed to detect, most did not show statistically significant CSTs and thus completely escaped the analysis.
The CSTfinder algorithm may provide a significant validation for novel genes not confirmed by transcript information or similarity to known proteins. Interestingly an application to a limited set of human–mouse ortholog gene regions significantly validated several genes, finding CSTs with high coding potential (CPS > 500) in correspondence to almost all the relevant coding exons (Table 2). More importantly, many of the genes present in databases lack reliable transcript or protein similarity information and have merely been computationally predicted by the GenScan (3) or TwinScan (6) software programs. Conversely, as many of the proteins collected in public repositories are hypothetical, being the result of computational translation of predicted ORFs, the validity of those not showing any potentially coding CST should be questioned.
Additionally, CSTfinder may help the identification of transcripts derived from alternative splicing in those cases where some variant isoforms are not represented in the sequence database. For example a high scoring CST found in the 5′-UTR of Rho guanine nucleotide exchange factor (GEF) 7 variant 1 mRNA (NM_003899) corresponds to a CDS portion in variant 2 mRNA encoded by the same gene (NM_145735).
If CSTs overlapping coding exons also cover non-coding regions (e.g. 5′-UTRs, 3′-UTRs and splice juctions) they usually show lower CPSs, sometimes below the cut-off threshold. In this case the coding-like evolutionary signature is obscured by the unconstrained evolutionary dynamics of the non-coding part. For some selected CSTs a window-based approach, also implemented in the CSTfinder algorithm, can be used to discriminate regions evolving under different evolutionary dynamics.
We have tested the effectiveness of the method at various evolutionary distances by sampling different groups of human–mouse orthologous CDSs. This means that the applicability of the method is not constrained by the evolutionary distance between species compared but rather by the sensitivity of identification of local alignment by a Blast-like similarity search. As species divergence increases, the number of CSTs detected decreases. However, the capacity of CSTfinder to differentiate between coding and non-coding regions within and among CSTs remains essentially constant.
The reliability of CST coding potential prediction is obviously dependent on the CPS threshold chosen, even if a large number of CTSs matching coding exons show a very high CPS (see Fig. 2 and green boxes in Fig. 3). The analysis carried out on the RANDOM dataset (see Materials and Methods) clearly shows that a CPS threshold of 500 is highly conservative, as only 1 out of the total of 6564 CSTs identified showed a CPS above this threshold. The analysis carried out on the SYNTENY dataset confirmed a low false positive rate (∼2%) if a CPS cut-off of 500 is chosen. However, some of these false positives could actually be missed exons. Indeed, 11 of the 44/1845 CSTs with CPS > 500 non-overlapping annotated genes showed significant matches with human ESTs, GenScan predicted exons or SwissProt entries. Our analyses suggest that it is worth considering CSTs showing a CPS > 30 for further analyses as many of these overlap coding exons (70% in the SYNTENY dataset) but at the same time also neighboring non-coding regions (see red box in Fig. 3B).
The application of CSTfinder, in association with other tools specifically designed for coding and non-coding gene detection, may thus represent a very valuable approach for large-scale detection of non-coding regulatory elements, validation of gene predictions and discovery of novel genes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank David Horner for helpful comments on the manuscript. This work has been supported by Telethon and MIUR.
REFERENCES
- 1.Delcher A.L., Harmon,D., Kasif,S., White,O. and Salzberg,S.L. (1999) Improved microbial gene identification with GLIMMER. Nucleic Acids Res., 27, 4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Besemer J., Lomsadze,A. and Borodovsky,M. (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res., 29, 2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burge C. and Karlin,S. (1997) Prediction of complete gene structures in human genomic DNA. J. Mol. Biol., 268, 78–94. [DOI] [PubMed] [Google Scholar]
- 4.Krogh A. (2000) Using database matches with for HMMGene for automated gene detection in Drosophila. Genome Res., 10, 523–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rogic S., Mackworth,A.K. and Ouellette,F.B. (2001) Evaluation of gene-finding programs on mammalian sequences. Genome Res., 11, 817–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korf I., Flicek,P., Duan,D. and Brent,M.R. (2001) Integrating genomic homology into gene structure prediction. Bioinformatics, 17 (suppl. 1), S140–S148. [DOI] [PubMed] [Google Scholar]
- 7.Pachter L., Alexandersson,M. and Cawley,S. (2002) Applications of generalized pair hidden Markov models to alignment and gene finding problems. J. Comput. Biol., 9, 389–399. [DOI] [PubMed] [Google Scholar]
- 8.Makalowski W. and Boguski,M.S. (1998) Evolutionary parameters of the transcribed mammalian genome: an analysis of 2,820 orthologous rodent and human sequences. Proc. Natl Acad. Sci. USA, 95, 9407–9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pennacchio L.A. and Rubin,E.M. (2001) Genomic strategies to identify mammalian regulatory sequences. Nature Rev. Genet., 2, 100–109. [DOI] [PubMed] [Google Scholar]
- 10.Badger J.H. and Olsen,G.J. (1999) CRITICA: coding region identification tool invoking comparative analysis. Mol. Biol. Evol., 16, 512–524. [DOI] [PubMed] [Google Scholar]
- 11.Wiehe T., Gebauer-Jung,S., Mitchell-Olds,T. and Guigo,R. (2001) SGP-1: prediction and validation of homologous genes based on sequence alignments. Genome Res., 11, 1574–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rivas E. and Eddy,S.R. (2001) Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics, 2, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 14.Makalowski W., Zhang,J. and Boguski,M. (1996) Comparative analysis of 1196 orthologous mouse and human full-length mRNA and protein sequences. Genome Res., 6, 846–857. [DOI] [PubMed] [Google Scholar]
- 15.Jareborg N., Birney,E. and Durbin,R. (1999) Comparative analysis of noncoding regions of 77 orthologous mouse and human gene pairs. Genome Res., 9, 815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pesole G., Liuni,S., Grillo,G., Licciulli,F., Mignone,F., Gissi,C. and Saccone,C. (2002) UTRdb and UTRsite: specialized databases of sequences and functional elements of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Update 2002. Nucleic Acids Res., 30, 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hubbard T., Barker,D., Birney,E., Cameron,G., Chen,Y., Clark,L., Cox,T., Cuff,J., Curwen,V., Down,T. et al. (2002) The Ensembl genome database project. Nucleic Acids Res., 30, 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins J.E., Goward,M.E., Cole,C.G., Smink,L.J., Huckle,E.J., Knowles,S., Bye,J.M., Beare,D.M. and Dunham,I. (2003) Reevaluating human gene annotation: a second-generation analysis of chromosome 22. Genome Res., 13, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dermitzakis E.T., Reymond,A., Lyle,R., Scamuffa,N., Ucla,C., Deutsch,S., Stevenson,B.J., Flegel,V., Bucher,P., Jongeneel,C.V. et al. (2002) Numerous potentially functional but non-genic conserved sequences on human chromosome 21. Nature, 420, 578–582. [DOI] [PubMed] [Google Scholar]
- 20.Dubchak I., Brudno,M., Loots,G.G., Pachter,L., Mayor,C., Rubin,E.M. and Frazer,K.A. (2000) Active conservation of noncoding sequences revealed by three-way species comparisons. Genome Res., 10, 1304–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]