Abstract
Understanding the role of ‘epigenetic’ changes such as DNA methylation and chromatin remodeling has now become critical in understanding many biological processes. In order to delineate the global methylation pattern in a given genomic DNA, computer software has been developed to create a virtual image of restriction landmark genomic scanning (Vi-RLGS). When using a methylation- sensitive enzyme such as NotI as the restriction landmark, the comparison between real and in silico RLGS profiles of the genome provides a methylation map of genomic NotI sites. A methylation map of the Arabidopsis genome was created that could be confirmed by a methylation-sensitive PCR assay. The method has also been applied to the mouse genome. Although a complete methylation map has not been completed, a region of methylation difference between two tissues has been tested and confirmed by bisulfite sequencing. Vi-RLGS in conjunction with real RLGS will make it possible to develop a more complete map of genomic sites that are methylated or demethylated as a consequence of normal or abnormal development.
INTRODUCTION
Changes in genomic DNA methylation occur during the course of normal development, and the epigenetic regulation of gene expression by methylation is thought to be an important mechanism in the determination of cell fate during embryogenesis (1–4). In contrast, aberrant DNA methylation is a hallmark of cancer in higher organisms. Most of the 5′-methylcytosine in mammalian DNA, however, resides in repeat sequences that represent 37.5% of the mouse genome, such as the long terminal repeats (LTRs), which are specialized intragenomic parasites (5,6). Repetitive sequences and some oncogenes have been found to be hypomethylated whereas tumor suppressor genes have been found to be hypermethylated and silenced in cancer (7). Current research indicates that DNA methylation is a complex process and is mechanistically connected to other aspects of chromatin structure. Recent studies have identified several new protein partners for the DNA methyltransferases (Dnmtases) and indicate how they can be targeted to specific genomic regions to methylate DNA and silence gene expression. DNA methyl transferases have been shown to associate with CpG binding proteins, histone deacetylases, histone methyltransferases and SNF-2-like ATPases, all of which have roles in altering chromatin structure (1,7,8). All of these phenomena have been implicated as mechanisms that contribute to the normal and abnormal development of the phenotype in higher organisms. The detection and interpretation of these DNA methylation patterns is expected to lead to a more complete and clearer understanding of the complex epigenetic biology (1,2).
Many approaches have been developed for the detection of DNA methylation, which can be divided into two categories. One approach involves gene-by-gene analysis. The other approach uses global analyses such as methylation-sensitive representational difference analysis (MS-RDA), methylation-sensitive arbitary primed PCR, methylation- specific oligonucleotide microarray and restriction landmark genomic scanning (RLGS) (9–13). Although there are currently enormous efforts underway to screen for DNA methylation in the genome, complementary genome-wide global approaches to identify the target methylated sequences are urgently needed. It is also very likely that the gene-by-gene approaches will not be able to identify all biologically important methylation sites, since the function of ∼50% of the genes is unknown (14) and relatively low frequency methylation polymorphisms, or sporadic de novo methyl ation, may cause biological phenomena. RLGS is a method for the two-dimensional display of end-labeled DNA restriction fragments (15) and has been shown to be an unbiased method for DNA methylation scanning to identify changes in methylation status (16–18). The two-dimensional DNA electrophoresis pattern has been predicted from clone sequence information (19) and applied to prototype virtual image (Vi)-RLGSs using human genome sequence data (20,21). If the restriction landmark enzyme used is methylation sensitive, RLGS in conjunction with Vi-RLGS can be used to compile a map of genomic sites that are methylated (referred to as a ‘methylation map’). Here we present a broadly applicable Vi-RLGS software package that was used to develop a ‘methylation map’ in the Arabidopsis genome and extended to the analysis of a mammalian genome.
MATERIALS AND METHODS
RLGS reaction
RLGS was performed according to published protocols (15). Briefly, non-specific broken ends of 1–3 µg of genomic DNA were blocked in a 10 µl reaction by the addition of nucleotide analogs (αS-dGTP, αS-dCTP, ddATP, ddTTP) with 2 U DNA polymerase I (37°C, 20 min) followed by enzyme inactivation (65°C, 30 min). The pH of the buffer was adjusted and the DNA digested (37°C, 2 h) with 20 U NotI (Promega). Sequenase (version 2.0, U.S.B.) was used to fill in the NotI ends with [α-32P]dGTP and [α-32P]dCTP (Amersham) by incubating for 30 min at 37°C. The labeled DNA was digested (37°C, 1 h) with 20 U of a second restriction enzyme and a portion separated by electrophoresis through a 60 cm long, 0.8% agarose tube gel (first-dimension separation). The agarose tube gel was then equilibrated in the third digestion buffer, and the DNA was digested in the gel with the third enzyme at 37°C for 2 h. The agarose gel was placed horizontally (rotated 90° relative to the first direction of electrophoresis) across the top of a non-denaturing 5% polyacrylamide gel, connected to the polyacryamide gel with molten agarose and the DNA separated by electrophoresis in the second dimension. The gels were dried and exposed to X-ray film in the presence of intensifying screens (Quanta III, DuPont) for 2–10 days. Duplicate analysis was performed for each sample.
Virtual image RLGS (Vi-RLGS)
The software is capable of managing 1010 bp of sequence data in GenBank or FASTA format retrieved through any database, picking up the first line of necessary information such as accession number and chromosomal location, calculating fragment lengths for the first and second dimensions, visualizing the spot on the two-D pattern, and presenting the fragment sequence after clicking on the spot. To compute the mobility of each DNA fragment, we used Southern’s method (22), in which the reciprocal of mobility (M) plotted against fragment length (L) is linear (L = 1 / M). In addition, five known data points were obtained from electrophoresis (RLGS) of the 1 kb ladder (TaKaRa 3412A; 6 and 12 kb) in the first dimension, and the 100 bp ladder (TaKaRa 3407A; 100, 800 and 1500 bp) in the second dimension. We also confirmed the precision of the mobility of those spots in the agarose and polyacrylamide gels by comparing the relative mobility obtained from other spots. The mobility of a given sequence length in X and Y dimensions are therefore calculated by using the above-mentioned reference standards and equations. Mouse sequences are obtained from Celera and public databases. Arabidopsis sequences are from the Arabidopsis Information Resource (TAIR). Vi-RLGS is freely available for the academic user through the web site at http://ftp.cancergenetics.roswellpark.org/nagase. Repetitive sequences were screened by Repeat Masker developed by A. F. A. Smit and P. Green. It is available at http://ftp.genome.washington.edu/RM/RepeatMasker.html.
Correspondence between real and virtual RLGS loci
DNA was eluted from RLGS spots in 0.5 M ammonium acetate, pH 8, 1 mM EDTA (150 µl) (37°C overnight), precipitated with 2.5 vol. of cold ethanol with 1 µg of glycogen (–70°C overnight) and resuspended in 10 µl of 10 mM Tris–HCl, pH 8.0, 1 mM EDTA buffer. After optimization of PCR conditions using C57BL/6J genomic DNAs, we amplified 1 µl of each DNA by PCR (35 cycles) using primers derived from the sequences predicted by Vi-RLGS. The identity between real and virtual ‘spot’ sequences was confirmed by molecular weight (2% agarose gel) and by sequence analysis of the PCR product.
Bisulfite sequencing
Briefly, 2.5 µg of DNA from normal tissue was digested with 5 U EcoRV and then denatured in 0.3 M NaOH (20 min at 42°C). Freshly prepared sodium bisulfite (pH 5.0, 208 µl, 3.6 M) and hydroquinone (12 µl, 10 mM) was added to the denatured DNA in a final volume of 240 µl and then incubated for 18 h at 55°C. DNA was purified using the MARRLIGEN Matrix Gel Extraction System and eluted in 100 µl of sterilized water. NaOH was added to a final concentration of 0.3 M and incubated (15 min at 37°C). After neutralizing the samples with 3 M ammonium acetate (pH 7), the DNA was precipitated and resuspended in 10 µl of sterile water. Duplicate PCRs and direct sequencings were performed using 1 µl of each bisulfite-treated DNA.
Data deposition
Arabidopsis genome sequence was obtained from TAIR (http://www.arabidopsis.org/). The August 2002 Celera Database scaffold sequences were commercially available at http://www.celera.com. C57BL/6J mouse sequences were from Ensembl Trace Server (http://trace.ensembl.org/). The February 2003 freeze of the University of California, Santa Cruz version of the draft assembly of human genome was available at http://genome.ucsc.edu. Mouse RLGS spot mapping data were available at http://genome.gsc.riken.go.jp/RLGS/RLGShome.html. Repetitive sequences were found by Repeat Masker at http://ftp.genome.washington.edu/RM/RepeatMasker.html.
RESULTS
The Arabidopsis genome as a model genome to test Vi-RLGS
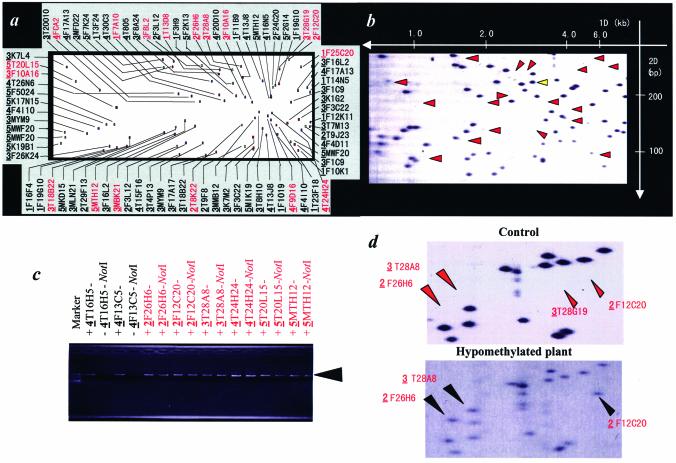
Arabidopsis thaliana is a small flowering plant that is widely used as a model organism in plant biology. The whole genome has been sequenced (23) and has a total size of 125 Mb, which is the size of an average-sized mammalian chromosome. We initially used the Arabidopsis genome sequence and NotI in combination with EcoRV and MboI to display a Vi-RLGS profile (see Supplementary Material, Fig. S1). An example of a Vi-RLGS pattern and the corresponding real RLGS pattern is shown in Figure 1. The sequence analysis of ‘spot DNA’ eluted from the RLGS gel demonstrated that the location of many spots was exactly as predicted using the Vi-RLGS algorithm (data not shown). There is, however, a difference in the number and the pattern of some spots between the real and virtual RLGS images. The red arrowheads in Figure 1b indicate spots that are present in the virtual RLGS profile but absent, or present as faint signals, in the real RLGS profile. To confirm the influence of 5′-methylcytosine on NotI landmark detection, methylation-sensitive PCR was performed on genomic DNA using primers specifying the region surrounding the NotI fragments indicated by arrows in Figure 1b. Figure 1c shows the PCR assay of eight loci. The loci corresponding to spots 4T16H5 and 4F13C5, that are present in the real RLGS pattern, were not amplified (since the NotI site was digested), and loci not visible or only weakly visible spots, 2F26H6, 3T28A8, 2F12C20, 4T24H24, 5T20L15 and 5MTH12, were amplified since the NotI site was methylated and therefore not cut. Sequence analysis of the cloned DNA fragments confirmed that they represented the NotI sites in the target DNA region predicted by Vi-RLGS.
Figure 1.
Application of Vi-RLGS to the Arabidopsis genome. (a) Vi-RLGS profile and (b) real RLGS profile of Arabidopsis nuclear DNA (NotI–EcoRV–MboI). Vi-RLGS spots are defined in the profile with the chromosome number (the first bold and underlined numerical number) and clone ID (the following five to six digit code). Red arrows in (b) indicate invisible or very faint spots in the real RLGS that were predicted from virtual RLGS. A yellow arrow indicates an example where no corresponding spot is seen in the Vi-RLGS. This may be due to incomplete sequence information. (c) PCR amplification designed for two unmethylated (in 4T16H5 and 4F13C5) and six methylated (in 2F26H6, 2F12C20, 3T28A8, 4T24H24, 5T20L15 and 5MTH12) NotI sites whose methylation status in the genome is predicted by subtraction between Vi-RLGS and real RLGS. The BAC/PAC ID numbers following the chromosome number are indicated on each lane. PCRs are performed for NotI digested (–NotI) and undigested (–) Arabidopsis genomic DNAs. Primers designed to amplify an ∼1-kb sequence containing the NotI sites predicted by Vi-RLGS were synthesized. If amplification products are produced (+) in NotI digested DNA, and the products have a NotI site, this means that the NotI sites are methylated and the virtual RLGS spot will not be detected in the real RLGS pattern. (d) Comparison between real RLGS patterns of normal (top) and hypomethylated (bottom) Arabidopsis genome. Three out of four invisible spots (red arrows in control) predicted from virtual RLGS appear on the real RLGS pattern of hypomethylated Arabidopsis genome DNA (black arrows).
The RLGS analysis was also carried out on Arabidopsis plants with hypomethylated DNA generated following growth in the presence of 5-aza-2′-deoxycytidine, which results in a 22% overall reduction in methylation. Spots 2F26H6, 3T28A8 and 2F12C20 (Fig. 1c), which are present in the virtual profile, but absent in the real RLGS profile from untreated plants, were present in the real RLGS pattern from the hypomethylated plants. These results further support the notion that their absence from the ‘real’ RLGS profile is due to DNA methylation.
Physical mapping of methylation sites in the whole Arabidopsis genome (methylation map)
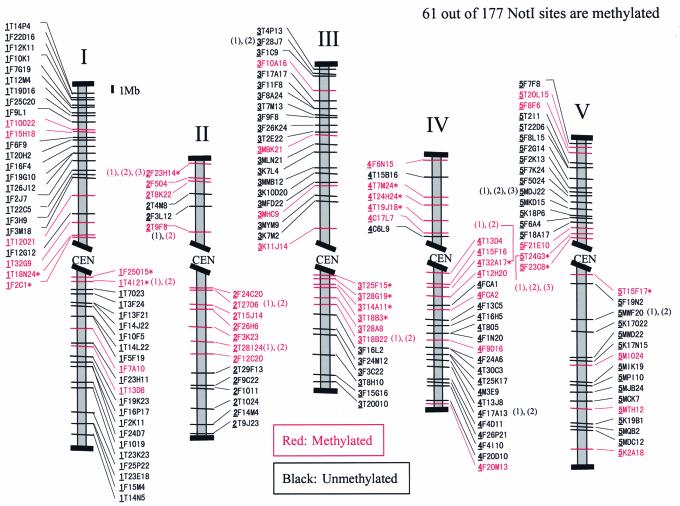
To find spots present in virtual profiles and absent in real RLGS patterns requires extensive analysis of methylated regions in Arabidopsis genomic DNA. Despite a few discrepancies in the centromere regions (data not shown), the comparison of virtual and real RLGS patterns and subsequent methylation-sensitive PCR assays demonstrated that 34.5% of all NotI sites in the Arabidopsis sequence database were methylated in the genome. Figure 2 shows the latest draft of the Arabidopsis methylation map for leaf DNA. We then analyzed the sequence of methylated regions using Repeat Masker. Almost half of the methylated NotI sites were within or close to repetitive sequences.
Figure 2.
A NotI physical map of Arabidopsis with the methylation status predicted by Vi-RLGS and real RLGS. Methylated spots were confirmed in triplicate by PCR assays. Loci with asterisks have not yet been confirmed by the PCR method. When there are two or three NotI sites within one BAC/PAC, they are numbered in order from centromere to telomere.
Application of Vi-RLGS to mammalian genomes
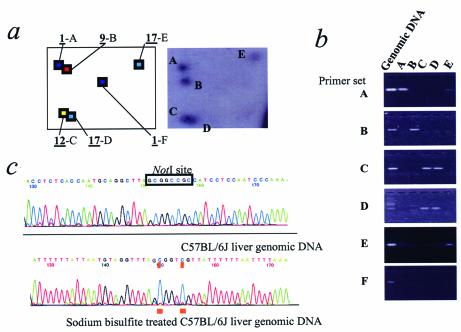
We next applied the Vi-RLGS software directly to the mouse genome, using a NotI–PstI–PvuII combination (see Fig. S2). Comparison of a sample field from C57BL/6J liver DNA identified approximately 1460 spots in real RLGS compared with 2170 spots in the same field of the virtual pattern. For instance, although the 1-F spot is not present in the real RLGS pattern (Fig. 3a), primer pairs for the predicted sequence still produce a PCR product from genomic DNA (Fig. 3b) in Figure 3. This spot sequence is highly homologous to LTR sequences detected by Repeat Masker. Bisulfite sequencing confirmed that the two CpGs in this NotI site were completely methylated in genomic DNA (Fig. 3c). Thus, this NotI site would not be cut or end-labeled and would therefore not appear in the real RLGS profile. The vast majority of the ‘extra spots’ in the virtual profile are derived from repetitive sequences, which would be expected to be methylated, and absent in the real profile. Consequently, 497 virtual spots predicted to be within repetitive DNA by Repeat Masker were removed from the profile (see Fig. S2). We confirmed the identity of 21 of 22 real spots in two regions of the RLGS profile (Figs 3a and 4) by sequence-specific PCR, using primers determined from the virtual sequence. One real RLGS spot (Fig. 4, arrow) could not be identified in the virtual RLGS patterns, before or after removing the repeat sequences. Presumably this is due to incomplete sequence information.
Figure 3.
Application of Vi-RLGS to the mouse genome. (a) A region corresponding to 6–8 kb in the first dimension, and 80–110 bp in the second dimension is shown. The chromosome number is underlined followed by the alphabetical identification. The virtual spots are color coordinated according to chromosome. The real profile is from C57BL/6J kidney. (b) PCR analysis using each sequence-specific primer set. Templates from C57BL/6J genomic DNA and eluted spot DNAs are indicated on each lane. Spots C and D overlap in the real RLGS profile and could not be purified separately. Therefore, the spot DNA was amplified by both sets of primers (C and D). (c) Sequence trace of C57BL/6J normal and bisulfite-treated genomic liver DNA. Bisulfite sequencing indicates that the two CpGs at NotI sites are highly methylated (underlined). BLAST analysis indicates that there is only one LTR with 100% homology to the LTR sequence-specific primers used for the PCR.
Figure 4.
Correspondence between mouse real (bottom) and virtual RLGS (top) profiles in the region corresponding to 4–8 kb in the first dimension and 110–180 bp in the second. The PCR results for 17 RLGS spots in four divided areas (a, b, c and d) are shown. C57BL/6J genomic DNA and eluted spot DNAs are amplified by each sequence-specific primer set in each area. The sequences of the PCR products are then confirmed by direct sequencing (data not shown). The black arrow indicates an unusual spot that is present in the real profile but is absent from the virtual profile (see text for explanation).
Tissue-specific methylation detection by real RLGS in conjunction with Vi-RLGS
Using Vi-RLGS, loci that have apparent tissue-specific differences in DNA methylation can be identified. A locus that shows reduced intensity in the C57BL/6J liver was identified from duplicate real RLGS NotI–PstI–PvuII patterns. The virtual sequence identified a genomic region around a NotI site located in a 3′ CpG island associated with the Adra1b gene. Bisulfite sequencing indicated that this region is methylated in liver but not in kidney (Fig. 5). Thus, the combination of virtual RLGS and real RLGS is a powerful technique to scan the methylation status at thousands of methylation-specific enzyme sites in mouse genomic DNAs.
Figure 5.
Differential methylation in tissue development. Real RLGS profiles from C57BL/6J liver and C57BL/6J kidney genomic DNA. The spot indicated by the arrow is very faint in liver genomic DNA. The spot DNA sequence was confirmed by PCR of spot DNA eluted from the kidney genomic DNA gel using primers determined from the virtual sequence. Bisulfite sequencing of the liver genomic DNA indicates that the two CpGs at NotI sites and two adjacent sites are highly methylated (underlined).
DISCUSSION
In this study, we successfully applied Vi-RLGS to determine the methylation status of all the NotI sites in the A.thaliana genome (23). Interestingly, methylation sites were not distributed uniformly at NotI sites in the Arabidopsis genome but rather tended to cluster close to the centromere. There are a substantial number of methylated NotI sites that do not correspond to repeat sequences and therefore are potential gene-regulation regions both in Arabidopsis and the mouse (data not shown). Thus, RLGS, in conjunction with in silico analysis, has the potential to screen the genome for sites of differential methylation and potentially identify gene regulatory regions in any given tissue. By using genomic sequence information and appropriate RLGS enzyme combinations, the methylation status of RLGS loci within specific target genes can be determined.
There are several ways to explain discrepancies in the number and pattern of spots between the real RLGS and virtual images: (i) the genome sequence is incomplete, (ii) spontaneous polymorphisms, (iii) DNA methylation, (iv) a double-strand DNA conformation which may result in aberrant migration in an acrylamide gel. The vast majority of differences between real and virtual patterns, however, appear to be due to either (iii) or (iv). Anomalous spot mobilities in polyacrylamide matrixes have been reported (24,25). Discrepancies between Vi-RLGS and real RLGS profiles, therefore, arise not only from the methylation differences but also from migration differences in the polyacrylamide gel. Approximately 20% of spots migrate faster or slower in RLGS gels than predicted by the molecular weight of the DNA. Since most of the differences between real and predicted mobility are relatively small, there is generally little problem in identifying the corresponding real and virtual RLGS spots if the spot pattern is not too dense (Fig. 1). It becomes more difficult, however, in the more complex mouse pattern shown in Figure S2. However, previous work has genetically mapped more than 2000 RLGS loci (26) that will greatly aid in the identification of the corresponding real and virtual RLGS loci. For these loci, the virtual sequence should physically map to the same chromosomal region as identified by the genetic mapping of real RLGS loci. (Tentative sequence information of mapped loci are available on request.) In addition, many landmark enzymes can be used for RLGS and Vi-RLGS can create any virtual RLGS pattern. Thus, this methodology has great potential for determining the DNA methylation status within the genome.
Epigenetic regulation of gene expression by means of DNA methylation has an important role in development (1). Transitions between silent and transcriptionally competent chromatin states are dependent on a balance between factors that sustain a silent state, such as HDACs, and those that promote a transcriptionally active state, such as HATs (27). Disturbances of any of these components may shift the balance between an active and a silent chromatin conformation, resulting in an altered transcriptional state. Genomic methylation status also undergoes changes, especially during development. In mouse, RLGS analysis has identified at least 48 spots with distinct intensity difference between kidney and liver genomic DNAs prepared from the same mouse. We confirmed that one spot corresponds to the 3′ non-coding region of the Adra1b gene (Fig. 5). Such tissue-specific methylation may induce altered transcriptional states between tissues and determine the cell fate. The application of whole genome methylation analysis by Vi-RLGS to the Arabidopsis and mouse genomes provides a novel method for identifying specific differences in DNA methylation associated with alterations in chromatin structure that are associated with important biological phenomena such as differentiation, proliferation, aging and diseases such as cancer.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs John Cowell and Michael Higgins for critical reading of the manuscript. We thank Reiko Kiuchi, Hisashi Kato, Arlene Morrow, Jun Igarashi, Haruhiko Okada and Jill Petruzzi for technical support. We also thank Joe Costello (UCSF) for the protocol for eluting spot DNA from RLGS gels. This work is supported by Roswell Park Alliance Foundation 55-5263-26, NCI grant CA68612 (W.A.H.), the NCI Core Center Grant CA16056 awarded to Roswell Park Cancer Institute, RIKEN Presidential Research Grant for Intersystem Collaboration, and the Research Project for the Study on Biological Cross-talk Functions of the Science and Technology Agency of the Japanese Government.
REFERENCES
- 1.Jaenisch R. and Bird,A. (2003) Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature Genet., 33 (Suppl.), 245–254. [DOI] [PubMed] [Google Scholar]
- 2.Jones P.A. and Takai,D. (2001) The role of DNA methylation in mammalian epigenetics. Science, 293, 1068–1070. [DOI] [PubMed] [Google Scholar]
- 3.Wolffe A.P. and Matzke,M.A. (1999) Epigenetics: regulation through repression. Science, 286, 481–486. [DOI] [PubMed] [Google Scholar]
- 4.Pennisi E. (2001) Behind the scenes of gene expression. Science, 293, 1064–1067. [DOI] [PubMed] [Google Scholar]
- 5.Yoder J.A., Walsh,C.P. and Bestor,T.H. (1997) Cytosine methylation and the ecology of intragenomic parasites. Trends Genet., 13, 335–340. [DOI] [PubMed] [Google Scholar]
- 6.Waterston R.H., Lindblad-Toh,K., Birney,E., Rogers,J., Abril,J.F., Agarwal,P., Agarwala,R., Ainscough,R., Alexandersson,M., An,P. et al. (2002) Initial sequencing and comparative analysis of the mouse genome. Nature, 420, 520–562. [DOI] [PubMed] [Google Scholar]
- 7.Baylin S.B., Herman,J.G., Graff,J.R., Vertino,P.M. and Issa,J.P. (1998) Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv. Cancer Res., 72, 141–196. [PubMed] [Google Scholar]
- 8.Ahringer J. (2000) NuRD and SIN3 histone deacetylase complexes in development. Trends Genet., 16, 351–356. [DOI] [PubMed] [Google Scholar]
- 9.Ushijima T., Morimura,K., Hosoya,Y., Okonogi,H., Tatematsu,M., Sugimura,T. and Nagao,M. (1997) Establishment of methylation-sensitive-representational difference analysis and isolation of hypo- and hypermethylated genomic fragments in mouse liver tumors. Proc. Natl Acad. Sci. USA, 94, 2284–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalgo M.L., Liang,G., Spruck,C.H.,III, Zingg,J.M., Rideout,W.M.,III and Jones,P.A. (1997) Identification and characterization of differentially methylated regions of genomic DNA by methylation-sensitive arbitrarily primed PCR. Cancer Res., 57, 594–599. [PubMed] [Google Scholar]
- 11.Gitan R.S., Shi,H., Chen,C.M., Yan,P.S. and Huang,T.H. (2002) Methylation-specific oligonucleotide microarray: a new potential for high-throughput methylation analysis. Genome Res., 12, 158–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan P.S., Chen,C.M., Shi,H., Rahmatpanah,F., Wei,S.H., Caldwell,C.W. and Huang,T.H. (2001) Dissecting complex epigenetic alterations in breast cancer using CpG island microarrays. Cancer Res., 61, 8375–8380. [PubMed] [Google Scholar]
- 13.Plass C., Shibata,H., Kalcheva,I., Mullins,L., Kotelevtseva,N., Mullins,J., Kato,R., Sasaki,H., Hirotsune,S., Okazaki,Y. et al. (1996) Identification of Grf1 on mouse chromosome 9 as an imprinted gene by RLGS-M. Nature Genet., 14, 106–109. [DOI] [PubMed] [Google Scholar]
- 14.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 15.Hatada I., Hayashizaki,Y., Hirotsune,S., Komatsubara,H. and Mukai,T. (1991) A genomic scanning method for higher organisms using restriction sites as landmarks. Proc. Natl Acad. Sci. USA, 88, 9523–9527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akama T.O., Okazaki,Y., Ito,M., Okuizumi,H., Konno,H., Muramatsu,M., Plass,C., Held,W.A. and Hayashizaki,Y. (1997) Restriction landmark genomic scanning (RLGS-M)-based genome-wide scanning of mouse liver tumors for alterations in DNA methylation status. Cancer Res., 57, 3294–3299. [PubMed] [Google Scholar]
- 17.Kondo T., Bobek,M.P., Kuick,R., Lamb,B., Zhu,X., Narayan,A., Bourc’his,D., Viegas-Pequignot,E., Ehrlich,M. and Hanash,S.M. (2000) Whole-genome methylation scan in ICF syndrome: hypomethylation of non- satellite DNA repeats D4Z4 and NBL2. Hum. Mol. Genet., 9, 597–604. [DOI] [PubMed] [Google Scholar]
- 18.Costello J.F., Fruhwald,M.C., Smiraglia,D.J., Rush,L.J., Robertson,G.P., Gao,X., Wright,F.A., Feramisco,J.D., Peltomaki,P., Lang,J.C. et al. (2000) Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nature Genet., 24, 132–138. [DOI] [PubMed] [Google Scholar]
- 19.Qiu P., Kupfer,K.C. and Garrard,W.T. (1997) A method for genome comparisons and hybridization studies using known megabase-scale DNA sequences as a reference. Genomics, 43, 307–315. [DOI] [PubMed] [Google Scholar]
- 20.Rouillard J.M., Erson,A.E., Kuick,R., Asakawa,J., Wimmer,K., Muleris,M., Petty,E.M. and Hanash,S. (2001) Virtual genome scan: a tool for restriction landmark-based scanning of the human genome. Genome Res., 11, 1453–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zardo G., Tiirikainen,M.I., Hong,C., Misra,A., Feuerstein,B.G., Volik,S., Collins,C.C., Lamborn,K.R., Bollen,A., Pinkel,D. et al. (2002) Integrated genomic and epigenomic analyses pinpoint biallelic gene inactivation in tumors. Nature Genet., 32, 453–458. [DOI] [PubMed] [Google Scholar]
- 22.Southern E.M. (1979) Measurement of DNA length by gel electrophoresis. Anal. Biochem., 100, 319–323. [DOI] [PubMed] [Google Scholar]
- 23.The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 24.Marini J.C., Levene,S.D., Crothers,D.M. and Englund,P.T. (1983) A bent helix in kinetoplast DNA. Cold Spring Harb. Symp. Quant. Biol., 47 (Pt 1), 279–283. [DOI] [PubMed] [Google Scholar]
- 25.Lerman L.S. and Frisch,H.L. (1982) Why does the electrophoretic mobility of DNA in gels vary with the length of the molecule? Biopolymers, 21, 995–997. [DOI] [PubMed] [Google Scholar]
- 26.Akiyoshi S., Kanda,H., Okazaki,Y., Akama,T., Nomura,K., Hayashizaki,Y. and Kitagawa,T. (2000) A genetic linkage map of the MSM Japanese wild mouse strain with restriction landmark genomic scanning (RLGS). Mamm. Genome, 11, 356–359. [DOI] [PubMed] [Google Scholar]
- 27.Jenuwein T. and Allis,C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.