Abstract
Topoisomerase II is a ubiquitous enzyme that removes knots and tangles from the genetic material by generating transient double-strand DNA breaks. While the enzyme cannot perform its essential cellular functions without cleaving DNA, this scission activity is inherently dangerous to chromosomal integrity. In fact, etoposide and other clinically important anticancer drugs kill cells by increasing levels of topoisomerase II-mediated DNA breaks. Cells rely heavily on recombination to repair double-strand DNA breaks, but the specific pathways used to repair topoisomerase II-generated DNA damage have not been defined. Therefore, Saccharomyces cerevisiae was used as a model system to delineate the recombination pathways that repair DNA breaks generated by topoisomerase II. Yeast cells that expressed wild-type or a drug-hypersensitive mutant topoisomerase II or overexpressed the wild-type enzyme were examined. Based on cytotoxicity and recombination induced by etoposide in different repair-deficient genetic backgrounds, double-strand DNA breaks generated by topoisomerase II appear to be repaired primarily by the single-strand invasion pathway of homologous recombination. Non-homologous end joining also was triggered by etoposide treatment, but this pathway was considerably less active than single-strand invasion and did not contribute significantly to cell survival in S.cerevisiae.
INTRODUCTION
DNA knots and tangles that form as a result of normal nuclear processes must be resolved in order for the cell to survive. The enzyme that catalyzes this essential function is known as topoisomerase II (1–8).
As part of its catalytic cycle, topoisomerase II generates transient protein-linked double-strand breaks in DNA (1–8). Although the enzyme cannot remove DNA knots or tangles without cleaving the genetic material, the creation of these breaks is inherently dangerous to chromosomal integrity (2,4–11). The potential cytotoxicity of topoisomerase II rises markedly when DNA tracking enzymes such as polymerases or helicases collide with covalent topoisomerase II-cleaved DNA complexes (12–16). These events have the ability to convert transient enzyme-associated breaks into permanent double-strand breaks in the genetic material. Consequently, while topoisomerase II is necessary for cell survival, it also has the capacity to fragment the genome.
Since topoisomerase II-mediated DNA breaks generally are short lived and are present at low levels, they do not present a problem to the cell under normal circumstances. However, conditions that significantly increase the physiological concentration of these DNA breaks trigger mutagenic events and can lead to the induction of programmed cell death pathways (10,17–20).
Because of its potentially lethal DNA cleavage reaction, topoisomerase II has emerged as one of the most successful targets currently used for cancer chemotherapy (2,5,8,21–25). Although drugs targeted to the enzyme come from several unrelated classes, they all share a common feature: every agent in clinical use kills cells by increasing levels of topoisomerase II-generated DNA breaks (2,5,8,21–25). Since these drugs convert topoisomerase II into a potent cellular toxin, they are referred to as ‘topoisomerase II poisons’, to distinguish them from catalytic inhibitors of the enzyme (2,5,23,25,26).
Despite the therapeutic benefits of topoisomerase II-targeted anticancer drugs, there is strong circumstantial evidence that the enzyme also can initiate leukemogenic events. For example, the use of topoisomerase II-active agents as part of some chemotherapeutic regimens has been associated with the development of specific secondary leukemias (9,11,27–29). The underlying genetic alteration in these cases involves translocations of the MLL oncogene at chromosomal band 11q23 (9,11,27–29). Furthermore, ∼80% of acute infant leukemias display 11q23 rearrangements (9,11,28), and these malignancies have been linked to the maternal consumption of foods that contain high levels of naturally occurring topoisomerase II poisons (30,31).
The genetic factors that contribute to the sensitivity of cells to topoisomerase II-targeted agents or predispose some patients to secondary or infant leukemias are not well characterized. However, a number of cellular alterations have been implicated. Changes in drug uptake or metabolism have been linked to resistance or increased risks of secondary leukemias, respectively (32–36). In addition, alterations in the expression, cellular localization or activity of topoisomerase II have dramatic effects on drug sensitivity in both the laboratory and clinical setting (33,35,37–43). Finally, the capacity to repair double-strand DNA breaks generated by type II topoisomerases can have striking consequences for cellular survival following treatment with antineoplastic drugs in both eukaryotic and bacterial systems (17,44–47). Deficiencies in recombination pathways lead to hypersensitivity to these agents.
Although cells rely heavily on recombination to repair double-strand DNA breaks (48–52), the specific pathways that are used to repair topoisomerase II-generated DNA damage have yet to be delineated. Non-homologous end joining appears to play an important role in mammalian species (48,52), but this process is inherently mutagenic and most likely mediates the chromosomal translocations observed in topoisomerase II-associated leukemias (11,28,53,54). In contrast, homologous recombination has the ability to reverse DNA breaks in a manner that does not compromise the genetic integrity of the cell. Recent work demonstrates the importance of homologous recombination for double-strand DNA break repair in both mouse and human cells (51,55–61). However, due to the redundancy of homologous processes in mammalian species (51,58,59,62), it has been difficult to ascribe roles for individual pathways in the reversal of topoisomerase II-generated DNA damage.
Therefore, the budding yeast, Saccharomyces cerevisiae, was used as the model system to delineate the recombination pathways that repair the double-strand DNA breaks generated by topoisomerase II. Yeast affords a number of advantages over other systems. First, homologous recombination is highly active and a number of specific pathways have been defined genetically (49,63). Second, modifications in topoisomerase II or recombination proteins can be engineered within isogenic backgrounds. Third, deletion of RAD52, which is involved in a number of recombinational repair pathways, increases the sensitivity of yeast cells to topoisomerase II-targeted drugs ∼100-fold (17).
Results indicate that double-strand DNA breaks generated by topoisomerase II are repaired in S.cerevisiae primarily by homologous recombination rather than non-homologous end joining. Furthermore, it appears that the major homologous recombination pathway used to repair these breaks is single-strand invasion.
MATERIALS AND METHODS
Negatively supercoiled bacterial plasmid pBR322 DNA was prepared as described (64). CP-115,953 and TOP-53 were generously provided by Pfizer Global Research and Taiho Pharmaceutical Co., respectively. Etoposide and camptothecin were obtained from Sigma. Drugs were prepared as 20 mM solutions in 100% DMSO and stored at 4°C. Restriction endonucleases were from New England Biolabs. Growth media were prepared using standard protocols. All other chemicals were analytical reagent grade.
Yeast strains and plasmids
Saccharomyces cerevisiae strains generated for this study (Table 1) were derivatives of JN362acc (MATa ura3-52 leu2 trp1 his7 ade1-2 ISE2 can1 cyh2). JN362acc was derived from JN362a (38) by deletion of the can1 and cyh2 genes. When appropriate, the TOP2 gene of JN362acc was replaced with top2S740W using pMJ2 top2S740W/URA3 in a two-step pop-in/pop-out gene replacement using 5-flouro-orotic acid as counterselection (65) to generate the strain MS001. Gene replacement was confirmed by sequencing a PCR-amplified fragment of genomic DNA containing the central portion of the TOP2 gene. Strains that constitutively overexpressed yeast topoisomerase II were generated by transforming yeast with pDED1TOP2 (38). Construction of repair-deficient yeast was carried out in parallel with both JN362acc and MS001 using one-step gene replacement (65) (see Table 1 for strains). Gene replacement was confirmed by PCR of genomic DNA and restriction enzyme digestion. Genomic DNA was prepared using a Wizard Genomic DNA Purification Kit (Promega). Construction of the recombination reporter plasmids YCpHR and YCpL2 has been described (66–68) (see Figs 3 and 6 for structures).
Table 1. Saccharomyces cerevisiae strains.
| Straina | Genotype | Plasmid for gene replacement | Strain origin |
|---|---|---|---|
| JN362acc | MATa ura3-52 leu2 trp1 his7 ade1-2 ISE2 can1 cyh2 | H. Ikeda, derived from Nitiss et al. (38) | |
| MS001 | top2S740W | pMJ2 top2S740W/URA3 (76) | This study |
| MS101-a,bb | Δrad50::hisG | pNKY83 (98) | This study |
| MS111-a,b | Δrad52::LEU2 | pSM20 Δrad52::LEU2 (17,38) | This study |
| MS121-a,b | Δrad54::KAN | pFA6aKanMX (99) | This study |
| MS131-a,b | Δrad1::URA3 | pCR2.1 Δrad1::URA3 | This study |
| MS141-a,b | Δku70::LEU2 | pGEM4ZS-H/LEU2 (100) | This study |
aAll strains are isogenic to JN362acc except where noted.
ba, using JN362acc; b, using MS001.
Figure 3.

Restriction digests of intact and recombined YCpHR homologous recombination reporter plasmids. A map of the homologous recombination reporter plasmid YCpHR is shown at the top. The four black lines along the backbone of the plasmid indicate PstI cut sites and the numbers internal to the plasmid correspond to the restriction fragment sizes as indicated on the gel at the bottom. Plasmids were rescued from TOP2 strains before (WT) and after (R1, R2 and R3) exposure to etoposide, CP-115,953 or camptothecin, respectively. Samples were digested with PstI and subjected to electrophoresis in an agarose gel that contained ethidium bromide. A corresponding digest of the original plasmid is also shown (Control). The sizes of restriction digest fragments are indicated to the left in kilobases. The restriction fragment representing the recombined portion of YCpHR is indicated at the right (*).
Figure 6.

Non-homologous end joining in YCpL2 triggered by topoisomerase II-generated DNA damage. The repair-proficient top2S740W strain was transformed with YCpL2, a reporter plasmid related to YCpHR but used to monitor non-homologous end joining. Cultures were treated with 0–170 µM etoposide for 5 h. Data represent the number of recombinant cells per 104 viable cells and are the average of two independent experiments plated in duplicate. Standard errors of the mean are represented by error bars. A map of YCpL2 is shown (not to scale).
Determination of etoposide sensitivity
Yeast strains were incubated in SC minimal medium with 0–170 µM etoposide for 0–24 h, plated in duplicate on YPDA medium solidified with 1.5% Bacto-agar and cultured for 3–4 days at 30°C. Drug sensitivity was quantitated by counting the number of surviving colonies.
Determination of recombination frequency
Yeast strains were transformed with either YCpHR or YCpL2 by selecting on SC-URA (except for Δrad-1::URA3 strains, in which transformants were selected for and subsequent analyses carried out on SC-LEU). Single colonies were grown overnight in SC-URA and diluted to 2–8 × 106 cells/ml. The diluted yeast were split into parallel cultures of 3 ml, to which were added 0–170 µM etoposide, 180 µM TOP-53, 50 µM CP-115,953, 140 µM camptothecin or an equivalent volume of DMSO as a no-drug control. Cultures were grown for 5 h and dilutions from each culture were plated in duplicate on SC-URA (or SC-LEU) for total cell viability, in duplicate either on SC-URA + 60 µg/ml canavanine for selection of yeast carrying recombined YCpHR (66) or on SC-URA + 60 µg/ml canavanine + 10 µg/ml cycloheximide for selection of yeast carrying recombined YCpL2 (67,68). Recombination frequencies were calculated for each drug treatment by dividing the number of recombinants/ml by the number of total viable cells/ml.
Analysis of recombinant plasmids
Single colonies that grew on SC-URA + canavanine or on SC-URA + canavanine + cycloheximide were picked to 5 ml SC-URA and grown to confluency. Plasmids were rescued into Escherichia coli using the EZ Yeast Plasmid Prep Kit (Geno Technology Inc.). The resulting E.coli transformants were picked in duplicate and plasmids were prepared for restriction enzyme digestion using the Strataprep Plasmid Miniprep Kit (Stratagene).
FACS analysis of yeast
At 0–24 h growth time in the absence or presence of 170 µM etoposide, 1 ml aliquots of yeast cultures were fixed and prepared for FACS analysis using Sytox Green (Molecular Probes, Eugene, OR) as described (69). Cells for flow cytometric analysis were fixed in cold 70% ethanol, sonicated for 15 s in 1 ml of 50 mM sodium citrate (pH 7.0) containing 0.25 mg/ml RNase A and incubated for 1 h at 50°C. Cells were then stained with 1 µM Sytox Green in 1 ml of 50 mM sodium citrate (pH 7.0) for 1 h in the dark. DNA content was measured on a Becton Dickinson FACSCalibur.
RESULTS
Repair of double-strand DNA breaks in yeast
The budding yeast, S.cerevisiae, can potentially use a number of DNA recombination pathways to repair double-strand breaks in its genetic material (reviewed in 49,50). The most common of these pathways are depicted in Figure 1.
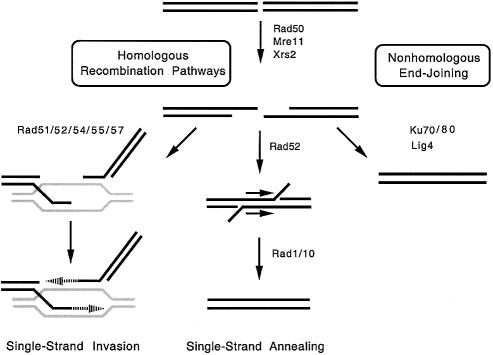
Figure 1.
Pathways of double-strand DNA break repair in S.cerevisiae. Components that play integral roles in homologous recombination (single-strand invasion and single-strand annealing) and non-homologous end joining pathways are shown.
The initial processing of double-strand DNA breaks generally relies on the heterotrimeric Rad50/Mre11/Xrs2 complex to generate single-stranded ends at the site of the break (49,50,70). This processed DNA can be shuttled into several different recombination pathways. First, the break can be repaired by the single-strand invasion pathway of homologous recombination (49,50). In this case, the processed break is recognized by Rad52, which recruits the RecA homolog, Rad51. Together with its accessory proteins (Rad54/Rad55/Rad57), Rad51 promotes strand invasion into homologous sequences in the genome, which are used as templates to regenerate the original nucleotide sequence. This pathway is capable of repairing the initial double-strand DNA break in an error-free manner. However, it can also result in loss of heterozygosity in diploids or can lead to gene duplications or deletions (49,50).
Second, the processed break can be repaired by the single-strand annealing pathway of homologous recombination (49,50). This pathway is dependent on the presence of direct repeats (or closely related sequences) proximal to and flanking the initial break site. It also relies on Rad52, but does not require any other protein component of the single-strand invasion pathway. Instead, Rad52 anneals the repeated sequences, followed by the actions of the Rad1/Rad10 endonuclease, which removes the non-annealed tails. Single-strand annealing is not an error-free pathway and deletes one of the repeated sequences, as well as the genetic information that is located between them.
Third, the processed break can be rejoined by the non-homologous end joining pathway (49,50,71,72). In this case, the DNA ends are recognized by Ku70/Ku80, which promotes ligation of the break by Lig4 (49,50,71,72). This is an error-prone pathway that results in the loss of sequences proximal to the original DNA break (49,50,71,72). If multiple breaks are present in the genome, non-homologous end joining can lead to the formation of chromosomal rearrangements or translocations (52,73).
Cytotoxicity of topoisomerase II-induced DNA damage in recombination-deficient yeast
In order to characterize the pathways utilized to repair topoisomerase II-mediated DNA breaks, a series of drug-permeable, isogenic haploid S.cerevisiae strains was constructed that contained single deletions of DNA recombination genes (Table 1). Included in this series were strains deficient in RAD50 or RAD52, both of which function in most recombination pathways; RAD54, which is unique to the single-strand invasion pathway; RAD1, which is required for the single-strand annealing pathway but not for the single-strand invasion pathway; or KU70, which has no role in homologous recombination but is necessary for non-homologous end joining.
Topoisomerase II-generated DNA damage was induced by treating yeast strains with the anticancer drug etoposide. This agent is a potent topoisomerase II poison that dramatically increases levels of topoisomerase II-mediated DNA breaks in treated cells (2,5,8,21–25). Etoposide appears to act primarily by inhibiting the DNA religation event mediated by the enzyme (74). In the drug concentration range utilized in the present study, topoisomerase II is the only significant cytotoxic target for etoposide in yeast (38,75).
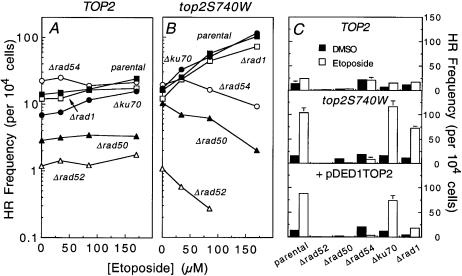
As seen in Figure 2A, yeast cells carrying allelic wild-type topoisomerase II (i.e. TOP2) treated with etoposide fell into two categories. The parental strain, as well as strains lacking RAD1 or KU70, displayed low sensitivity to etoposide at all drug concentrations employed. At the highest drug exposure examined (170 µM etoposide for 24 h), cell survival was ∼80% that of the control drug-free cultures. In marked contrast, strains lacking RAD50, RAD52 or RAD54 were hypersensitive to etoposide. Less than 1% survival was observed in these cultures at 170 µM drug.
Figure 2.
Cytotoxicity of etoposide to repair-compromised yeast strains. The effects of etoposide on cell survival of yeast deficient in a series of repair genes are shown. The mutations were made in three different strain backgrounds: the original strain which expressed wild-type TOP2 (TOP2) (A), a mutant strain in which the chromosomal copy of TOP2 was replaced with the etoposide hypersensitive top2S740W allele (top2S740W) (B) and the TOP2 strain which overexpressed wild-type TOP2 from the plasmid pDED1TOP2 (pDED1TOP2) (C). The mutations introduced in these strains were Δrad1 (open squares), Δku70 (closed circles), Δrad54 (open circles), Δrad50 (closed triangles) and Δrad52 (open triangles). Repair-proficient parental strains (parental) are represented by closed squares. Data were calculated relative to cell growth at 24 h in drug-free cultures, such that the level of growth for each strain in the absence of etoposide was set to 100%. Data represent the average of two or more independent experiments, each plated in duplicate.
Although repair-competent yeast strains display some sensitivity to topoisomerase II poisons, appreciable levels of cell kill require high concentrations of enzyme-associated DNA breaks. Several alternatives were considered in an effort to increase cellular levels of topoisomerase II-mediated DNA cleavage. Raising the drug concentration was rejected due to the possibility that there might be targets other than topoisomerase II at higher drug levels. Increasing the duration of etoposide exposure also was rejected due to concerns regarding the metabolic breakdown of the drug over prolonged time courses.
Therefore, a third alternative was chosen: the wild-type chromosomal TOP2 gene was replaced by top2S740W. This mutant allele encodes a topoisomerase II protein in which Ser740 has been replaced by a tryptophan residue. Ser740 in yeast topoisomerase II is equivalent to Ser83 in the A subunit of E.coli DNA gyrase (76). This conversion of serine to tryptophan in DNA gyrase is one of the most common causes of drug resistance in clinical isolates (77–80). It renders E.coli ∼10-fold resistant to antibacterial quinolones and abolishes any detectable drug binding to the gyrase–DNA complex (81). While the S740W mutation in yeast topoisomerase II confers resistance to antineoplastic quinolones, the resulting enzyme also is several-fold hypersensitive to etoposide (76). Moreover, the in vitro DNA cleavage phenotype of top2S740W (i.e. etoposide hypersensitivity and quinolone resistance) parallels the drug cytotoxicity profile of yeast strains expressing the mutant allele (76). Finally, the catalytic activity of top2S740W is similar to that of wild-type yeast topoisomerase II (76).
Results with top2S740W strains are shown in Figure 2B. These strains displayed a sensitivity towards etoposide that was ∼10-fold greater than observed in the TOP2 strains. However, their response to recombination defects was qualitatively the same as those described for yeast expressing wild-type topoisomerase II. Once again, cells lacking RAD1 or KU70 displayed little or no additional sensitivity to etoposide, while strains lacking RAD50, RAD52 or RAD54 were approximately two orders of magnitude more susceptible to the drug.
top2S740W is believed to be hypersensitive to etoposide because the drug inhibits DNA religation to a greater extent (>10-fold) than observed with the wild-type enzyme (76,82). Thus, the drug-induced DNA breaks generated in yeast cells expressing top2S740W probably are more stable than those generated in Top2-containing strains.
Recent studies with mutant type I topoisomerases indicate subtle, but potentially important, differences in the repair pathways and/or checkpoints that are triggered by enzymes with faster rates of cleavage as compared to those with decreased rates of religation (83). To control for this possibility with the type II enzyme, cytotoxicity studies were repeated with strains that constitutively overexpressed (5- to 10-fold) wild-type yeast topoisomerase II from the pDED1TOP2 plasmid (38) (Fig. 2C). Although overexpression of the enzyme renders yeast cells hypersensitive to anticancer drugs by increasing the number of chromosomal DNA breaks in vivo, it does not affect the stability of these breaks. Similar to top2S740W, pDED1TOP2 conferred repair-competent cells with an ∼10-fold hypersensitivity to etoposide. Results with repair-deficient pDED1TOP2 lines were similar to those observed with the corresponding TOP2 and top2S740W strains. However, Δrad50, Δrad52 or Δrad54 strains that contained pDED1TOP2 displayed drug sensitivity that was intermediate to comparable strains in the TOP2 and top2S740W backgrounds.
In addition to the above recombination pathways, it is possible that a repair process specific for topoisomerase II-induced DNA damage exists. To this point, a tyrosyl-DNA phosphodiesterase (Tdp1) that removes trapped topoisomerase I from the 3′-ends of single-strand DNA breaks (84) has been characterized. However, no equivalent activity for the removal of topoisomerase II from the 5′-ends of cleaved DNA has been identified. Furthermore, it is unlikely that endonucleolytic pathways that require Rad27 (the yeast homolog of the 5′-flap endonuclease FEN-1) (49,85–88) are essential, since deletion of the RAD27 gene did not confer hypersensitivity to etoposide in any of the topoisomerase II backgrounds examined (data not shown).
Two conclusions can be drawn from the above findings. First, drug sensitization within a given topoisomerase II background was observed only in strains that lacked RAD50, RAD52 or RAD54. Thus, it appears that homologous recombination, more specifically the single-strand invasion pathway, is primarily responsible for repairing topoisomerase II-mediated DNA breaks that occur following treatment with etoposide. Second, since defects in RAD1 or KU70 did not affect drug sensitivity, it is unlikely that single-strand annealing or non-homologous end joining plays a major role in the repair of topoisomerase II-generated DNA damage in yeast.
Homologous recombination triggered by topoisomerase II
Because of the findings of the cytotoxicity experiments, the effects of topoisomerase II-generated DNA damage on homologous recombination were examined directly. A plasmid-based homologous recombination reporter system was employed for these studies. In this system, the strains utilized for cytotoxicity experiments were transformed with YCpHR, a plasmid that contains the canavanine sensitivity gene, CAN1, flanked on either side by a copy of the LEU2 gene (66) (Fig. 3). Because of the two direct repeats of LEU2, this plasmid can serve as a recombination substrate for both the single-strand invasion and single-strand annealing pathways.
Homologous recombination between the two LEU2 genes by either pathway results in a deletion of the CAN1 gene (Fig. 3). Since the chromosomal allele of the CAN1 gene is disrupted in the parental yeast strain, recombination was scored by the ability of cells to grow in the presence of canavanine.
To verify that yeast colonies that scored as canavanine-resistant resulted from a homologous recombination event between the two LEU2 genes on YCpHR (rather than a microdeletion or point mutation in CAN1), plasmids were rescued from canavanine-resistant yeast and analyzed by restriction enzyme digestion. Representative digests are shown in Figure 3 (bottom). Based on restriction fragments generated by treatment with PstI, the loss of canavanine sensitivity was accompanied by a deletion of ∼6 kb in >99% of the isolates. This length corresponds to the size of the predicted CAN1 fragment that would be lost following homologous recombination between the two LEU2 genes.
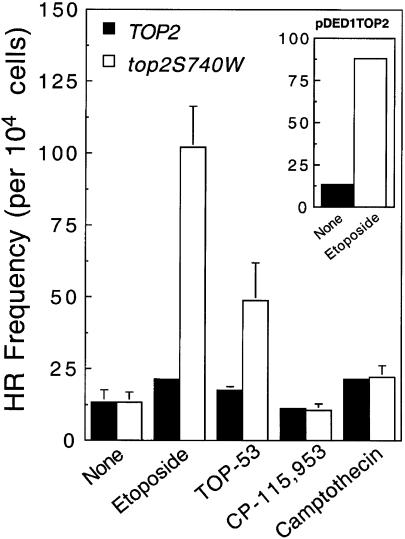
As seen in Figure 4, relatively little increase in homologous recombination was observed in the repair-competent parental TOP2 strain following exposure to etoposide, the etoposide derivative TOP-53 or the quinolone CP-115,953, all of which have been demonstrated to target topoisomerase II in yeast (37,38,75,89). Thus, it appears that the level of topoisomerase II-generated DNA damage induced by drugs in the TOP2 strain was insufficient to increase the levels of homologous recombination in the YCpHR reporter plasmid above drug-independent levels.
Figure 4.
Homologous recombination in YCpHR triggered by drugs. Repair-proficient yeast expressing either wild-type (TOP2, closed bars) or mutant (top2S740W, open bars) topoisomerase II transformed with YCpHR were treated with 170 µM etoposide, 180 µM TOP-53, 50 µM CP-115,953 or 140 µM camptothecin for 5 h. Data represent the number of recombinant cells per 104 viable cells and are the average of two or more independent experiments, each plated in duplicate. Standard deviations are indicated by error bars. (Inset) Levels of etoposide-triggered homologous recombination in YCpHR in repair-proficient yeast that also carried the topoisomerase II overexpression plasmid (pDED1TOP2).
However, when either the top2S740W strain or the pDED1TOP2 strain (that overexpressed wild-type topoisomerase II) was treated with etoposide, significant increases in homologous recombination in YCpHR were observed (Fig. 4 and inset, respectively). A 4-fold increase in homologous recombination also was observed upon exposure of the top2S740W strain to TOP-53 and, consistent with the quinolone-resistant phenotype of the mutant enzyme, no increase in homologous recombination was observed following treatment with CP-115,953.
As a control, both the TOP2 and top2S740W strains were treated with camptothecin, a drug that specifically poisons topoisomerase I. As expected for a drug that induces DNA strand breaks, levels of homologous recombination were elevated (∼2-fold) in the presence of the drug. However, recombination levels were similar regardless of which topoisomerase II allele was expressed in the strain. With the above results, this finding confirms the topoisomerase II specificity of the recombination reporter system.
The effects of etoposide on the frequency of homologous recombination of YCpHR in the repair-deficient TOP2 strains are shown in Figure 5A. As reported previously, baseline levels of recombination decreased significantly in Δrad50 and Δrad52 strains (49,90). However, as expected from the data shown in Figure 4, the YCpHR reporter system was relatively insensitive to etoposide in strains that contained a wild-type TOP2 allele.
Figure 5.
Homologous recombination in YCpHR triggered by topoisomerase II-generated DNA damage. Repair-deficient strains carrying the YCpHR reporter plasmid were exposed to 0–170 µM etoposide for 5 h. Data represent the number of recombinant cells per 104 viable cells. Repair-deficient strains generated in the TOP2 and top2S740W backgrounds are shown in (A) and (B), respectively. Strains are labeled as in Figure 2. Data generated at 0 and 170 µM etoposide for the TOP2, top2S740W and pDED1TOP2 (which overexpresses topoisomerase II) strains are shown in (C) (standard errors of the mean are represented by error bars). Data represent the average of two or more independent experiments, each plated in duplicate.
Therefore, the effects of the drug on the frequency of homologous recombination of YCpHR in the strains that contained the etoposide-hypersensitive top2S740W allele were determined (Fig. 5B). Marked differences were observed for the repair-deficient strains. Levels of etoposide-induced homologous recombination increased ∼7-fold in the parental top2S740W strain and also increased significantly in strains that carried the Δrad1 or Δku70 genotype. These data strongly suggest that the homologous recombination pathways that lead to survival following treatment with etoposide are still intact in these strains. Therefore, it is unlikely that single-strand annealing or non-homologous end joining plays a major role in processing etoposide-induced topoisomerase II-mediated DNA damage.
In contrast, no increase in recombination was observed in strains that carried the Δrad50, Δrad52 or Δrad54 genotype. [The apparent recombination observed for these strains is most likely due to events unrelated to homologous recombination. To this point, the majority of plasmids recovered from canavanine-resistant colonies in these repair-deficient backgrounds had abnormal (i.e. not recombinant) digestion patterns or carried point mutations in CAN1 (data not shown).] In addition, levels of recombination fell in Δrad50, Δrad52 or Δrad54 strains at higher concentrations of etoposide. Although this decrease is not well understood at the present time, it may indicate that alternative repair pathways are being induced in these recombination- compromised strains. This latter issue aside, results of these experiments strongly suggest that topoisomerase II-mediated DNA breaks that result from treatment of yeast cells with etoposide induce the single-strand invasion pathway of homologous recombination.
To control for the increased stability of DNA breaks generated by top2S740W, the effects of etoposide on homologous recombination were determined in pDED1TOP2 strains. Results for 170 µM etoposide are compared to those of the parental TOP2 and mutant top2S740W strains in Figure 5C. Once again, drug-induced changes in DNA recombination in strains that overexpressed topoisomerase II paralleled those described for strains that expressed the mutant etoposide-hypersensitive enzyme.
Non-homologous end joining triggered by topoisomerase II-generated DNA damage
Deletion of KU70 had no appreciable effect on cell survival in the presence of etoposide (see Fig. 2). However, due to the highly active homologous recombination in S.cerevisiae, the non-homologous end joining pathway generally plays a lesser role in the repair of double-strand DNA breaks as compared to other organisms (72). To determine whether etoposide has any effect on non-homologous end joining in yeast, the top2S740W strain was transformed with YCpL2, a reporter plasmid related to YCpHR that is used to monitor non-homologous events (67,68) (Fig. 6). YCpL2 carries the cycloheximide sensitivity gene, CYH2 (in the opposite orientation to the CAN1 gene) in place of one of the two LEU2 genes of YCpHR. Non-homologous recombination was scored by the ability to grow in the presence of both cycloheximide and canavanine. As determined by restriction analysis, rescued plasmids contained deletions that ranged in size from ∼0.5 to ∼5.5 kb (data not shown).
Baseline non-homologous recombination monitored by YCpL2 in the repair-competent TOP2 (not shown) or top2S740W (Fig. 6) strain was approximately three orders of magnitude lower than seen for homologous recombination in the same genetic background. Furthermore, levels of non-homologous recombination decreased an additional order of magnitude in an isogenic Δku70 strain (data not shown). This finding confirms that events scored by the YCpL2 reporter plasmid represent (at least in part) the non-homologous end joining pathway.
As assessed by YCpL2, etoposide stimulated non-homologous recombination in the repair-competent top2S740W strain ∼3-fold over drug-independent levels (Fig. 6). Therefore, the lack of effect of Ku70 activity on the cellular sensitivity to etoposide (see Fig. 2) probably reflects the comparatively low activity of the non-homologous end joining pathway in yeast rather than a lack of stimulation by topoisomerase II-mediated double-strand DNA breaks.
Effects of topoisomerase II-generated DNA damage on cell cycle distribution and progression in repair-deficient strains
To further investigate the cellular processes that are altered by the accumulation of topoisomerase II-mediated DNA breaks, the effects of etoposide on cell cycle distribution and progression were characterized in repair-competent and repair-deficient S.cerevisiae strains.
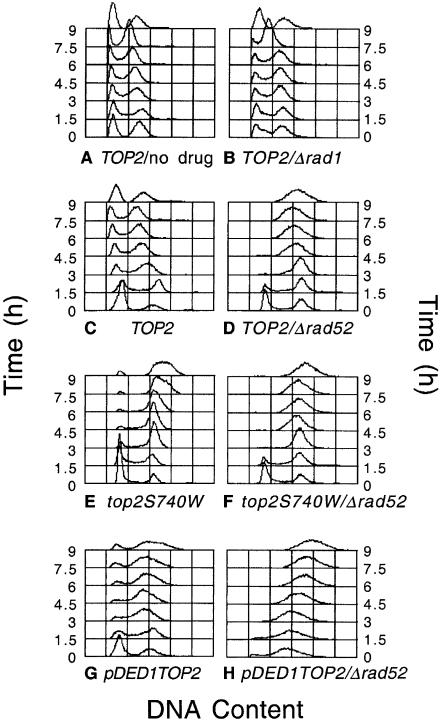
In the absence of drug, gene deletions in TOP2 strains had minimal effects on cell morphology, cell doubling times (<30% difference between strains, 1.8–2.3 h) and cell cycle distribution of asynchronous populations. Following treatment with etoposide, however, TOP2 cells displayed two distinct terminal phenotypes that paralleled their cytotoxicity phenotypes (FACS analyses from representative strains are shown in Fig. 7). Strains whose drug sensitivity was similar to that of the parental strain (Δrad1 or Δku70; see Figs 2 and 9) exhibited a cell cycle distribution at 24 h that was virtually indistinguishable from that of drug-free cultures. In contrast, recombination-deficient strains that displayed increased sensitivity to etoposide (Δrad50, Δrad52 or Δrad54; see Figs 2 and 9) exhibited an abnormal cell cycle distribution at 24 h with a single broad peak centered at a DNA content of 2N–3N.
Figure 7.

Terminal phenotype of TOP2 strains in the presence of etoposide. The following yeast strains were grown for 24 h in the presence of 170 µM etoposide (green) or an equivalent amount of drug solvent (DMSO, red): TOP2 (A), TOP2Δrad1 (B) and TOP2Δrad52 (C). Peaks representing haploid (1N) and diploid (2N) DNA contents are indicated. Aliquots of 1 ml were removed and used for FACS analysis with Sytox Green as the DNA stain.
Figure 9.
Effects of etoposide on the growth of yeast. Simultaneous to taking samples for the FACS analyses shown in Figure 8, yeast were diluted and plated in duplicate to YPDA to determine the number of viable cells at each time point. Cell growth is plotted relative to the number of cells at 0 h. Cultures were grown in the absence of drug (TOP2/no drug, closed squares) or in the presence of 170 µM etoposide as follows: TOP2/Δrad1 (open squares), TOP2 (closed circles), TOP2/Δrad52 (open circles), top2S740W (closed triangles), top2S740W/Δrad52 (open triangles), pDED1TOP2 (closed diamonds), pDED1TOP2/Δrad52 (open diamonds).
To extend the above data, time courses for cell cycle progression (as determined by FACS analysis) (Fig. 8) and cell growth (Fig. 9) of TOP2 strains were examined in parallel in the absence of drug, in the presence of etoposide or in the presence of etoposide in a Δrad52 background. Consistent with the unaltered terminal phenotype of the parental strain following a 24 h exposure to etoposide, there were no obvious effects of drug on the progression of the cell cycle in repair-competent cells or in strains such as Δrad1, whose drug sensitivity is similar to the parental strain (Fig. 8A–C). These data parallel the minor effects of etoposide on the growth rate of the parental TOP2 strain (Fig. 9).
Figure 8.
Cell cycle progression of yeast during exposure to etoposide. The indicated strains were grown for 24 h in the absence of drug (TOP2/no drug) (A) or in the presence of 170 µM etoposide as follows: TOP2/Δrad1 (B), TOP2 (C), TOP2/Δrad52 (D), top2S740W (E), top2S740W/Δrad52 (F), pDED1TOP2 (G), pDED1TOP2/Δrad52 (H). Aliquots of 1 ml were removed every 1.5 h from 0–9 h and used for FACS analysis with Sytox Green as the DNA stain.
In contrast, etoposide had a dramatic effect on the cell cycle of the TOP2/Δrad52 strain (Fig. 8D). An immediate block at G2/M was observed and cells never re-entered a normal cell cycle. Rather, populations slowly proceeded to the abnormal 2N–3N terminal phenotype described above. Once again, cell cycle results were consistent with the effects of etoposide on the growth of the TOP2/Δrad52 strain (Fig. 9). Even at the earliest times, no growth of cultures was observed and the drug was cytotoxic at every time point examined.
As observed with TOP2 strains, gene deletions in top2S740W cells had minimal physiological consequences in the absence of drug (not shown). However, the effects of etoposide on cell cycle and cell growth were much more pronounced in these drug hypersensitive strains (Figs 8E and F and 9). Even in a repair-proficient background, the FACS analysis, 2N–3N terminal phenotype and growth curve of top2S740W cells resembled those of the TOP2/Δrad52 strain. Furthermore, the combination of the top2S740W mutation and the Δrad52 repair deficiency resulted in the most rapid loss of cell viability of any of the strains examined.
The effects of etoposide on pDED1TOP2 strains (which overexpressed topoisomerase II) closely resembled those seen with the drug-hypersensitive point mutant of yeast topoisomerase II (Figs 8G and H and 9). Even in a repair-proficient genetic background, etoposide was cytostatic over the time course of the cytotoxicity experiment (Fig. 9). One difference was noted between repair-deficient pDED1TOP2 strains and the other strains examined. In backgrounds that impaired the single-strand invasion pathway (Δrad50, Δrad52 or Δrad54), pDED1TOP2 cells displayed a decreased proportion of G1 cells in the absence of drug (Fig. 8H and data not shown). This alteration in cell cycle probably reflects the higher baseline levels of enzyme-mediated DNA cleavage in strains that express increased levels of topoisomerase II.
DISCUSSION
The ability to cleave and religate DNA is critical to all of the catalytic functions of topoisomerase II (1–8). However, the covalent enzyme-cleaved DNA intermediate that is formed during this process also has the potential to create permanent and cytotoxic double-strand breaks in the genetic material (2,4–7,10,11,91). Several very successful anticancer agents take advantage of this aspect of the topoisomerase II mechanism and kill malignant cells by increasing levels of enzyme-mediated DNA scission (2,5,8,21–25). Despite the importance of topoisomerase II-mediated DNA cleavage to cell life and death, little is known regarding the physiological pathways that process the double-strand DNA breaks generated by the enzyme. Therefore, the role of recombination in the cellular response to topoisomerase II-generated DNA damage was investigated.
Saccharomyces cerevisiae cells were treated with the anticancer drug etoposide in order to increase physiological levels of topoisomerase II-mediated DNA breaks. Three isogenic strains were used for these studies. The first contained its original chromosomal TOP2 allele. The second strain contained the mutant top2S740W in place of the wild-type TOP2 chromosomal allele. The resulting mutant enzyme displays catalytic activity that is comparable to wild-type topoisomerase II, but is hypersensitive to etoposide due to a greater drug-induced inhibition of enzyme-mediated DNA religation (76,82). The third strain constitutively overexpressed a plasmid-based wild-type topoisomerase II (pDED1TOP2) (38) that conferred drug hypersensitivity by increasing the number, but not the stability, of enzyme-mediated DNA breaks.
Results with a series of repair-compromised cell lines strongly suggest that the major pathway that protects yeast from the consequences of topoisomerase II-mediated DNA breaks is the single-strand invasion pathway of homologous recombination. This is the main pathway used to repair double-stranded DNA breaks in S.cerevisiae (49,50). While etoposide treatment also triggers Ku-dependent non- homologous end joining, levels are ∼3 orders of magnitude lower than observed for homologous recombination and appear to contribute little to cell survival.
Qualitatively, the top2S740W and pDED1TOP2 strains were similar. Therefore, at least for the events examined in the present study, it appears that the overall number of topoisomerase II-mediated DNA breaks (as opposed to the stability of the breaks) determines the cellular response to etoposide. However, quantitative differences between the two strains suggest that long-lived DNA breaks are more lethal to cells, especially in repair-compromised backgrounds. Thus, precise relationships between the longevity of topoisomerase II-mediated DNA breaks and downstream processes remain an open question.
The lethality of etoposide appears to correlate with a G2/M block, followed by a lack of re-entry into a normal cell cycle and the gradual appearance of an abnormal 2N–3N terminal phenotype. It is not clear what this terminal phenotype represents. As determined by microscopy, cells with the 2N–3N DNA complement are larger than normal cells and have multiple buds (data not shown). Thus, these cells may represent a small population that continued to synthesize DNA despite a failure to properly segregate daughter chromosomes.
The results of the present study are consistent with the DNA damage checkpoint activation and G2/M cell cycle arrest that have been observed in human cells following exposure to etoposide (92–94). Furthermore, human cells with recombination defects due to mutations in the WRN or ATM genes are hypersensitive to etoposide (95,96) and Rad51 foci are induced in primary human fibroblasts following treatment with the drug (97). Thus, while the complexity of repair pathways increases in higher eukaryotes and humans, it appears that the fundamental events triggered by the formation of topoisomerase II-associated DNA breaks are similar between lower and higher eukaryotic species.
In summary, topoisomerase II-generated DNA damage appears to be repaired in S.cerevisiae primarily by the single-strand invasion pathway of homologous recombination. Recent studies indicate that similar homologous recombination pathways, in addition to non-homologous end joining, are induced in mammalian cells by the presence of double-strand chromosomal breaks (48,51,52,55–61). Results with genetically defined systems, such as yeast, provide a framework for the formulation of testable hypotheses that may help to define the cellular response to topoisomerase II-generated DNA damage in human cells.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Michele Nadaf for assistance with the FACS analysis, to Dr Mary-Ann Bjornsti for helpful discussions and insights and to Kennneth D. Bromberg and Renier Vélez-Cruz for critical reading of the manuscript. This work was supported by grants GM33944 and GM53960 (to N.O.), CA82313 (to J.L.N.) and CA52814 (to J.L.N.) from the National Institutes of Health, support from the American Lebanese Syrian Associated Charities (ALSAC) (to J.L.N.) and Grants-in-Aid for Scientific Research (B) and Scientific Research on Priority Areas (B) from the Ministry of Education, Science, Sports and Culture of Japan (to H.I.). M.S. was a trainee under National Institutes of Health Grant 5 T32 CA09582.
REFERENCES
- 1.Wang J.C. (1996) DNA topoisomerases. Annu. Rev. Biochem., 65, 635–692. [DOI] [PubMed] [Google Scholar]
- 2.Burden D.A. and Osheroff,N. (1998) Mechanism of action of eukaryotic topoisomerase II and drugs targeted to the enzyme. Biochim. Biophys. Acta, 1400, 139–154. [DOI] [PubMed] [Google Scholar]
- 3.Nitiss J.L. (1998) Investigating the biological functions of DNA topoisomerases in eukaryotic cells. Biochim. Biophys. Acta, 1400, 63–81. [DOI] [PubMed] [Google Scholar]
- 4.Wang J.C. (1998) Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys., 31, 107–144. [DOI] [PubMed] [Google Scholar]
- 5.Fortune J.M. and Osheroff,N. (2000) Topoisomerase II as a target for anticancer drugs: when enzymes stop being nice. Prog. Nucleic Acid Res. Mol. Biol., 64, 221–253. [DOI] [PubMed] [Google Scholar]
- 6.Champoux J.J. (2001) DNA topoisomerases: structure, function and mechanism. Annu. Rev. Biochem., 70, 369–413. [DOI] [PubMed] [Google Scholar]
- 7.Sabourin M. and Osheroff,N. (2002) Topoisomerases. In Creighton,T.E. (ed.), Encyclopedia of Molecular Medicine. John Wiley & Sons, New York, NY, Vol. 5, pp. 3192–3197. [Google Scholar]
- 8.Wilstermann A.M. and Osheroff,N. (2003) Stabilization of eukaryotic topoisomerase II-DNA cleavage complexes. Curr. Top. Med. Chem., 3, 321–338. [DOI] [PubMed] [Google Scholar]
- 9.Rowley J.D. (1998) Chromosome translocations: dangerous liaisons. J. Lab. Clin. Med., 132, 244–250. [DOI] [PubMed] [Google Scholar]
- 10.Baguley B.C. and Ferguson,L.R. (1998) Mutagenic properties of topoisomerase-targeted drugs. Biochim. Biophys. Acta, 1400, 213–222. [DOI] [PubMed] [Google Scholar]
- 11.Felix C.A. (2001) Leukemias related to treatment with DNA topoisomerase II inhibitors. Med. Pediatr. Oncol., 36, 525–535. [DOI] [PubMed] [Google Scholar]
- 12.Holm C., Covey,J.M., Kerrigan,D. and Pommier,Y. (1989) Differential requirement of DNA replication for the cytotoxicity of DNA topoisomerase I and II inhibitors in Chinese hamster DC3F cells. Cancer Res., 49, 6365–6368. [PubMed] [Google Scholar]
- 13.Corbett A.H. and Osheroff,N. (1993) When good enzymes go bad: conversion of topoisomerase II to a cellular toxin by antineoplastic drugs. Chem. Res. Toxicol., 6, 585–597. [DOI] [PubMed] [Google Scholar]
- 14.Howard M.T., Neece,S.H., Matson,S.W. and Kreuzer,K.N. (1994) Disruption of a topoisomerase-DNA cleavage complex by a DNA helicase. Proc. Natl Acad. Sci. USA, 91, 12031–12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Arpa P. (1994) Determinants of cellular sensitivity to topoisomerase-targeting antitumor drugs. Adv. Pharmacol., 29B, 127–143. [DOI] [PubMed] [Google Scholar]
- 16.Chen A.Y. and Liu,L.F. (1994) DNA topoisomerases: essential enzymes and lethal targets. Annu. Rev. Pharmacol. Toxicol., 34, 191–218. [DOI] [PubMed] [Google Scholar]
- 17.Nitiss J. and Wang,J.C. (1988) DNA topoisomerase-targeting antitumor drugs can be studied in yeast. Proc. Natl Acad. Sci. USA, 85, 7501–7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersson H.C. and Kihlman,B.A. (1989) The production of chromosomal alterations in human lymphocytes by drugs known to interfere with the activity of DNA topoisomerase II. I. m-AMSA. Carcinogenesis, 10, 123–130. [DOI] [PubMed] [Google Scholar]
- 19.Han Y.H., Austin,M.J., Pommier,Y. and Povirk,L.F. (1993) Small deletion and insertion mutations induced by the topoisomerase II inhibitor teniposide in CHO cells and comparison with sites of drug-stimulated DNA cleavage in vitro. J. Mol. Biol., 229, 52–66. [DOI] [PubMed] [Google Scholar]
- 20.Kaufmann S.H. (1998) Cell death induced by topoisomerase-targeted drugs: more questions than answers. Biochim. Biophys. Acta, 1400, 195–211. [DOI] [PubMed] [Google Scholar]
- 21.Pommier Y., Fesen,M.R. and Goldwasser,F. (1996) Topoisomerase II inhibitors: the epipodophyllotoxins, m-AMSA and the ellipticine derivatives. In Chabner,B.A. and Longo,D.L. (eds), Cancer Chemotherapy and Biotherapy: Principles and Practice, 2nd Edn. Lippincott-Raven, Philadelphia, PA, pp. 435–461. [Google Scholar]
- 22.Pommier Y. (1997) DNA topoisomerase II inhibitors. In Teicher,B.A. (ed.), Cancer Therapeutics: Experimental and Clinical Agents. Humana Press, Totowa, NJ, Vol. I, pp. 153–174. [Google Scholar]
- 23.Hande K.R. (1998) Clinical applications of anticancer drugs targeted to topoisomerase II. Biochim. Biophys. Acta, 1400, 173–184. [DOI] [PubMed] [Google Scholar]
- 24.Li T.K. and Liu,L.F. (2001) Tumor cell death induced by topoisomerase-targeting drugs. Annu. Rev. Pharmacol. Toxicol., 41, 53–77. [DOI] [PubMed] [Google Scholar]
- 25.Walker J.V. and Nitiss,J.L. (2002) DNA topoisomerase II as a target for cancer chemotherapy. Cancer Invest., 20, 570–589. [DOI] [PubMed] [Google Scholar]
- 26.Kreuzer K.N. and Cozzarelli,N.R. (1979) Escherichia coli mutants thermosensitive for deoxyribonucleic acid gyrase subunit A: effects on deoxyribonucleic acid replication, transcription and bacteriophage growth. J. Bacteriol., 140, 424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Felix C.A. (1998) Secondary leukemias induced by topoisomerase-targeted drugs. Biochim. Biophys. Acta, 1400, 233–255. [DOI] [PubMed] [Google Scholar]
- 28.Rowley J.D. (1998) The critical role of chromosome translocations in human leukemias. Annu. Rev. Genet., 32, 495–519. [DOI] [PubMed] [Google Scholar]
- 29.Smith M.A., Rubinstein,L., Anderson,J.R., Arthur,D., Catalano,P.J., Freidlin,B., Heyn,R., Khayat,A., Krailo,M., Land,V.J., Miser,J., Shuster,J. and Vena,D. (1999) Secondary leukemia or myelodysplastic syndrome after treatment with epipodophyllotoxins. J. Clin. Oncol., 17, 569–577. [DOI] [PubMed] [Google Scholar]
- 30.Ross J.A., Potter,J.D., Reaman,G.H., Pendergrass,T.W. and Robison,L.L. (1996) Maternal exposure to potential inhibitors of DNA topoisomerase II and infant leukemia (United States): a report from the Children’s Cancer Group. Cancer Causes Control, 7, 581–590. [DOI] [PubMed] [Google Scholar]
- 31.Ross J.A. (1998) Maternal diet and infant leukemia: a role for DNA topoisomerase II inhibitors? Int. J. Cancer, 11 (suppl.), 26–28. [PubMed] [Google Scholar]
- 32.Takagi E., Kawatsu,H., Shimokata,K., Nishiyama,Y., Kojima,K. and Yoshida,S. (1988) Establishment and characterization of resistant cells to etoposide (VP16) from a mouse breast cancer cell line, FM3A. Jpn. J. Cancer Res., 79, 938–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beck W.T. (1990) Mechanisms of multidrug resistance in human tumor cells. The roles of P-glycoprotein, DNA topoisomerase II and other factors [Review]. Cancer Treat. Rev., 17, 11–20. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y.N., Mickley,L.A., Schwartz,A.M., Acton,E.M., Hwang,J.L. and Fojo,A.T. (1990) Characterization of adriamycin-resistant human breast cancer cells which display overexpression of a novel resistance-related membrane protein. J. Biol. Chem., 265, 10073–10080. [PubMed] [Google Scholar]
- 35.Larsen A.K. and Skladanowski,A. (1998) Cellular resistance to topoisomerase targeted drugs: from drug uptake to cell death. Biochim. Biophys. Acta, 1400, 257–274. [DOI] [PubMed] [Google Scholar]
- 36.Felix C.A., Walker,A.H., Lange,B.J., Williams,T.M., Winick,N.J., Cheung,N.K., Lovett,B.D., Nowell,P.C., Blair,I.A. and Rebbeck,T.R. (1998) Association of CYP3A4 genotype with treatment-related leukemia. Proc. Natl Acad. Sci. USA, 95, 13176–13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elsea S.H., Osheroff,N. and Nitiss,J.L. (1992) Cytotoxicity of quinolones toward eukaryotic cells. Identification of topoisomerase II as the primary cellular target for the quinolone CP-115,953 in yeast. J. Biol. Chem., 267, 13150–13153. [PubMed] [Google Scholar]
- 38.Nitiss J.L., Liu,Y.X., Harbury,P., Jannatipour,M., Wasserman,R. and Wang,J.C. (1992) Amsacrine and etoposide hypersensitivity of yeast cells overexpressing DNA topoisomerase II. Cancer Res., 52, 4467–4472. [PubMed] [Google Scholar]
- 39.Beck W.T., Danks,M.K., Wolverton,J.S., Kim,R. and Chen,M. (1993) Drug resistance associated with altered DNA topoisomerase II. Adv. Enzyme Regul., 33, 113–127. [DOI] [PubMed] [Google Scholar]
- 40.Feldhoff P.W., Mirski,S.E., Cole,S.P. and Sullivan,D.M. (1994) Altered subcellular distribution of topoisomerase II alpha in a drug-resistant human small cell lung cancer cell line. Cancer Res., 54, 756–762. [PubMed] [Google Scholar]
- 41.Withoff S., De Jong,S., De Vries,E.G.E. and Mulder,N.H. (1996) Human DNA topoisomerase II: biochemistry and role in chemotherapy resistance. Anticancer Res., 16, 1867–1880. [PubMed] [Google Scholar]
- 42.Dingemans A.M.C., Pinedo,H.M. and Giaccone,G. (1998) Clinical resistance to topoisomerase-targeted drugs. Biochim. Biophys. Acta, 1400, 275–288. [DOI] [PubMed] [Google Scholar]
- 43.Mirski S.E., Sparks,K.E., Yu,Q., Lang,A.J., Jain,N., Campling,B.G. and Cole,S.P. (2000) A truncated cytoplasmic topoisomerase II alpha in a drug-resistant lung cancer cell line is encoded by a TOP2A allele with a partial deletion of exon 34. Int. J. Cancer, 85, 534–539. [DOI] [PubMed] [Google Scholar]
- 44.McDaniel L.S., Rogers,L.H. and Hill,W.E. (1978) Survival of recombination-deficient mutants of Escherichia coli during incubation with nalidixic acid. J. Bacteriol., 134, 1195–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lewin C.S., Howard,B.M., Ratcliffe,N.T. and Smith,J.T. (1989) 4-Quinolones and the SOS response. J. Med. Microbiol., 29, 139–144. [DOI] [PubMed] [Google Scholar]
- 46.Woodworth D.L. and Kreuzer,K.N. (1996) Bacteriophage T4 mutants hypersensitive to an antitumor agent that induces topoisomerase-DNA cleavage complexes. Genetics, 143, 1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neece S.H., Carles-Kinch,K., Tomso,D.J. and Kreuzer,K.N. (1996) Role of recombinational repair in sensitivity to an antitumour agent that inhibits bacteriophage T4 type II DNA topoisomerase. Mol. Microbiol., 20, 1145–1154. [DOI] [PubMed] [Google Scholar]
- 48.Lieber M.R. (1999) The biochemistry and biological significance of nonhomologous DNA end joining: an essential repair process in multicellular eukaryotes. Genes Cells, 4, 77–85. [DOI] [PubMed] [Google Scholar]
- 49.Paques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haber J.E. (2000) Partners and pathways repairing a double-strand break. Trends Genet., 16, 259–264. [DOI] [PubMed] [Google Scholar]
- 51.Johnson R.D. and Jasin,M. (2001) Double-strand-break-induced homologous recombination in mammalian cells. Biochem. Soc. Trans, 29, 196–201. [DOI] [PubMed] [Google Scholar]
- 52.Jackson S.P. (2002) Sensing and repairing DNA double-strand breaks. Carcinogenesis, 23, 687–696. [DOI] [PubMed] [Google Scholar]
- 53.Pedersen-Bjergaard J. and Rowley,J.D. (1994) The balanced and the unbalanced chromosome aberrations of acute myeloid leukemia may develop in different ways and may contribute differently to malignant transformation. Blood, 83, 2780–2786. [PubMed] [Google Scholar]
- 54.Lovett B.D., Lo Nigro,L., Rappaport,E.F., Blair,I.A., Osheroff,N., Zheng,N., Megonigal,M.D., Williams,W.R., Nowell,P.C. and Felix,C.A. (2001) Near-precise interchromosomal recombination and functional DNA topoisomerase II cleavage sites at MLL and AF-4 genomic breakpoints in treatment-related acute lymphoblastic leukemia with t(4;11) translocation. Proc. Natl Acad. Sci. USA, 98, 9802–9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sargent R.G., Brenneman,M.A. and Wilson,J.H. (1997) Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol. Cell. Biol., 17, 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liang F., Han,M., Romanienko,P.J. and Jasin,M. (1998) Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl Acad. Sci. USA, 95, 5172–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takata M., Sasaki,M.S., Sonoda,E., Morrison,C., Hashimoto,M., Utsumi,H., Yamaguchi-Iwai,Y., Shinohara,A. and Takeda,S. (1998) Homologous recombination and non-homologous end-joining pathways of DNA doublestrand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J., 17, 5497–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thacker J. (1999) The role of homologous recombination processes in the repair of severe forms of DNA damage in mammalian cells. Biochimie, 81, 77–85. [DOI] [PubMed] [Google Scholar]
- 59.Thompson L.H. and Schild,D. (1999) The contribution of homologous recombination in preserving genome integrity in mammalian cells. Biochimie, 81, 87–105. [DOI] [PubMed] [Google Scholar]
- 60.Haber J.E. (2000) Recombination: a frank view of exchanges and vice versa. Curr. Opin. Cell Biol., 12, 286–292. [DOI] [PubMed] [Google Scholar]
- 61.Richardson C. and Jasin,M. (2000) Frequent chromosomal translocations induced by DNA double-strand breaks. Nature, 405, 697–700. [DOI] [PubMed] [Google Scholar]
- 62.Morrison C. and Takeda,S. (2000) Genetic analysis of homologous DNA recombination in vertebrate somatic cells. Int. J. Biochem. Cell Biol., 32, 817–831. [DOI] [PubMed] [Google Scholar]
- 63.Aguilera A., Chavez,S. and Malagon,F. (2000) Mitotic recombination in yeast: elements controlling its incidence. Yeast, 16, 731–754. [DOI] [PubMed] [Google Scholar]
- 64.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 65.Rothstein R. (1991) Targeting, disruption, replacement and allele rescue: integrative DNA transformation in yeast. In Guthrie,C. and Fink,G.R. (eds), Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego, CA, Vol. 194, pp. 281–301. [DOI] [PubMed] [Google Scholar]
- 66.Yamagata K., Kato,J., Shimamoto,A., Goto,M., Furuichi,Y. and Ikeda,H. (1998) Bloom’s and Werner’s syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc. Natl Acad. Sci. USA, 95, 8733–8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsukamoto Y., Kato,J. and Ikeda,H. (1996) Hdf1, a yeast Ku-protein homologue, is involved in illegitimate recombination, but not in homologous recombination. Nucleic Acids Res., 24, 2067–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tsukamoto Y., Kato,J. and Ikeda,H. (1996) Effects of mutations of RAD50, RAD51, RAD52 and related genes on illegitimate recombination in Saccharomyces cerevisiae. Genetics, 142, 383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Burns C.G., Ohi,R., Mehta,S., O’Toole,E.T., Winey,M., Clark,T.A., Sugnet,C.W., Ares,M.,Jr and Gould,K.L. (2002) Removal of a single alpha-tubulin gene intron suppresses cell cycle arrest phenotypes of splicing factor mutations in Saccharomyces cerevisiae. Mol. Cell. Biol., 22, 801–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hopfner K.P., Putnam,C.D. and Tainer,J.A. (2002) DNA double-strand break repair from head to tail. Curr. Opin. Struct. Biol., 12, 115–122. [DOI] [PubMed] [Google Scholar]
- 71.Moore J.K. and Haber,J.E. (1996) Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 2164–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lewis L.K. and Resnick,M.A. (2000) Tying up loose ends: nonhomologous end-joining in Saccharomyces cerevisiae. Mutat. Res., 451, 71–89. [DOI] [PubMed] [Google Scholar]
- 73.Pierce A.J., Stark,J.M., Araujo,F.D., Moynahan,M.E., Berwick,M. and Jasin,M. (2001) Double-strand breaks and tumorigenesis. Trends Cell Biol., 11, S52–S59. [DOI] [PubMed] [Google Scholar]
- 74.Osheroff N. (1989) Effect of antineoplastic agents on the DNA cleavage/religation reaction of eukaryotic topoisomerase II: inhibition of DNA religation by etoposide. Biochemistry, 28, 6157–6160. [DOI] [PubMed] [Google Scholar]
- 75.Nitiss J.L., Liu,Y.X. and Hsiung,Y. (1993) A temperature sensitive topoisomerase II allele confers temperature dependent drug resistance on amsacrine and etoposide: a genetic system for determining the targets of topoisomerase II inhibitors. Cancer Res., 53, 89–93. [PubMed] [Google Scholar]
- 76.Hsiung Y., Elsea,S.H., Osheroff,N. and Nitiss,J.L. (1995) A mutation in yeast TOP2 homologous to a quinolone-resistant mutation in bacteria. Mutation of the amino acid homologous to Ser83 of Escherichia coli gyrA alters sensitivity to eukaryotic topoisomerase inhibitors. J. Biol. Chem., 270, 20359–20364. [DOI] [PubMed] [Google Scholar]
- 77.Wolfson J.S. and Hooper,D.C. (1989) Bacterial resistance to quinolones: mechanisms and clinical importance. Rev. Infect. Dis., 11, S960–968. [DOI] [PubMed] [Google Scholar]
- 78.Reece R.J. and Maxwell,A. (1991) DNA gyrase: structure and function. Crit. Rev. Biochem. Mol. Biol., 26, 335–375. [DOI] [PubMed] [Google Scholar]
- 79.Hooper D.C. (1998) Bacterial topoisomerases, anti-topoisomerases and anti-topoisomerase resistance. Clin. Infect. Dis., 27, S54-S63. [DOI] [PubMed] [Google Scholar]
- 80.Hooper D.C. (1993) Quinolone mode of action—new aspects. Drugs, 3, 8–14. [DOI] [PubMed] [Google Scholar]
- 81.Willmott C.J. and Maxwell,A. (1993) A single point mutation in the DNA gyrase A protein greatly reduces binding of fluoroquinolones to the gyrase-DNA complex. Antimicrob. Agents Chemother., 37, 126–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Strumberg D., Nitiss,J.L., Dong,J., Kohn,K.W. and Pommier,Y. (1999) Molecular analysis of yeast and human type II topoisomerases. Enzyme-DNA and drug interactions. J. Biol. Chem., 274, 28246–28255. [DOI] [PubMed] [Google Scholar]
- 83.Reid R.J., Fiorani,P., Sugawara,M. and Bjornsti,M.A. (1999) CDC45 and DPB11 are required for processive DNA replication and resistance to DNA topoisomerase I-mediated DNA damage. Proc. Natl Acad. Sci. USA, 96, 11440–11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pouliot J.J., Yao,K.C., Robertson,C.A. and Nash,H.A. (1999) Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science, 286, 552–555. [DOI] [PubMed] [Google Scholar]
- 85.Reagan M.S., Pittenger,C., Siede,W. and Friedberg,E.C. (1995) Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J. Bacteriol., 177, 364–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sommers C.H., Miller,E.J., Dujon,B., Prakash,S. and Prakash,L. (1995) Conditional lethality of null mutations in RTH1 that encodes the yeast counterpart of a mammalian 5′- to 3′-exonuclease required for lagging strand DNA synthesis in reconstituted systems. J. Biol. Chem., 270, 4193–4196. [DOI] [PubMed] [Google Scholar]
- 87.Wu X. and Wang,Z. (1999) Relationships between yeast Rad27 and Apn1 in response to apurinic/apyrimidinic (AP) sites in DNA. Nucleic Acids Res., 27, 956–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu X., Wilson,T.E. and Lieber,M.R. (1999) A role for FEN-1 in nonhomologous DNA end joining: the order of strand annealing and nucleolytic processing events. Proc. Natl Acad. Sci. USA, 96, 1303–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Byl J.A., Cline,S.D., Utsugi,T., Kobunai,T., Yamada,Y. and Osheroff,N. (2001) DNA topoisomerase II as the target for the anticancer drug TOP-53: mechanistic basis for drug action. Biochemistry, 40, 712–718. [DOI] [PubMed] [Google Scholar]
- 90.You J.C. (2000) The effects of RAD52 epistasis group genes on various types of spontaneous mitotic recombination in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun., 270, 112–118. [DOI] [PubMed] [Google Scholar]
- 91.Rowley J.D. (1994) 1993 Robert R. de Villiers Lecture. Chromosome translocations: dangerous liaisons. Leukemia, 8, S1–S6. [PubMed] [Google Scholar]
- 92.Drewinko B. and Barlogie,B. (1976) Survival and cycle-progression delay of human lymphoma cells in vitro exposed to VP-16-213. Cancer Treat. Rep., 60, 1295–1306. [PubMed] [Google Scholar]
- 93.Kalwinsky D.K., Look,A.T., Ducore,J. and Fridland,A. (1983) Effects of the epipodophyllotoxin VP-16-213 on cell cycle traverse, DNA synthesis and DNA strand size in cultures of human leukemic lymphoblasts. Cancer Res., 43, 1592–1597. [PubMed] [Google Scholar]
- 94.Kaufmann W.K. and Kies,P.E. (1998) DNA signals for G2 checkpoint response in diploid human fibroblasts. Mutat. Res., 400, 153–167. [DOI] [PubMed] [Google Scholar]
- 95.Elli R., Chessa,L., Antonelli,A., Petrinelli,P., Ambra,R. and Marcucci,L. (1996) Effects of topoisomerase II inhibition in lymphoblasts from patients with progeroid and “chromosome instability” syndromes. Cancer Genet. Cytogenet., 87, 112–116. [DOI] [PubMed] [Google Scholar]
- 96.Pichierri P., Franchitto,A., Mosesso,P., Proietti de Santis,L., Balajee,A.S. and Palitti,F. (2000) Werner’s syndrome lymphoblastoid cells are hypersensitive to topoisomerase II inhibitors in the G2 phase of the cell cycle. Mutat. Res., 459, 123–133. [DOI] [PubMed] [Google Scholar]
- 97.Raderschall E., Golub,E.I. and Haaf,T. (1999) Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl Acad. Sci. USA, 96, 1921–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Alani E., Subbiah,S. and Kleckner,N. (1989) The yeast RAD50 gene encodes a predicted 153-kD protein containing a purine nucleotide-binding domain and two large heptad-repeat regions. Genetics, 122, 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Longtine M.S., McKenzie,A.,III, Demarini,D.J., Shah,N.G., Wach,A., Brachat,A., Philippsen,P. and Pringle,J.R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast, 14, 953–961. [DOI] [PubMed] [Google Scholar]
- 100.Feldmann H. and Winnacker,E.L. (1993) A putative homologue of the human autoantigen Ku from Saccharomyces cerevisiae. J. Biol. Chem., 268, 12895–12900. [PubMed] [Google Scholar]