Abstract
FSH, a key regulator of gonadal function, contains a β-subunit (FSHβ) that is transcriptionally induced by activin, a member of the TGFβ-superfamily. This study used 4.7 kb of the ovine FSHβ-promoter linked to luciferase (oFSHβLuc) plus a well-characterized activin-responsive construct, p3TPLuc, to investigate the hypothesis that Smad3, TGFβ-activated kinase 1 (TAK1), or both cause activin-mediated induction of FSH. Overexpression of either Smad3 or TAK1 induced oFSHβLuc in gonadotrope-derived LβT2 cells as much as activin itself. Induction of p3TPLuc by activin is known to require Smad3 activation in many cell types, and this was true in LβT2 cells, where 10-fold induction by activin (2–8 h after activin treatment) was blocked more than 90% by two dominant negative (DN) inhibitors of Smad3 [DN-Smad3 (3SA) and DN-Smad3 (D407E)]. By contrast, 6.5-fold induction of oFSHβLuc by ac-tivin (10–24 h after activin treatment) was not blocked by either DN-Smad inhibitor, suggesting that activation of Smad3 did not trigger induction of oFSHβLuc. By contrast, inhibition of TAK1 by a DN-TAK1 construct led to a 50% decrease in activin-mediated induction of oFSHβLuc, and a specific inhibitor of TAK1 (5Z-7-Oxozeanol) blocked induction by 100%, indicating that TAK1 is necessary for activin induction of oFSHβLuc. Finally, inhibiting p38-MAPK (often activated by TAK1) blocked induction of oFSHβLuc by 60%. In conclusion, the data presented here indicate that activation of TAK1 (and probably p38-MAPK), but not Smad3, is necessary for triggering induction of oFSHβ by activin.
Abbreviations: BMP, Bone morphogenetic protein; Ca-ActRIB, constitutively active activin receptor IB; DN, dominant negative; αGSU, α-glycoprotein subunit; oFSHβLuc, ovine FSHβ-promoter linked to luciferase; JNK, c-Jun N-terminal kinase; MAPKKK, MAPK kinase kinase; SBE, Smad binding element; TAB, TAK1-binding protein; TAK1, TGFβ-activated kinase 1
FSH IS ESSENTIAL for female folliculogenesis and stimulates spermatogenesis in males (1). It is an α/β heterodimer made only in pituitary gonadotropes where transcription of its unique β-subunit (FSHβ) is paramount in determining overall FSH production. Transcription of FSHβ is controlled primarily by GnRH (2, 3) and gonadal- or pituitary-derived activins, inhibins, and follistatin (2–10).
Activin, a major inducer of FSHβ-transcription, seems central to FSH expression in vivo. It comprises three isoforms that are homo- or heterodimers of activin βA or βB chains (7). Activin A (homodimer of βA subunits) is the most potent inducer known for FSHβ (11), although other members of the TGFβ family have been shown to induce transcription of the FSHβ-subunit (5, 8). Recent studies suggest that activin action not only induces FSHβ but is also important for GnRH-mediated induction of FSHβ expression as well (12, 13).
Activin exerts its biological effects by binding to activin type II serine/threonine kinase receptors (ActRII or ActRIIB), which then bind and phosphorylate the activin type I receptor (ActRIB, also known as ALK4) (14–20). Once this occurs, the signal can be transmitted downstream by phosphorylating and activating Smad2 and/or Smad3. One or more of these Smads associate with a common signaling molecule, Smad4 (Co-Smad), to form a Smad2/4 or Smad3/4 complex that translocates to the nucleus, where it binds a specific DNA sequence known as a Smad binding element (SBE). The affinity of Smad complexes for DNA is low, but binding is usually stabilized by interaction(s) with adjacent tissue- and cell-specific transcription factors (21–28).
Individual or palindromic SBEs have been identified in both ovine and rat FSHβ-promoters, and they appear to be important for activin-mediated induction of FSHβ. Several are reported to bind Smad4. Also, the tale homeodomain of Pbx1 and Prep1 has been shown to partner with Smad4 within a DNA binding complex in the ovine FSHβ-promoter, and the same was shown for Pitx2c in the rat FSHβ-promoter (29, 30). Furthermore, overexpression of Smad3 (but not Smad2) has been shown to increase basal and activin-induced expression of FSHβ in LβT2 cells. Nevertheless, the binding of Smad4 to SBEs in the ovine and rat promoters of FSHβ is not significantly increased after treatment of cells with activin (29–31). Therefore, whereas Smad4 and its associated transcription factors appear to play pivotal roles in expression of FSHβ, no data actually prove that activin triggers FSHβ induction through the activation of Smad3 and subsequent binding to Smad4 and one or more FSHβ-SBEs.
Recently a Smad-independent pathway has been associated with the actions of several TGFβ family members [TGFβ, bone morphogenetic proteins (BMPs), activins] that use TGFβ-activated kinase 1 (TAK1), a member of the MAPK kinase kinase (MAPKKK) family (32). Activation of TAK1 requires the association of TAK1-binding protein (TAB)1, which induces a conformational change in TAK1, allowing it to autophosphorylate its activation domain (33). The activated TAK1/TAB1 complex also binds TAB2 or TAB3 proteins, which modulate the regulatory effects of TAK1/TAB1 (34–38). Activation of TAK1 initiates MAP kinase cascades that have been shown to phosphorylate downstream c-Jun N-terminal kinase (JNK), ERK, and p38 (34, 39–46).
The studies presented here focused initially on the hypothesis that Smad3 induces expression of an ovine FSHβ-promoter-luciferase construct (oFSHβLuc) that contains all the regulatory sequences needed for normal cell-specific expression and regulation of FSHβ in mouse gonadotropes (47–50). The transformed gonadotrope cell line LβT2 was used as a model to study the signaling pathway mediating activin induction of FSHβ expression (51). Studies included comparisons with a well-characterized activin-responsive construct known to require Smad3 for induction, p3TPLuc (48). When it appeared that Smad3 was important for basal expression of oFSHβ but could not be proven to trigger activin-mediated induction of oFSHβLuc, the studies focused on the hypothesis that a novel Smad-independent pathway uses TAK1 activation to induce oFSHβLuc.
Materials and Methods
Reagents
Recombinant human activin A was obtained from R&D Systems, Inc. (Minneapolis, MN). MAPK inhibitors SB203580, SP600125, and PD98059 were purchased from Calbiochem (San Diego, CA). Recombinant human follistatin 288 was obtained from Dr. A. F. Parlow at the National Hormone and Pituitary Program (Torrance, CA), and 5Z-7-Oxozeanol (TAK1 inhibitor) was obtained from Chugai Pharmaceutical Co., Ltd. (Tokyo, Japan).
Reporter plasmids and expression vectors
The ovine reporter plasmid oFSHβLuc has been previously described (52). Smad3 and dominant negative (DN) Smad3 [DN-Smad3 (D407E)] were obtained from Dr. Mitsuyasu Kato (University of Tsukuba, Tsukuba, Japan) (53). DN-Smad3 (3SA) was obtained from Dr. Theresa A. Guise (University of Texas, San Antonio, TX) (54). Expression constructs for TAK1 (55), TAB1 (55), TAB2 (36), TAB3 (34), and DN-TAK1-KN (56) were obtained from Dr. Jun Tsuji (North Carolina State University, Raleigh, NC). Constitutively active activin receptor IB (Ca-ActRIB) was obtained from Dr. J. Wrana (Mount Sinai Hospital, Ontario, Canada) (18). Mock plasmid (pCMV DNA), containing the CMV promoter with no luciferase reporter gene, was obtained from Promega Corp. (Madison, WI). Control plasmid (pGL3-control) containing the GL3 luciferase reporter driven by an SV40 promoter was purchased from Promega.
Culture and transient transfection of LβT2 gonadotropes
LβT2 cells (51) were grown at 37 C in DMEM (Life Technologies, Inc., Gaithersburg, MD) containing 10% fetal bovine serum (Hyclone Laboratories, Inc., Logan, UT), 100 U/ml penicillin G, and 100 μg/ml streptomycin under 95% air-5% CO2. Cells were grown in 150-cm2 flasks until they were confluent and then transferred to 96-multiwell plates (Falcon; Fisher Scientific, Raleigh, NC) at a concentration of 25,000 cells/well. Cells were cotransfected, 24 h later, in triplicate with 50 ng oFSHβLuc, p3TPLuc, pGL3-control and 50–150 ng pCMV using Fugene6 (Roche Molecular Biochemicals, Basel, Switzerland). After transfection (24 h), cells were treated with fresh media, with or without activin (100 ng/ml), for an additional 24 h. Cells were then lysed in 50 μl passive lysis buffer, and 20 μl was assayed for luciferase activity.
Real-time RT-PCR
Total RNA from LβT2 cells was isolated and converted to cDNA as reported (5). Oligonucleotides for taqman real-time PCR were designed for mouse FSHβ, LHβ, and the α-glycoprotein subunit (αGSU) common to both LH and FSH, using software from Integrated DNA Technologies, Inc. (Coralville, IA). Mouse 18s ribosomal RNA served as the internal control for quantifying all molecules as reported earlier (57). The taqman probe for FSHβ was reported previously (57), and the primers and probes for LHβ and αGSU were: LHβ forward, 5′-AATCCCGCTCCA-CACAGTACATGA-3′; LHβ reverse, 5′-TCAGCTCAGGAGGTGTC-CATTGTT-3′; LHβ probe, 5′-TGCCTTCCTCTTCAATGAGCTCAAAG-GCGA-3′; αGSU forward, 5′-AGATCGACAATCACCTGCCCAGAA-3′; αGSU reverse, 5′AGGAACATGGACAGCATGACCAGA-3′; αGSU probe, 5′-TCCAGAGCTTGCAGAAGAGCTATGGA-3′.
Real-time PCR was performed in duplicate on triplicate cDNA samples from LβT2 cells using an i-Cycler (Bio-Rad Laboratories, Inc., Hercules, CA). Samples were incubated at 95 C for 3 min, and then for 40 complete cycles (95 C for 30 sec, 55 C for 30 sec, and 72 C for 30 sec). There was a final extension step of 72 C for 3 min. Threshold cycle values were determined with Bio-Rad software and used for relative quantization with the 2−ΔΔCt method.
Luciferase assay
Cells were harvested by adding 1 × passive lysis buffer (Promega, Madison, WI) as described above for LβT2 cells. Luciferase activity was measured in 40% of the cell lysate (20 μl) by adding 100 μl luciferase assay system from Promega. Luciferase activity was measured for 20 sec using a Monolight 2010 single tube luminometer (Analytical Luminescence Laboratory, San Diego, CA) or automated Victor-Light microplate luminometer (PerkinElmer, Boston, MA).
Antibodies
The rabbit antimouse TAK1 (554–579) and rabbit antimouse TAB1 (480–500) polyclonal antibodies (Upstate Biotechnology, Inc., Lake Placid, NY) were also used to detect endogenous TAK1 and TAB1 in LβT2 cells. Secondary antibody used for detection was antirabbit IgG (Calbiochem).
Western blotting
Analysis of TAK1 and TAB1 expression by Western blot was performed as previously described (55). Briefly, LβT2 cells were cultured in 6-well tissue culture plates with 1 million cells/well for 24 h. Cells were pretreated with follistatin-288 (250 ng/ml) for 16 h to minimize FSHβ expression (unpublished results by Dr. H. J. Huang, this laboratory), presumably by eliminating any effects of endogenously produced activin or activin-like activist (5). Cells were then washed and treated with or without activin A (100 ng/ml) for the indicated time points. Cells were washed once with PBS, pH 7.4 (PBS; 4 C) and collected in 1 ml PBS. Cells were centrifuged at 1000 × g for 5 min (4 C) and lysed in 50 μl of 0.5% Triton X-100 lysis buffer (20 mM HEPES, pH 7.4; 150 mM NaCl; 12.5 mM β-glycerophosphate; 1.5 mM MgCl2; 2 mM EGTA; 10 mM NaF; 2 mM dithiothreitol; 1 mM sodium orthovanadate; 1 mM phenylmethylsulfonylfluoride; and 20 mM aprotinin). Cells were sonicated for 5 sec and centrifuged at 10,000 × g for 5 min, and the cleared lysates were fractionated on a 7% SDS-polyacrylamide gel. Proteins were transferred to Hybond-P membranes (Amersham Pharmacia Biotech, Piscataway, NJ) and incubated with antibodies, and antibody localization was visualized with horseradish peroxidase-conjugated antibodies to rabbit IgG using the enhanced chemiluminescence Western blotting system (ECL; Amersham).
Statistics
Data (see Fig. 7; Western blot) were obtained two times, and the clearest blot is shown. Data from all other experiments were replicated at least three times, and all samples were assayed in triplicate. Means ± SEM values are shown in all figures; data in all (with one exception; see Fig. 7) were analyzed using one-way ANOVA with Tukey’s multiple comparison test according to the Prism version 4 (GraphPad Software, Inc., San Diego, CA).
Fig. 7.
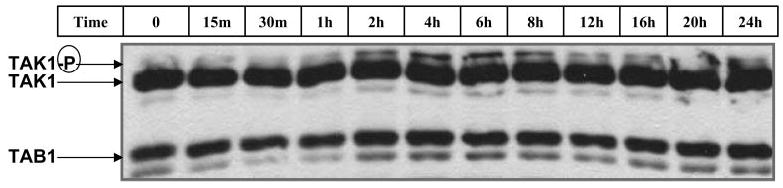
Activin phosphorylated TAK1 within 2 h and maintained TAK1 activation for 24 h. LβT2 cells were plated at 1 million cells per well in 6-well plates. Cells were pretreated with follistatin-288 (250 ng/ml; 16 h) and washed with culture media. Cells were then treated with activin (100 ng/ml) for 0, 15, or 30 min or 1, 2, 4, 6, 8, 12, 16, 20, or 24 h. Phosphorylation of endogenous TAK1 was detected by Western blot analysis as described in Materials and Methods.
Results
Transfected Smad3 increased basal and activin-induced expression of oFSHβLuc equally
To investigate the role of Smad3 in mediating oFSHβ induction by activin, LβT2 cells were cotransfected with oFSHβLuc and increasing amounts of Smad3 DNA (25, 50, 75, 100, and 125 ng) (Fig. 1). Cotransfection with Smad3 increased basal expression of oFSHβLuc by 89, 116, 206, 240, and 312%, respectively. Activin treatment of cultures with transfected Smad3 increased oFSHβLuc induction above control cultures. However, increased induction paralleled increased basal expression at all concentrations of transfected Smad3, with an average increase of 4.5 ± 0.3. The ratios of induced/basal expression at each level of Smad3 were: 4.6 ± 2.1, 5.6 ± 1.5, 4.2 ± 1.1, 4.6 ± 1.2, and 3.7 ± 1.0, respectively, for the 25- to 125-ng treatments. None of these were significantly different from each other, nor were they different from the 5.7 ± 1.6 ratio observed in control cultures not transfected with Smad3 (Fig. 1).
Fig. 1.
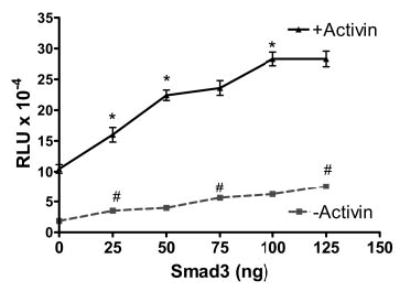
Overexpression of Smad3 equally stimulated basal and activin-induced expression of oFSHβLuc. LβT2 cells were plated at 25,000 cells/well in 96-well tissue culture plates. After 24 h, they were cotransfected with 50 ng oFSHβLuc and increasing amounts of Smad3 expression construct (25, 50, 75, 100, and 125 ng). DNA amounts were kept constant at 200 ng DNA transfected per well using pCMV (mock plasmid) to balance amounts of DNA. After transfection (24 h), cells were treated with or without activin (100 ng/ml) for an additional 24 h and assayed for luciferase activity. Ratios of induced/basal were not significantly different from the 5.7 ± 1.6 ratio observed in control cultures not transfected with Smad3 (P > 0.05). One-way ANOVA/Tukey’s was used to show that increasing levels of Smad3 increased luciferase expression; a significant increase between points (P < 0.05) was designated: #, for basal expression; *, for activin-stimulated expression. RLU, Relative light units.
Smad3 activation is required for induction of p3TPLuc but not for oFSHβLuc
To determine the functional significance of endogenous Smad3 signaling, two DN inhibitors of Smad3 [Smad3 (3SA) and Smad3 (D407E)] were tested. First, LβT2 cells were co-transfected with p3TPLuc, which can be induced by activin through a Smad3 pathway. Then p3TPLuc was cotransfected with one of two DN-Smad3 expression vectors to block the actions of endogenous Smad3 (see Fig. 2) (49, 53). Basal expression of p3TPLuc was not inhibited by either DN-Smad, but both inhibitors blocked 7-fold induction by activin by 87–96%. These data showed that DN-Smad3 (3SA) and DN-Smad3 (D407E) were effective inhibitors of activin-mediated activation of Smad3 in LβT2 cells.
Fig. 2.
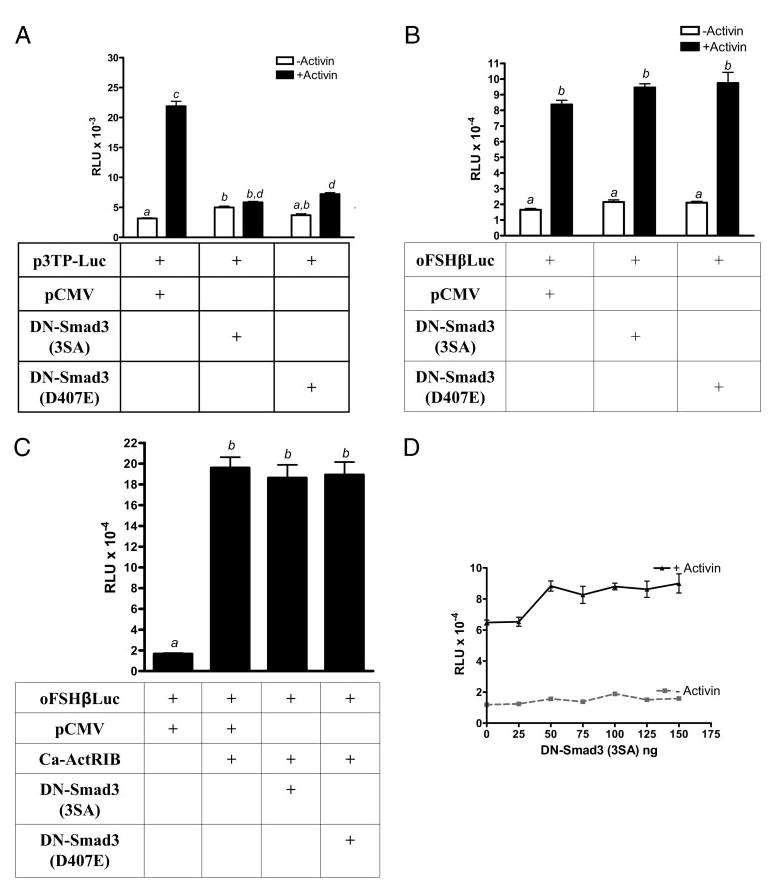
Activin did not require activated Smad3 to induce oFSHβLuc expression. LβT2 cells were prepared and plated as in Fig. 1 and then treated as follows: A, Cells were cotransfected with 50 ng p3TPLux plus 50 ng pCMV, DN-Smad3 (3SA), or DN-Smad3 (D407E). After transfection (24 h), cells were treated with or without activin (100 ng/ml) for an additional 6 h and assayed for luciferase activity. Means with different letters are significantly different (P < 0.05) (P < 0.001 for c vs. d). B, Cells were treated as in A except that oFSHβLuc was used instead of p3TPLux. Means with different letters are significantly different (P < 0.05). C, Cells were transfected with 50 ng oFSHβLuc plus 100 ng pCMV (column 1) or 50 ng pCMV and 50 ng Ca-ActRIB (column 2). Columns 3 and 4 were transfected with 50 ng of either DN-Smad3 (SA) or DN-Smad3 (D407E) in place of pCMV. Cells were collected 24 h after transfection and assayed for luciferase activity. Means with different letters are significantly different (P < 0.05). D, Cells were cotransfected with 50 ng oFSHβLuc and increasing amounts of DN-Smad3 (3SA) expression construct (25, 50, 75, 100, 125, and 150 ng). Total DNA amounts were kept constant using mock plasmid pCMV. Twenty-four hours after transfection, cells were treated with or without activin (100 ng/ml) for an additional 24 h. Ratios of induced/basal were not significantly different from the 5.5 ± 0.22 ratio observed in control cultures not transfected with DN-Smad3 (3SA) (P > 0.05).
Data (see Fig. 2B) show that DN-Smad3 (3SA) or DN-Smad3 (D407E) did not block basal or activin-mediated induction of oFSHβLuc. Activin induced oFSHβLuc expression by 5.1-fold, but neither Smad3 inhibitor altered the action of activin.
Subsequent results (see Fig. 2C) are similar, except that Ca-ActRIB was used to induce oFSHβLuc in LβT2 cells. The results show that cotransfection of Ca-ActRIB along with oFSHβLuc increased expression of oFSHβLuc by 9.5-fold. The DN inhibitors of Smad3, however, had no effect on this induction, just as observed for induction of oFSHβLuc with activin. These data show that the constitutively active activin receptor induced oFSHβLuc but that activation of Smad3 was not necessary for induction to occur.
Data indicate (see Fig. 2D) that there was no tendency for DN-Smad3 (3SA) to inhibit either basal or activin-mediated induction of oFSHβLuc over a range of concentrations (25–150 ng) higher and lower than the 50-ng amount used in Fig. 2, A–C. Activin increased expression of oFSHβLuc by 5.8-fold over basal expression, with or without DN-Smad3 (Fig. 2D).
Activin induction of oFSHβLuc and p3TPLuc have very different kinetics
The data in Fig. 3 show time-course data (24 h) for activin induction of oFSHβLuc and p3TPLuc. In Fig. 3A, activin A (100 ng/ml) had no observable effect on oFSHβLuc expression for 6 h. It then had a small, inductive effect from 8–12 h (~100% increase) before it increased expression linearly up to full induction at 22 h (520% increase). When DN-Smad3 (3SA) was cotransfected, the same pattern was observed, and there was no inhibition by DN-Smad3 (3SA) at any time during the 24-h period.
Fig. 3.
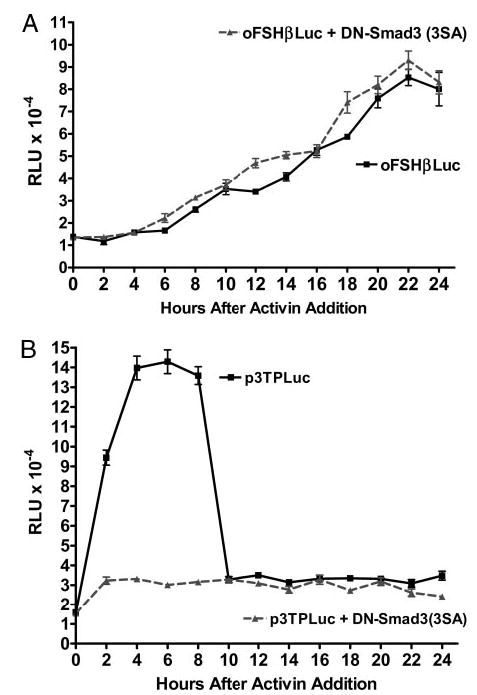
Different kinetics for oFSHβLuc and p3TPLux induction by activin. LβT2 cells were prepared and plated as in Fig. 1. A, Cells were cotransfected with 50 ng oFSHβLuc plus either 50 ng pCMV (▪) or 50 ng Smad3 (3SA) (▴). After transfection (24 h), triplicate cultures were sequentially treated with activin (100 ng/ml) every 2 h for 24 h. There was no significant increase in expression for the first 6 h of activin treatment for either control or DN-Smad3 (3SA)-transfected cells. Cells treated with activin for 8 h or longer showed significant increases in expression, but there was no significant difference at any time point between cultures transfected with pCMV (control) or DN-Smad3 (3SA). B, Cells were treated as in A, except p3TPLux was used instead of oFSHβLuc. All cultures treated with activin showed, at least, a significant doubling of expression at all time points (P < 0.05). Expression increased further (P < 0.001), up to 9.3-fold, between 2 and 8 h, in the absence of DN-Smad3 (3SA), but DN-Smad3 totally blocked this increase.
The data in Fig. 3B show that p3TPLuc was induced 6-fold at 2 h and 9-fold at 6 h and dropped to 2-fold by 10 h. Therefore, induction by activin was initiated well before 2 h and down-regulated shortly after 8 h. When cultures were cotransfected with DN-Smad3 (3SA), most of the induction between 2–10 h was blocked. Results shown with the DN-Smad3 in this time-course format indicated that activin induced expression of oFSHβ through a pathway that did not depend on Smad3 activation.
Altering expression of oFSHβLuc with TAK1, TAB1, TAB2, TAB3, and DN-TAK1
Next, the role of TAK1 as a potential mediator of activin induction was tested. LβT2 cells were transfected with either oFSHβLuc alone or with oFSHβLuc plus TAK1 and its binding proteins, TAB1, TAB2, or TAB3. Preliminary studies showed that overexpression of TAK1/TAB1 induced oFSHβLuc 45-fold, but this appeared to be non-specific induction because it also increased expression of the SV40-driven pGL3Luc control construct and the minimal thymidine kinase luciferase construct, T109Luc, that are not activin responsive (data not shown). However, cotransfection of TAK1 with either TAB2 or TAB3 did increase expression of oFSHβLuc, just like activin (6.3-fold), and activin was unable to augment this induction much above 6.3-fold when added along with TAB2 or TAB3. Thus, transfections with TAK1 and either TAB2 or TAB3 nearly substituted for activin induction of oFSHβLuc (Fig. 4A), suggesting they could be in the signaling pathway used by activin.
Fig. 4.
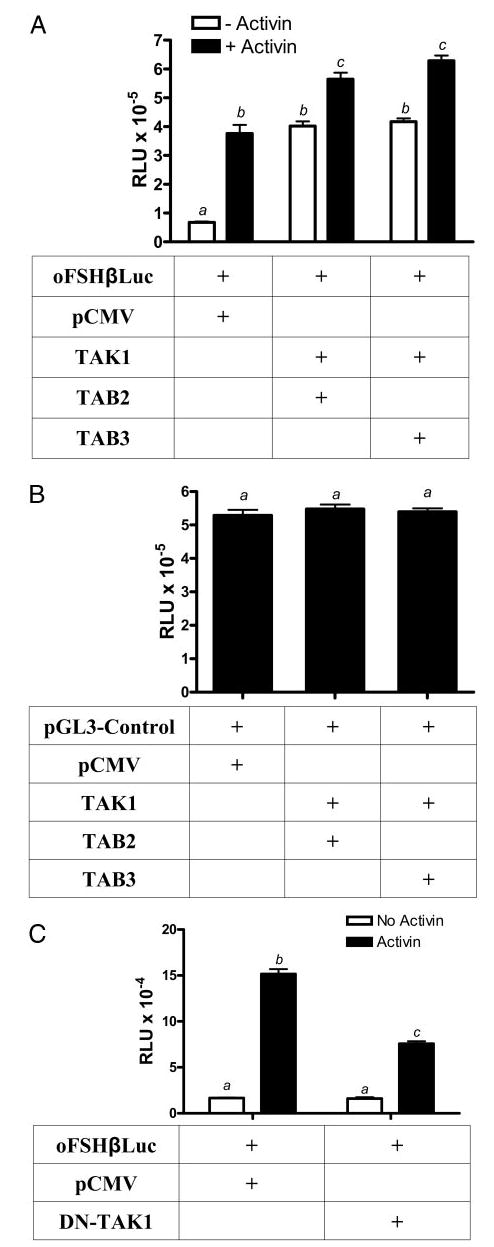
TAB2 or TAB3 partner with TAK1 to induce expression of oFSHβLuc like activin. LβT2 cells were prepared and plated as in Fig. 1. A, Cells were transiently cotransfected with 50 ng oFSHβLuc and 150 ng pCMV as mock DNA, or oFSHβLuc and 25 ng TAK1, and 125 ng of either TAB2 or TAB3. Twenty-four hours after transfection, cells were treated with or without activin (100 ng/ml), for an additional 24 h before analysis. Means with different letters are significantly different (P < 0.05). B, Cells were transiently cotransfected with 50 ng pGL3-control and either 150 ng mock plasmid pCMV or 25 ng TAK1 plus 125 ng TAB2 or TAB3. After transfection (24 h), cells were assayed for luciferase activity. The results of these treatments were not significantly different (P > 0.05). C, Cells were transiently cotransfected with 50 ng oFSHβLuc and 50 ng of either mock plasmid pCMV or expression vector encoding the TAK1 DN (DN-TAK1-KN). After 24 h of transfection, cells were treated with or without activin (100 ng/ml) for an additional 24 h. Means with different letters are significantly different (P < 0.01).
Transfections with TAK1/TAB2 or TAK1/TAB3 were tested with the pGL3Luc control expression vector, and the data in Fig. 4B show that its expression was not altered. Thus, overexpression of TAK1 in the presence of TAB2 or TAB3 was specific for inducing activin-responsive genes only.
To determine whether endogenous TAK1 was important for activin action, LβT2 cells were cotransfected with oFSHβLuc and a DN TAK1 mutant (DN-TAK1), which lacks the active site required for its kinase activity (32). This DN molecule (DN-TAK1) did suppress activin induction of oFSHβLuc by 50% (Fig. 4C). This finding was consistent with the concept that endogenous TAK1 is, at least partly, responsible for activin-mediated induction of oFSHβ.
Inhibiting activin induction of oFSHβLuc with a TAK1 inhibitor (5Z-7-Oxozeanol)
To further characterize the role of TAK1 in activin-mediated induction of oFSHβLuc, LβT2 cells were transfected with oFSHβLuc and then treated with increasing concentrations of 5Z-7-Oxozeanol, a newly discovered inhibitor of TAK1 that works by blocking the ATP binding site required for TAK1 kinase activity (55). Cells were treated with activin (100 ng/ml) and with or without 5Z-7-Oxozeanol for 24 h. Activin alone induced oFSHβLuc expression 11-fold, and addition of 5Z-7-Oxozeanol inhibited 100% of this induction in a dose-dependent manner, with an ED50 of 1.2 μM (Fig. 5A). There was no effect of 5Z-7-Oxozeanol on basal expression of oFSHβLuc (Fig. 5A).
Fig. 5.
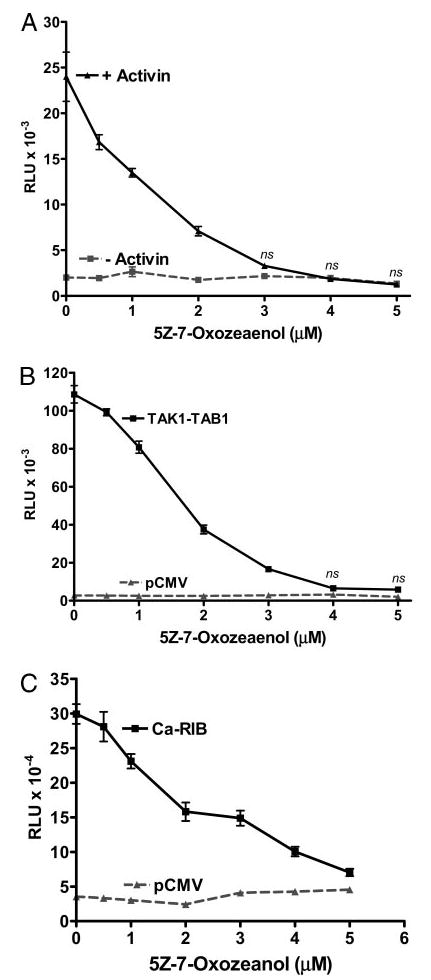
Inhibition of TAK1 with 5Z-7-Oxozeanol fully blocked activin induction of oFSHβLuc expression. LβT2 cells were prepared and plated as in Fig. 1. A, Cells were transiently transfected with 50 ng oFSHβLuc promoter construct for 24 h. Cells were then treated with or without 5Z-7-Oxozeanol at the indicated concentrations for 2 h before treatment with (▴) or without activin (▪) at 100 ng. Cells were incubated another 24 h before analysis for luciferase activity. Because 5Z-7-Oxozeanol is reported to be somewhat labile, halfway through the 24-h incubation, fresh media was added containing the same concentrations of 5Z-7-Oxozeanol and activin. Treatment with 5Z-7-Oxozeanol significantly inhibited induction of oFSHβLuc at all concentrations (P < 0.05) and completely inhibited it between 3–5 μM (ns, not significantly different from unstimulated control cultures). B, Cells were cotransfected with 50 ng oFSHβLuc and 50 ng pCMV mock DNA (▴) or 50 ng oFSHβLuc, 25 ng TAK1, and 25 ng TAB1 expression constructs (▪). After transfection (24 h), cells were treated with increasing concentrations of 5Z-7-Oxozeanol as in A above. Treatment with 5Z-7-Oxozeanol significantly inhibited induction of oFSHβLuc at all concentrations (P < 0.05) and completely inhibited it at 4 μM and above (ns). C, Cells were transiently cotransfected with 50 ng oFSHβLuc and 50 ng of either mock plasmid pCMV (▴) or expression vector encoding the Ca-ActRIB (▪). After transfection (24 h), cells were treated with increasing concentrations of 5Z-7-Oxozeanol as in A above. Treatment with 5Z-7-Oxozeanol significantly inhibited induction of oFSHβLuc at all concentrations (P < 0.05).
The data in Fig. 5B are similar to those in Fig. 5A, but cotransfection of TAK1/TAB1, which constitutively activates TAK1, was used to induce oFSHβLuc instead of activin (Fig. 4A). The potency of 5Z-7-Oxozeanol was tested on LβT2 cells cotransfected with TAK1/TAB1 to compare the ED50 values obtained for inhibition of endogenous TAK1. Expression of oFSHβLuc was increased 35-fold by TAK1/TAB1, and 5Z-7-Oxozeanol inhibited this induction by 99% in a dose-dependent manner, with an ED50 of 1.5 μM. As in Fig. 5A, basal expression of oFSHβLuc was unaffected by the chemical inhibitor. These results are consistent with the concept that 5Z-7-Oxozeanol strongly inhibited the extra TAK1 produced by transfecting LβT2 cells with a TAK1 expression construct.
The effects of TAK1 inhibitor 5Z-7-Oxozeanol were tested on the activin signaling pathway using Ca-ActRIB (T206D) to induce oFSHβLuc as in Fig. 2C. Cotransfection of Ca-ActRIB (T206D) stimulated oFSHβLuc by 8.4-fold (Fig. 5C), and addition of 5Z-7-Oxozeanol inhibited this expression by 95%, with an ED50 of 1.6 μM.
Specificity of 5Z-7-Oxozeanol inhibition
To confirm that 5Z-7-Oxozeanol was a specific inhibitor of oFSHβLuc, the effects of 5Z-7-Oxozeanol were tested on endogenous FSHβ and the two other closely related gonadotropin subunit genes expressed in LβT2 cells. LβT2 cells were treated with activin, with or without 5Z-7-Oxozeanol, and total RNA was isolated and analyzed using real-time RT-PCR for mouse FSHβ-mRNA (Fig. 6A). In addition, analyses were performed for αGSU subunit mRNA (Fig. 6B) and LHβ mRNA (Fig. 6C). The data show that 24 h of activin treatment increased mRNA for mouse FSHβ by 922-fold and that 5Z-7-Oxozeanol inhibited this induction by more than 99%. Although activin is often reported to be neutral toward expression of the αGSU, activin decreased mRNA for αGSU by 92% in this series of experiments, and 5Z-7-Oxozeanol reversed most of this inhibition. Likewise, mRNA for LHβ was slightly inhibited by activin (30% inhibition), and 5Z-7-Oxozeanol reversed this inhibition, to create an actual increase in LHβ mRNA of 60%. These data indicated that 5Z-7-Oxozeanol reversed the effects of activin on the expression of every glycoprotein hormone subunit produced in LβT2 cells. Because gene expression of αGSU and LHβ were stimulated, it was clear that 5Z-7-Oxozeanol did not inhibit protein synthesis or cellular function in general.
Fig. 6.
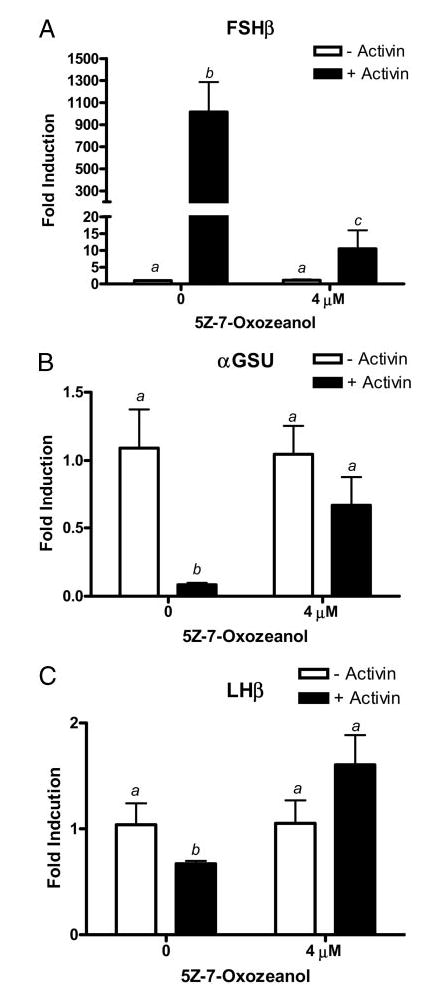
LβT2 cells were plated in 6-well plates at 1 million cells per well. The cells were treated with or without activin (100 ng/ml) and treated with or without 4 μM 5Z-7-Oxozeanol. Cells were incubated for 24 h and then washed with PBS, and total RNA was extracted using Tri-Zol reagent. Threshold cycle (Ct) values for (A) FSHβ, (B) αGSU, and (C) LHβ were normalized by subtracting their respective 18s rRNA Ct values. Neither activin nor 5Z-7-Oxozeanol altered 18s rRNA levels because Ct values for 18s rRNA did not change significantly (P > 0.05). Normalized Ct values were averaged and used to compare FSHβ, αGSU, and LHβ in the absence or presence of activin using the 2−(ΔΔCt) method for quantitation. The data are plotted as fold-induction of mRNA levels above basal expression. Means with different letters are significantly different (P < 0.05).
TAK1 is present in LβT2 cells and is phosphorylated 2–24 h after activin treatment
To establish the existence of TAK1 in LβT2 cells and its time-course of phosphorylation by activin, LβT2 cells were pretreated with follistatin for 16 h to bioneutralize any endogenously made activin and then treated with activin for 2–24 h. Cell extracts were prepared and analyzed by Western blot techniques to visualize TAK1, phophorylated TAK1, and TAB1 simultaneously. The data in Fig. 7 show that LβT2 cells expressed both TAK1 and TAB1 abundantly. TAK1 migrated with a molecular mass of 70 kDa, and TAB1 migrated with a molecular mass of 55 kDa, as expected (33). The immuno-blot revealed that activin phosphorylated TAK1 between 1–2 h (Fig. 7). Activation of TAK1 by activin was greatest at 4, 6, and 8 h but was maintained at higher-than-control levels up to 24 h. There was no change in overall expression of TAK1 or TAB1 during activin treatment.
p38 MAPK participates in activin-mediated induction of oFSHβLuc
TAK1 is a member of the MAPKKK family that can phosphorylate and activate members of the MAPK family, including the ERK, JNK, and p38 MAPKs. To determine whether MAPKs are involved in activin-mediated induction of oFSHβ, cultures of LβT2 cells were treated with or without specific chemical inhibitors of ERK, JNK, and p38 MAPKs with or without activin to determine whether the inhibitors would specifically block activin induction of oFSHβLuc expression. A chemical inhibitor of JNK (JNK II inhibitor) had no effect on induction of oFSHβLuc by activin (Fig. 8A). An ERK-specific chemical inhibitor (PD98059) did not block activin induction of oFSHβLuc, but it did increase basal expression, suggesting that ERK plays an inhibitory role in basal expression of the oFSHβ-gene (Fig. 8B). Finally, a well-known inhibitor of p38 (SB203580) inhibited activin action by 60% (Fig. 8C). These results suggest that p38-MAPK is downstream of the TAK1 pathway and that activation of p38-MAPK is important for activin induction of oFSHβLuc.
Fig. 8.
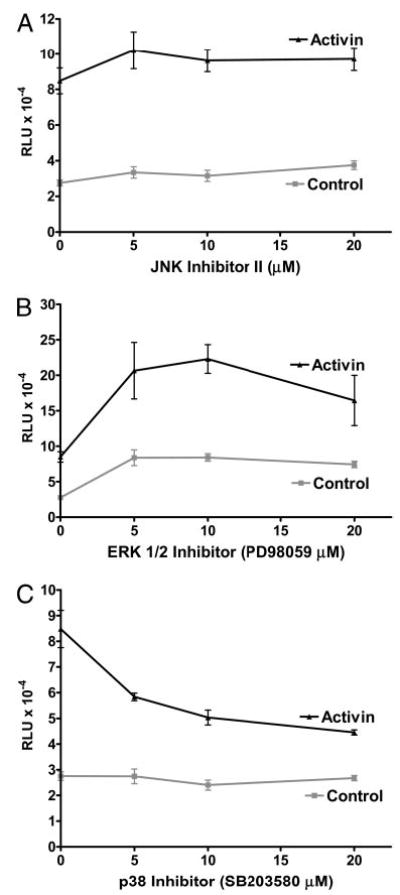
Inhibition of p38-MAPK partially blocks activin induction of oFSHβ. LβT2 cells were prepared and plated as in Fig. 1A and transiently transfected with 50 ng oFSHβLuc. A, After transfection (24 h), cells were treated with or without activin (100 ng/ml) and with JNK inhibitor II (0, 5, 10, and 20 μM); B, the same as A but using the ERK inhibitor; C, the same as A but using the p38 MAPK inhibitor (0, 5, 10, and 20 μM). Cultures were treated for 24 h with or without inhibitors and then analyzed for luciferase activity. Only SB203580 significantly decreased the ratio of induction/basal expression (P < 0.05).
Discussion
It is well known that activin can increase the synthesis (and associated secretion) of FSH by inducing transcription of its β-subunit (FSHβ) (11, 58). It is also known that activin classically transmits its signal intracellularly by phosphorylating Smad2 or Smad3, and it was recently reported that expression of extra Smad3 (but not Smad2) increases basal and activin-induced expression of rat (30) and ovine FSHβ-subunits in LβT2 cells (29). Initial studies in our laboratory focused on the role of Smad3 as a potential mediator activated by activin; however, the data showed that activin is able to stimulate FSHβ expression independent of Smad3 activation. The studies reported here identified that a TAK1 pathway is critical for mediating activin induction of FSHβ expression in gonadotropes.
Transfection of Smad3 is known to increase basal expression of Smad-dependent genes to levels comparable with induction by activin (59). Preliminary studies showed that Smad3 overexpression stimulated both basal and activin-induced expression of the FSHβ-gene. However, overexpression of Smad3 did not increase the ratio of induction/basal expression of oFSHβLuc over a wide range of Smad3 concentrations, including those that were maximally effective. One interpretation is that overexpression of Smad3 simply stimulates basal expression of oFSHβLuc. If this is all that occurs, the increased induction by activin observed in the presence of extra Smad3 might occur simply because transcription started at a higher level. This explanation does not imply that activin works through the activation of Smad3. Therefore, the data in Fig. 1 fail to prove that Smad3 participates in activin-mediated induction of the FSHβ-subunit.
In this report, it is shown, for the first time, that inhibiting activation of endogenous Smad3 does not affect activin’s ability to induce expression of the FSHβ-subunit. Two well-characterized Smad3 mutants [Smad3 (3SA) and Smad3 (D407E)] were used to block possible activation of endogenous Smad3 by activin. Smad3 (3SA) has three serines required for activation mutated to three alanines (60). Smad3 (D407E) has an aspartate mutated to glutamate, which allows it to bind tightly to the activin type I receptor’s (ActRIB) active site, therefore blocking the activation of Smad2 and Smad3 signaling pathways (53). The results show that neither of these inhibitors blocked activin-induced expression of oFSHβLuc (Fig. 2, B–D). Furthermore, both DN inhibitors were shown to be fully effective in LβT2 cells, because they blocked the ability of activin to induce p3TPLuc, which is known to depend on Smad3 activation in other cell types (59). These data can only be interpreted to mean that activin does not require the activation of endogenous Smad3 to induce FSHβ-gene. Furthermore, because DN-Smad3 (D407E) blocks activation of Smad2 along with Smad3 in other cell systems (53), its failure to block activin-mediated induction of oFSHβLuc strongly suggests that activation of neither Smad2 nor Smad3 is required for activin-mediated induction of FSHβ-subunit expression.
The results were surprising because both rat and ovine FSHβ-promoters contain consensus Smad binding sites (29, 30) at positions that are important for activin-mediated induction, and data show Smad4 binding to several of these positions. However, it should be noted that previous publications using LβT2 cells have shown Smad4 binding is observed under basal conditions and is not enhanced due to activin treatment (61). The role of Smads in binding to FSHβ-promoter elements is still unclear. It might be possible for Smads to play a role at the basal transcription complex independent of activin, where Smad3 or Smad4 are continuously moving into the nucleus to bind SBE(s) that drives basal transcription as shown in other systems (61, 62). This remains an unsolved question of activin action.
Another important finding is the different activation kinetics observed between oFSHβLuc and Smad-dependent construct (p3TPLuc) in response to activin treatment. In this report, the time-course of activin induction of Smad-dependent p3TPLuc showed very different kinetics compared with induction of oFSHβLuc (Fig. 3). Induction of p3TPLuc was relatively rapid (2–8 h), whereas induction of oFSHβLuc was observed only after 8 h of treatment. Essentially all previous reports using FSHβLuc in LβT2 cells measured activin induction at 24 h (51), which matched the optimal time observed in the studies reported here. Because Smad2 and Smad3 are normally activated within 1–4 h (49), the data in Fig. 4 provide further evidence that neither Smad2 nor Smad3 is the immediate trigger for activin induction of oFSHβLuc. Based on kinetics, however, Smad3 could be the immediate trigger for induction of p3TPLuc. These data even suggest the possibility that oFSHβLuc induction depends on one or more late genes that might be under control of one or more early response genes affected by activin, but the early genes, themselves, should not depend on Smad3 for this action because DN-Smad3 inhibitors do not block activin induction of oFSHβLuc. Multigene analysis could be necessary to study this possibility further.
The evidence showing that endogenous Smad3 (or Smad2) is not the trigger for activin-mediated induction of oFSHβLuc led to experiments that explored a Smad-independent pathway involving TAK1 (63, 64). Activin, TGFβ, and BMP were shown in multiple studies to signal through activation of TAK1 (MAPKKK), initiating a MAPK signaling cascade; therefore the initial results directed the focus of the study to the possible role of TAK1 in mediating activin stimulation of FSHβ-gene. Initially, a series of experiments with TAK1 and TAB1 showed that their dual overexpression cause robust stimulation of the oFSHβLuc, as previously shown in other systems of TAK1 (55). However, overexpression of TAK1 along with TAB1 results in constitutive kinase activation of TAK1, resulting in activating other genes including the control constructs. TAB2 and TAB3 were also identified as TAK1 binding partners, but they differ from TAK1 by their ability to bind to upstream regulatory elements of the signaling pathway. It has been recently shown that TAK1 binds TAB1 and can either partner with TAB2, forming a TAK1/TAB1/TAB2 complex, or partner with TAB3 to form a TAK1/TAB1/TAB3 complex. Therefore, experiments with TAK1 and TAB2 and TAB3 showed that overexpression of TAK1 with either TAB2 or TAB3 specifically induced oFSHβLuc. Furthermore, this induction substituted almost entirely for activin, which is consistent with TAK1 being an essential member of the activin signaling pathway. In addition, it was found that a DN inhibitor of endogenous TAK1 blocked activin-mediated induction of oFSHβLuc by 50%. This, too, was consistent with a physiological role for TAK1 in oFSHβLuc expression.
Supplementary studies with a newly available TAK1 inhibitor, 5Z-7-Oxozeanol, established that TAK1 was required for activin-mediated expression of oFSHβ. 5Z-7-Oxozeanol is a resorcylic acid lactone that is currently being investigated for its potential as a therapeutic agent for allergic cutaneous disorders. It has been shown to specifically inhibit the catalytic activity of TAK1 without affecting any of the other MAPKKK family members (55). The use of 5Z-7-Oxozeanol to inhibit endogenous TAK1 activation in LβT2 cells showed that: first, the TAK1 inhibitor was shown to fully block activin induction of oFSHβLuc in a dose-dependent manner (Fig. 5); then, it was shown that 5Z-7-Oxozeanol blocked induction of endogenous mouse FSHβ-mRNA in LβT2 cells by 92%, and specificity was established by showing that 5Z-7-Oxozeanol did not decrease mRNAs for endogenous LHβ and αGSU (Fig. 6). In fact, the TAK1 inhibitor actually increased levels of these latter mRNAs by reversing the effects of activin on these gonadotropin subunit mRNAs. Although regulation of FSHβ in LβT2 cells by activin is well established (51, 52), surprisingly activin down-regulated αGSU expression. It might be that activin regulates αGSU differently in primary and transformed LβT2 gonadotropes, or it could be that proper regulation of αGSU subunit requires paracrine factors provided by other pituitary cell types. Finally, 5Z-7-Oxozeanol fully inhibited oFSHβLuc expression in LβT2 cells cotransfected with the constitutively active activin receptor (Fig. 5B). All of these data showed that TAK1 was necessary for activin-mediated induction of the FSHβ-subunit in both the sheep and mouse in transformed LβT2 gonadotropes.
Because oFSHβLuc was induced relatively slowly by activin (after 8 h of treatment), the time-course for TAK1 phosphorylation was investigated and found to be compatible with induction of both p3TPLuc and oFSHβLuc. That is, TAK1 was abundant in LβT2 cells, and its phosphorylation was strongest from 2–8 h but continued to be elevated through 24 h. TAB1 was abundant in LβT2 cells and, presumably, TAB2 and/or TAB3 were also. Further study is needed on TAK1 activation and its relationships to its binding proteins to determine how it brings specificity of action to activin-mediated events.
Finally, TAK1 is reported to activate p38 MAPK in other signaling pathways (64). Therefore, inhibitors of ERK, JNK, and p38 MAPKs were used to determine whether any of the major MAPKs were involved in activin-mediated induction of oFSHβLuc. As expected, the inhibitor of p38 MAPK linked it with activin-mediated induction, but none of the other MAPKs were associated with activin action. These data do not prove that TAK1 is responsible for p38 activation, but the results are consistent with TAK1 activation of p38, which then activates transcription factors that directly or indirectly induce expression of the FSHβ-subunit.
In summary, it has been shown that activin stimulates induction of FSHβ through a Smad-independent pathway initiated by the activation of TAK1. Furthermore, p38 MAPK, known to be a downstream target of TAK1, was shown to be involved in activin induction of oFSHβ-gene.
Acknowledgments
We thank Xiao-Fan Wang (Duke University, Durham, NC) for discussions and information about Smad signaling, Joan Massague (Howard Hughes Medical Institute, New York, NY) for p3TPLuc constructs, Mitsuyasu Kato for DN-Smad constructs, and Theresa Guise for DN-Smad constructs. Special thanks to Chugai Pharmaceutical Co., Ltd. for providing a sample of TAK1 chemical inhibitor. Finally, we thank Dr. Pamela Mellon (University of California, San Diego, CA) for providing us the LβT2 cells.
Footnotes
This work was supported by the North Carolina State University Agricultural Research Service and National Institutes of Health Grant R01 HD-045429.
References
- 1.Kumar TR, Wang Y, Lu N, Matzuk MM. Follicle stimulating hormone is required for ovarian follicle maturation but not male fertility. Nat Genet. 1997;15:201–204. doi: 10.1038/ng0297-201. [DOI] [PubMed] [Google Scholar]
- 2.Besecke LM, Guendner MJ, Schneyer AL, Bauer-Dantoin AC, Jameson JL, Weiss J. Gonadotropin-releasing hormone regulates follicle-stimulating hormone-β gene expression through an activin/follistatin autocrine or para-crine loop. Endocrinology. 1996;137:3667–3673. doi: 10.1210/endo.137.9.8756531. [DOI] [PubMed] [Google Scholar]
- 3.Dalkin AC, Haisenleder DJ, Ortolano GA, Ellis TR, Marshall JC. The frequency of gonadotropin-releasing-hormone stimulation differentially regulates gonadotropin subunit messenger ribonucleic acid expression. Endocrinology. 1989;125:917–924. doi: 10.1210/endo-125-2-917. [DOI] [PubMed] [Google Scholar]
- 4.Haisenleder DJ, Ortolano GA, Dalkin AC, Ellis TR, Paul SJ, Marshall JC. Differential regulation of gonadotropin subunit gene expression by gonadotropin-releasing hormone pulse amplitude in female rats. Endocrinology. 1990;127:2869–2875. doi: 10.1210/endo-127-6-2869. [DOI] [PubMed] [Google Scholar]
- 5.Huang HJ, Wu JC, Su P, Zhirnov O, Miller WL. A novel role for bone morphogenetic proteins in the synthesis of follicle-stimulating hormone. Endocrinology. 2001;142:2275–2283. doi: 10.1210/endo.142.6.8159. [DOI] [PubMed] [Google Scholar]
- 6.Krummen LA, Woodruff TK, DeGuzman G, Cox ET, Baly DL, Mann E, Garg S, Wong WL, Cossum P, Mather JP. Identification and characterization of binding proteins for inhibin and activin in human serum and follicular fluids. Endocrinology. 1993;132:431–443. doi: 10.1210/endo.132.1.7678220. [DOI] [PubMed] [Google Scholar]
- 7.Ling N, Ying SY, Ueno N, Esch F, Denoroy L, Guillemin R. Isolation and partial characterization of a Mr 32,000 protein with inhibin activity from porcine follicular fluid. Proc Natl Acad Sci USA. 1985;82:7217–7221. doi: 10.1073/pnas.82.21.7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otsuka F, Shimasaki S. A novel function of bone morphogenetic protein-15 in the pituitary: selective synthesis and secretion of FSH by gonadotropes. Endocrinology. 2002;143:4938–4941. doi: 10.1210/en.2002-220929. [DOI] [PubMed] [Google Scholar]
- 9.Rivier J, Spiess J, McClintock R, Vaughan J, Vale W. Purification and partial characterization of inhibin from porcine follicular fluid. Biochem Biophys Res Commun. 1985;133:120–127. doi: 10.1016/0006-291x(85)91849-2. [DOI] [PubMed] [Google Scholar]
- 10.Vale W, Rivier J, Vaughan J, McClintock R, Corrigan A, Woo W, Karr D, Spiess J. Purification and characterization of an FSH releasing protein from porcine ovarian follicular fluid. Nature. 1986;321:776–779. doi: 10.1038/321776a0. [DOI] [PubMed] [Google Scholar]
- 11.Carroll RS, Corrigan AZ, Gharib SD, Vale W, Chin WW. Inhibin, activin, and follistatin: regulation of follicle-stimulating hormone messenger ribonucleic acid levels. Mol Endocrinol. 1989;3:1969–1976. doi: 10.1210/mend-3-12-1969. [DOI] [PubMed] [Google Scholar]
- 12.Norwitz ER, Xu S, Jeong KH, Bedecarrats GY, Winebrenner LD, Chin WW, Kaiser UB. Activin A augments GnRH-mediated transcriptional activation of the mouse GnRH receptor gene. Endocrinology. 2002;143:985–997. doi: 10.1210/endo.143.3.8663. [DOI] [PubMed] [Google Scholar]
- 13.Norwitz ER, Xu S, Xu J, Spiryda LB, Park JS, Jeong KH, McGee EA, Kaiser UB. Direct binding of AP-1 (Fos/Jun) proteins to a SMAD binding element facilitates both gonadotropin-releasing hormone (GnRH)- and activin-mediated transcriptional activation of the mouse GnRH receptor gene. J Biol Chem. 2002;277:37469–37478. doi: 10.1074/jbc.M206571200. [DOI] [PubMed] [Google Scholar]
- 14.Attisano L, Wrana JL, Cheifetz S, Massague J. Novel activin receptors: distinct genes and alternative mRNA splicing generate a repertoire of serine/threonine kinase receptors. Cell. 1992;68:97–108. doi: 10.1016/0092-8674(92)90209-u. [DOI] [PubMed] [Google Scholar]
- 15.Mathews LS, Vale WW. Expression cloning of an activin receptor, a predicted transmembrane serine kinase. Cell. 1991;65:973–982. doi: 10.1016/0092-8674(91)90549-e. [DOI] [PubMed] [Google Scholar]
- 16.ten Dijke P, Ichijo H, Franzen P, Schulz P, Saras J, Toyoshima H, Heldin CH, Miyazono K. Activin receptor-like kinases: a novel subclass of cell-surface receptors with predicted serine/threonine kinase activity. Oncogene. 1993;8:2879–2887. [PubMed] [Google Scholar]
- 17.Attisano L, Carcamo J, Ventura F, Weis FM, Massague J, Wrana JL. Identification of human activin and TGFβ type I receptors that form heteromeric kinase complexes with type II receptors. Cell. 1993;75:671–680. doi: 10.1016/0092-8674(93)90488-c. [DOI] [PubMed] [Google Scholar]
- 18.Attisano L, Wrana JL, Montalvo E, Massague J. Activation of signalling by the activin receptor complex. Mol Cell Biol. 1996;16:1066–1073. doi: 10.1128/mcb.16.3.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carcamo J, Weis FM, Ventura F, Wieser R, Wrana JL, Attisano L, Massague J. Type I receptors specify growth-inhibitory and transcriptional responses to transforming growth factor β and activin. Mol Cell Biol. 1994;14:3810–3821. doi: 10.1128/mcb.14.6.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathews LS, Vale WW, Kintner CR. Cloning of a second type of activin receptor and functional characterization in Xenopus embryos. Science. 1992;255:1702–1705. doi: 10.1126/science.1313188. [DOI] [PubMed] [Google Scholar]
- 21.Chen X, Rubock MJ, Whitman M. A transcriptional partner for MAD proteins in TGF-β signalling. Nature. 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- 22.Hua X, Liu X, Ansari DO, Lodish HF. Synergistic cooperation of TFE3 and smad proteins in TGF-β-induced transcription of the plasminogen activator inhibitor-1 gene. Genes Dev. 1998;12:3084–3095. doi: 10.1101/gad.12.19.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labbe E, Silvestri C, Hoodless PA, Wrana JL, Attisano L. Smad2 and Smad3 positively and negatively regulate TGFβ-dependent transcription through the forkhead DNA-binding protein FAST2. Mol Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- 24.Massague J. TGF-β signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 25.Pouponnot C, Jayaraman L, Massague J. Physical and functional interaction of SMADs and p300/CBP. J Biol Chem. 1998;273:22865–22868. doi: 10.1074/jbc.273.36.22865. [DOI] [PubMed] [Google Scholar]
- 26.Shi Y, Wang YF, Jayaraman L, Yang H, Massague J, Pavletich NP. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-β signaling. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- 27.Wotton D, Lo RS, Lee S, Massague J. A Smad transcriptional corepressor. Cell. 1999;97:29–39. doi: 10.1016/s0092-8674(00)80712-6. [DOI] [PubMed] [Google Scholar]
- 28.Zhou S, Zawel L, Lengauer C, Kinzler KW, Vogelstein B. Characterization of human FAST-1, a TGFβ and activin signal transducer. Mol Cell. 1998;2:121–127. doi: 10.1016/s1097-2765(00)80120-3. [DOI] [PubMed] [Google Scholar]
- 29.Bailey JS, Rave-Harel N, McGillivray SM, Coss D, Mellon PL. Activin regulation of the follicle-stimulating hormone β-subunit gene involves Smads and the TALE homeodomain proteins Pbx1 and Prep1. Mol Endocrinol. 2004;18:1158–1170. doi: 10.1210/me.2003-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suszko MI, Lo DJ, Suh H, Camper SA, Woodruff TK. Regulation of the rat follicle-stimulating hormone β-subunit promoter by activin. Mol Endocrinol. 2003;17:318–332. doi: 10.1210/me.2002-0081. [DOI] [PubMed] [Google Scholar]
- 31.Gregory SJ, Lacza CT, Detz AA, Xu S, Petrillo LA, Kaiser UB. Synergy between activin A and gonadotropin-releasing hormone in transcriptional activation of the rat follicle-stimulating hormone-β gene. Mol Endocrinol. 2005;19:237–254. doi: 10.1210/me.2003-0473. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi K, Shirakabe K, Shibuya H, Irie K, Oishi I, Ueno N, Taniguchi T, Nishida E, Matsumoto K. Identification of a member of the MAPKKK family as a potential mediator of TGF-β signal transduction. Science. 1995;270:2008–2011. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]
- 33.Shibuya H, Yamaguchi K, Shirakabe K, Tonegawa A, Gotoh Y, Ueno N, Irie K, Nishida E, Matsumoto K. TAB1: an activator of the TAK1 MAPKKK in TGF-β signal transduction. Science. 1996;272:1179–1182. doi: 10.1126/science.272.5265.1179. [DOI] [PubMed] [Google Scholar]
- 34.Cheung PC, Nebreda AR, Cohen P. TAB3, a new binding partner of the protein kinase TAK1. Biochem J. 2004;378:27–34. doi: 10.1042/BJ20031794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin G, Klika A, Callahan M, Faga B, Danzig J, Jiang Z, Li X, Stark GR, Harrington J, Sherf B. Identification of a human NF-κB-activating protein, TAB3. Proc Natl Acad Sci USA. 2004;101:2028–2033. doi: 10.1073/pnas.0307314101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, Chiu YH, Deng L, Chen ZJ. TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 37.Ishitani T, Takaesu G, Ninomiya-Tsuji J, Shibuya H, Gaynor RB, Matsumoto K. Role of the TAB2-related protein TAB3 in IL-1 and TNF signaling. EMBO J. 2003;22:6277–6288. doi: 10.1093/emboj/cdg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munoz-Sanjuan I, Bell E, Altmann CR, Vonica A, Brivanlou AH. Gene profiling during neural induction in Xenopus laevis: regulation of BMP signaling by post-transcriptional mechanisms and TAB3, a novel TAK1-binding protein. Development. 2002;129:5529–5540. doi: 10.1242/dev.00097. [DOI] [PubMed] [Google Scholar]
- 39.Qiao B, Padilla SR, Benya PD. TGF-β activated kinase 1 (TAK1) mimics and mediates TGF-β-induced stimulation of type II collagen synthesis in chondrocytes independent of Col2a1 transcription and Smad3 signaling. J Biol Chem. 2005 doi: 10.1074/jbc.M500646200. [DOI] [PubMed] [Google Scholar]
- 40.Hanafusa H, Ninomiya-Tsuji J, Masuyama N, Nishita M, Fujisawa J, Shibuya H, Matsumoto K, Nishida E. Involvement of the p38 mitogen-activated protein kinase pathway in transforming growth factor-β-induced gene expression. J Biol Chem. 1999;274:27161–27167. doi: 10.1074/jbc.274.38.27161. [DOI] [PubMed] [Google Scholar]
- 41.Sano Y, Harada J, Tashiro S, Gotoh-Mandeville R, Maekawa T, Ishii S. ATF-2 is a common nuclear target of Smad and TAK1 pathways in transforming growth factor-β signaling. J Biol Chem. 1999;274:8949–8957. doi: 10.1074/jbc.274.13.8949. [DOI] [PubMed] [Google Scholar]
- 42.Schnabl B, Bradham CA, Bennett BL, Manning AM, Stefanovic B, Brenner DA. TAK1/JNK and p38 have opposite effects on rat hepatic stellate cells. Hepatology. 2001;34:953–963. doi: 10.1053/jhep.2001.28790. [DOI] [PubMed] [Google Scholar]
- 43.Akiyama S, Yonezawa T, Kudo TA, Li MG, Wang H, Ito M, Yoshioka K, Ninomiya-Tsuji J, Matsumoto K, Kanamaru R, Tamura S, Kobayashi T. Activation mechanism of c-Jun amino-terminal kinase in the course of neural differentiation of P19 embryonic carcinoma cells. J Biol Chem. 2004;279:36616 –36620. doi: 10.1074/jbc.M406610200. [DOI] [PubMed] [Google Scholar]
- 44.Goswami M, Uzgare AR, Sater AK. Regulation of MAP kinase by the BMP-4/TAK1 pathway in Xenopus ectoderm. Dev Biol. 2001;236:259–270. doi: 10.1006/dbio.2001.0338. [DOI] [PubMed] [Google Scholar]
- 45.Dowdy SC, Mariani A, Janknecht R. HER2/Neu- and TAK1-mediated up-regulation of the transforming growth factor β inhibitor Smad7 via the ETS protein ER81. J Biol Chem. 2003;278:44377–44384. doi: 10.1074/jbc.M307202200. [DOI] [PubMed] [Google Scholar]
- 46.Sakurai H, Nishi A, Sato N, Mizukami J, Miyoshi H, Sugita T. TAK1-TAB1 fusion protein: a novel constitutively active mitogen-activated protein kinase kinase kinase that stimulates AP-1 and NF-κB signaling pathways. Biochem Biophys Res Commun. 2002;297:1277–1281. doi: 10.1016/s0006-291x(02)02379-3. [DOI] [PubMed] [Google Scholar]
- 47.Chen Y, Lebrun JJ, Vale W. Regulation of transforming growth factor β-and activin-induced transcription by mammalian Mad proteins. Proc Natl Acad Sci USA. 1996;93:12992–12997. doi: 10.1073/pnas.93.23.12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Datto MB, Frederick JP, Pan L, Borton AJ, Zhuang Y, Wang XF. Targeted disruption of Smad3 reveals an essential role in transforming growth factor β-mediated signal transduction. Mol Cell Biol. 1999;19:2495–2504. doi: 10.1128/mcb.19.4.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yingling JM, Datto MB, Wong C, Frederick JP, Liberati NT, Wang XF. Tumor suppressor Smad4 is a transforming growth factor β-inducible DNA binding protein. Mol Cell Biol. 1997;17:7019–7028. doi: 10.1128/mcb.17.12.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner M, Kleeff J, Lopez ME, Bockman I, Massaque J, Korc M. Transfection of the type I TGF-β receptor restores TGF-β responsiveness in pancreatic cancer. Int J Cancer. 1998;78:255–260. doi: 10.1002/(sici)1097-0215(19981005)78:2<255::aid-ijc21>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 51.Pernasetti F, Vasilyev VV, Rosenberg SB, Bailey JS, Huang HJ, Miller WL, Mellon PL. Cell-specific transcriptional regulation of follicle-stimulating hormone-β by activin and gonadotropin-releasing hormone in the LβT2 pituitary gonadotrope cell model. Endocrinology. 2001;142:2284–2295. doi: 10.1210/endo.142.6.8185. [DOI] [PubMed] [Google Scholar]
- 52.Huang HJ, Sebastian J, Strahl BD, Wu JC, Miller WL. The promoter for the ovine follicle-stimulating hormone-β gene (FSHβ) confers FSHβ-like expression on luciferase in transgenic mice: regulatory studies in vivo and in vitro. Endocrinology. 2001;142:2260–2266. doi: 10.1210/endo.142.6.8202. [DOI] [PubMed] [Google Scholar]
- 53.Goto D, Yagi K, Inoue H, Iwamoto I, Kawabata M, Miyazono K, Kato M. A single missense mutant of Smad3 inhibits activation of both Smad2 and Smad3, and has a dominant negative effect on TGF-β signals. FEBS Lett. 1998;430:201–204. doi: 10.1016/s0014-5793(98)00658-9. [DOI] [PubMed] [Google Scholar]
- 54.Kakonen SM, Selander KS, Chirgwin JM, Yin JJ, Burns S, Rankin WA, Grubbs BG, Dallas M, Cui Y, Guise TA. Transforming growth factor-β stimulates parathyroid hormone-related protein and osteolytic metastases via Smad and mitogen-activated protein kinase signaling pathways. J Biol Chem. 2002;277:24571–24578. doi: 10.1074/jbc.M202561200. [DOI] [PubMed] [Google Scholar]
- 55.Ninomiya-Tsuji J, Kajino T, Ono K, Ohtomo T, Matsumoto M, Shiina M, Mihara M, Tsuchiya M, Matsumoto K. A resorcylic acid lactone, 5Z-7-oxozeaenol, prevents inflammation by inhibiting the catalytic activity of TAK1 MAPK kinase kinase. J Biol Chem. 2003;278:18485–18490. doi: 10.1074/jbc.M207453200. [DOI] [PubMed] [Google Scholar]
- 56.Ono K, Ohtomo T, Ninomiya-Tsuji J, Tsuchiya M. A dominant negative TAK1 inhibits cellular fibrotic responses induced by TGF-β. Biochem Biophys Res Commun. 2003;307:332–337. doi: 10.1016/s0006-291x(03)01207-5. [DOI] [PubMed] [Google Scholar]
- 57.Wu JC, Su P, Safwat NW, Sebastian J, Miller WL. Rapid, efficient isolation of murine gonadotropes and their use in revealing control of follicle-stimulating hormone by paracrine pituitary factors. Endocrinology. 2004;145:5832–5839. doi: 10.1210/en.2004-0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weiss J, Guendner MJ, Halvorson LM, Jameson JL. Transcriptional activation of the follicle-stimulating hormone β-subunit gene by activin. En-docrinology. 1995;136:1885–1891. doi: 10.1210/endo.136.5.7720634. [DOI] [PubMed] [Google Scholar]
- 59.Wang EY, Ma EY, Woodruff TK. Activin signal transduction in the fetal rat adrenal gland and in human H295R cells. J Endocrinol. 2003;178:137–148. doi: 10.1677/joe.0.1780137. [DOI] [PubMed] [Google Scholar]
- 60.Liu X, Sun Y, Constantinescu SN, Karam E, Weinberg RA, Lodish HF. Transforming growth factor β-induced phosphorylation of Smad3 is required for growth inhibition and transcriptional induction in epithelial cells. Proc Natl Acad Sci USA. 1997;94:10669–10674. doi: 10.1073/pnas.94.20.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu C, Gaca MD, Swenson ES, Vellucci VF, Reiss M, Wells RG. Smads 2 and 3 are differentially activated by transforming growth factor-β (TGF-β) in quiescent and activated hepatic stellate cells. Constitutive nuclear localization of Smads in activated cells is TGF-β-independent. J Biol Chem. 2003;278:11721–11728. doi: 10.1074/jbc.M207728200. [DOI] [PubMed] [Google Scholar]
- 62.Inagaki Y, Mamura M, Kanamaru Y, Greenwel P, Nemoto T, Takehara K, Ten Dijke P, Nakao A. Constitutive phosphorylation and nuclear localization of Smad3 are correlated with increased collagen gene transcription in activated hepatic stellate cells. J Cell Physiol. 2001;187:117–123. doi: 10.1002/1097-4652(2001)9999:9999<00::AID-JCP1059>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 63.Zhang L, Deng M, Parthasarathy R, Wang L, Mongan M, Molkentin JD, Zheng Y, Xia Y. MEKK1 transduces activin signals in keratinocytes to induce actin stress fiber formation and migration. Mol Cell Biol. 2005;25:60 – 65. doi: 10.1128/MCB.25.1.60-65.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ogihara T, Watada H, Kanno R, Ikeda F, Nomiyama T, Tanaka Y, Nakao A, German MS, Kojima I, Kawamori R. p38 MAPK is involved in activin A- and hepatocyte growth factor-mediated expression of pro-endocrine gene neurogenin 3 in AR42J-B13 cells. J Biol Chem. 2003;278:21693–21700. doi: 10.1074/jbc.M302684200. [DOI] [PubMed] [Google Scholar]


