Abstract
Previous studies have established that the Snf2h-containing chromatin remodeling complex NoRC mediates epigenetic silencing of a subset of rRNA genes (rDNA) by recruiting enzymatic activities that modify histones and methylate DNA. Here we have analyzed nucleosome positions at the murine rDNA promoter and show that active and silent rDNA copies are characterized not only by specific epigenetic marks but also by differently positioned nucleosomes. At active genes the promoter-bound nucleosome covers nucleotides from −157 to −2, whereas at silent genes the nucleosome is positioned 25 nucleotides further downstream. We provide evidence that NoRC is the molecular machine that shifts the promoter-bound nucleosome downstream of the transcription start site into a translational position that is unfavorable for transcription complex formation.
Keywords: differentiation, NoRC, nucleosome positioning, RNA polymerase I, TIP5
Introduction
Chromatin has an essential role both in packaging the genome and regulating its function at specific genes. This dual role is brought about by a wide variety of enzymes that covalently modify histones and/or DNA, or affect chromatin structure by disrupting histone–DNA contacts. The biochemical properties of chromatin-modifying enzymes are well characterized, and links between histone- and DNA-modifying enzymes and ATP-dependent chromatin remodeling complexes have been established. Chromatin remodeling enzymes use the energy of ATP to alter the structure or positioning of nucleosomes, thus modulating the access of DNA binding proteins. Hence chromatin remodeling complexes are involved in the regulation of DNA-dependent processes, like transcription, replication, repair and recombination. One unifying view is that the ATPase subunit of the Swi/Snf2 subfamily acts as a molecular motor that facilitates dynamic changes in chromatin structure at both active and inactive genes. Chromatin remodeling complexes have been classified into Swi2-, ISWI-, CHD- or INO80-containing complexes and shown to play multiple roles in chromosome organization, DNA replication, transcription activation and repression (Corona and Tamkun, 2004).
Several epigenetic characteristics distinguish potentially active from inactive mammalian rRNA genes (rDNA), including differential DNA methylation and specific histone modifications (Grummt and Pikaard, 2003; McStay, 2006). On active gene copies, the promoter is unmethylated and histone tails are demarcated by euchromatic modifications, such as acetylation of histone H4 and methylation of histone H3 at lysine 4. In contrast, the promoter of silent genes exhibits heterochromatic features (Santoro and Grummt, 2005). In exploring the epigenetic control mechanism by which the active or silent state of rDNA is inherited, we have identified NoRC, a remodeling complex consisting of Snf2h and TIP5 that localizes within nucleoli (Strohner et al, 2001). NoRC recruits DNA methyltransferase and histone-modifying activities to the rDNA promoter, thereby mediating heterochromatin formation and rDNA silencing (Santoro et al, 2002). Previous studies have revealed a hierarchy of epigenetic events that trigger rDNA silencing, starting with NoRC being recruited to rDNA by TTF-I bound to the promoter-proximal terminator T0 (Nemeth et al, 2004; Strohner et al, 2004; Santoro and Grummt, 2005). NoRC interacts with the Sin3 corepressor complex and DNA methyltransferases, which deacetylate nucleosomes and methylate rDNA, respectively. Methylation of CpG-133 impairs UBF binding to chromatin and inhibits transcription complex formation (Santoro and Grummt, 2001). However, despite these advances in understanding single steps by which the silent state of rDNA is established, the function of the ATP-dependent chromatin remodeling activity of NoRC remained elusive.
In this study, we have analyzed the role of NoRC in nucleosome positioning at the rDNA promoter in vivo. We have identified two distinct translational nucleosome positions that characterize active and silent rDNA repeats, respectively. Whereas the ratio of ‘active' versus ‘silent' nucleosome positions is approximately 1:1 in NIH3T3 cells, this ratio changes to at least 1:5 in differentiated cells or in cells overexpressing NoRC. The results reveal that NoRC—in addition to its established role in recruiting histone-modifying enzymes and DNA methyltransferases to rDNA—plays an active role in nucleosome dynamics at the rDNA promoter, and specific nucleosome positions determine the transcriptional readout of rRNA genes.
Results
Mapping of nucleosome positions at the rDNA promoter
To examine nucleosome positions at the mouse rDNA promoter in vivo, crosslinked cells were permeabilized with lysolecithin, chromatin was digested with MNase and mononucleosome-sized DNA was subjected to LM-PCR using a linker- and an rDNA-specific primer (Figure 1A). The positions of MNase cleavage sites were mapped from both directions using different rDNA-specific primers that amplify the top or bottom strand of DNA. The 5′ border of the nucleosomes was determined by using two reverse primers harboring rDNA sequences from −1 to −21 (primer A) and from −20 to −39 (primer B). The 3′ ends of nucleosomal DNA fragments were mapped with the forward primer C (from −105 to −87). Amplification of the reaction products with each of the rDNA primers yielded two DNA fragments (Figure 1B), suggesting the presence of two rDNA populations that are discriminated by the position of promoter-bound nucleosomes. The lengths of the two DNA fragments differ by 20–30 bp, indicating that the two nucleosome positions are 20–30 bp apart from each other.
Figure 1.
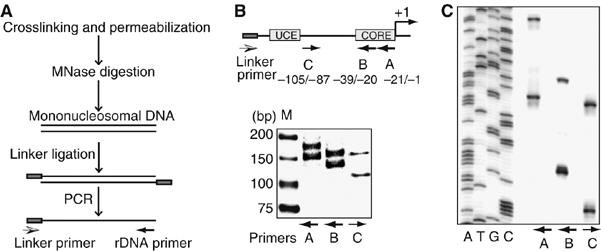
Mapping of nucleosome positions at the rDNA promoter. (A) Schematic representation of the method used to map nucleosome positions. The black lines represent mononucleosomal genomic DNA. The box indicates staggered linker. The arrows represent the primers used for LM-PCR mapping. (B) Mapping of nucleosome positions at the murine rDNA promoter. Mononucleosome-sized DNA from crosslinked NIH3T3 cells was subjected to LM-PCR using the linker primer and either of the rDNA-specific primers shown at the top. 32P-labeled PCR products were analyzed by PAGE. Lane M refers to the DNA marker. The scheme on top indicates the location of the core promoter and the UCE relative to the transcription start site at +1. The horizontal arrows represent the position of primers used for LM-PCR mapping. (C) Sequencing gel used to determine the lengths of LM-PCR products. Lanes A, T, G and C show products of sequencing reactions. The adjacent lanes show the LM-PCR products obtained with the linker primer and rDNA-specific primers A, B and C as indicated.
To map the MNase cleavage sites at the nucleotide level of resolution, the LM-PCR products were separated on a denaturing gel along with a sequencing reaction of murine rDNA (Figure 1C). After subtraction of the 22 bp linker DNA, the lengths of LM-PCR fragments place the 5′ edge of the translationally positioned nucleosomes to nucleotides −157 and −132 with respect to the initiation site, that is, within the upstream control element (UCE) and the core promoter. The 3′ edges of the two translationally positioned nucleosomes are at nucleotides −2 and +22, respectively. Thus, the two positioned nucleosomes extend from −157 to −2 and from −132 to +22. The observation that the two nucleosome positions differ by 25 bp suggests that regulatory DNA elements that are exposed to the surface of the histone octamer in one translational frame should be inaccessible within the other translational frame. Of note, previous mapping of nucleosome positions on in vitro assembled nucleosomal arrays revealed a dominant nucleosome position with the 3′ end at position +22, if both NoRC and TTF-I were present in the remodeling assays (Strohner et al, 2004). The finding that the 3′ boundary of the more downstream nucleosome is identical to the position that is triggered by TTF-I and NoRC on reconstituted chromatin implies that this nucleosome position represents the inactive rDNA population. Apparently, NoRC recognizes a specific sequence and/or structural feature of the rDNA promoter and shifts the 3′ edge of the promoter-bound nucleosome to a preferred position downstream of the transcription start site.
Different nucleosome positions at active and silent rDNA promoters
The finding that there are two distinct nucleosome positions at the rDNA promoter supports the view that the different accessibility and transcriptional potential of active and silent rDNA copies is not only due to the establishment of specific heterochromatic features at the promoter but also to different nucleosomal positions at active and silent rDNA arrays. Active and silent rRNA genes can be distinguished by the degree of promoter methylation, active copies being unmethylated and therefore sensitive to HpaII digestion, whereas silent ones are methylated and resistant to HpaII digestion (Santoro and Grummt, 2001). If nucleosomes were differently positioned at active and silent rDNA repeats, then the DNA covered by the nucleosome should either be sensitive or resistant to HpaII digestion, depending on whether or not the HpaII site at −143 was methylated. As shown in Figure 2A, the upper fragment (U) that corresponds to the more upstream positioned nucleosome was not amplified, indicating that the nucleosomal DNA was unmethylated and therefore cleaved by HpaII. This demonstrates that at transcriptionally active rRNA genes, a positioned nucleosome covering sequences from −157 to −2 places the core promoter (CORE) and the UCE at the edges of the nucleosome.
Figure 2.
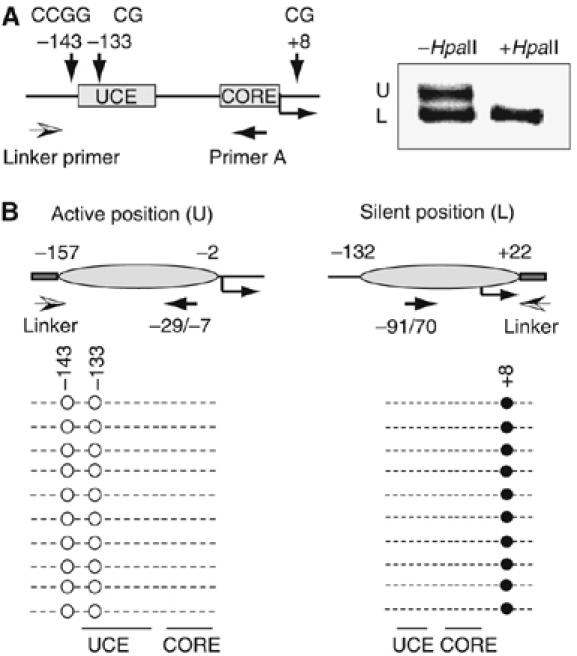
Nucleosomes are differently positioned at the promoter of active and silent rRNA genes. (A) Nucleosome positions at the promoter of methylated and unmethylated rDNA repeats. Nucleosomal DNA was incubated in the absence or presence of HpaII before amplification by LM-PCR. The transcription start site, the HpaII site at −143, the CpG residues at −133 and +8 and the primers used for LM-PCR are illustrated above. (B) Methylation analysis of specific CpG residues. The scheme on top shows the position of CpG residues within the murine rDNA promoter. The methylation status of each CpG was determined by bisulfite treatment of mononucleosomal DNA, cloning of the upper and lower LM-PCR fragment and sequencing of 10 randomly selected clones. Each row represents the sequence of an individual clone. The open and filled circles denote unmethylated and methylated CpGs, respectively, the arrows indicate the primers used for LM-PCR and the ellipses show the position of nucleosomes at the promoter of active and silent rDNA copies.
The smaller PCR product covers sequences from −132 to +22 and does not contain the CCGG site at −143. Therefore, it is not surprising that this fragment was amplified after treatment with HpaII. In order to monitor the methylation status of the shorter PCR product, we performed bisulfite sequencing of both LM-PCR fragments. The murine rDNA promoter contains CpG base pairs at −143, −133 and +8. Previous studies have established that all three CpG residues are unmethylated in active genes and methylated in silent gene copies (Santoro and Grummt, 2001). In the experiment in Figure 2B, mononucleosomal DNA was treated with bisulfite before LM-PCR amplification and cloning of the upper and lower fragment. Sequencing of 10 selected clones revealed that bisulfite treatment has converted the CpG residues at −143 and −133 in the upper fragment to uracils that were amplified as thymine during PCR, that is, they were unmethylated. This is consistent with the observed HpaII sensitivity and supports the notion that a positioned nucleosome spanning sequences from −157 to −2 covers the promoter of active rRNA genes. In contrast, all clones derived from the faster migrating DNA fragment contained sequences from −132 to +22, and the only CpG residue at +8 remained unconverted. This indicates that the DNA fragment covered by the more downstream positioned nucleosome was methylated, that is, represents the fraction of silent rDNA copies. Noteworthy, in this ‘inactive' position, the critical CpG dinucleotide at −133 is at the 5′ boundary of the positioned nucleosome.
Overexpression of TIP5 increases the number of nucleosomes in the ‘silent' position
Given that NoRC is required for rDNA silencing, the ratio of ‘active' versus ‘silent' nucleosome positions at the murine rDNA promoter should be decreased if the level of cellular NoRC was elevated. To test this, we used a cell line, termed 3T3/TIP5, which stably overexpresses TIP5, the large subunit of NoRC (Li et al, 2005). In 3T3/TIP5 cells, the level of pre-rRNA was decreased, the size and number of nucleoli was reduced and cell growth was impaired. Moreover, in parental NIH3T3 cells, ∼40% of rDNA was methylated, that is, was transcriptionally silent, whereas the level of methylated copies was increased to 80–90% in 3T3/TIP5 cells. Significantly, the ratio of the nucleosome positions at the rDNA promoter was changed upon overexpression of TIP5. As shown in Figure 3A, in NIH3T3 cells, the ratio of the two PCR fragments was similar, ∼60% of nucleosomes being in the more upstream and ∼40% in the more downstream position. In 3T3/TIP5 cells, on the other hand, the ratio of the two PCR fragments changed to 15:85%, consistent with enhanced NoRC levels increasing the proportion of downstream nucleosome positions at the expense of upstream ones. The intriguing correlation between increased rDNA methylation and elevated amounts of the downstream nucleosome in 3T3/TIP5 cells supports the view that at silent genes the promoter-bound nucleosome is located further downstream, covering sequences from −132 to +22. This indicates that transcriptional silencing involves shifting of a translationally positioned nucleosome at the rDNA promoter 25 bp further downstream into a different position, and suggests that NoRC-mediated changes in nucleosome positioning may be an important step in the establishment of a repressed rDNA promoter architecture.
Figure 3.
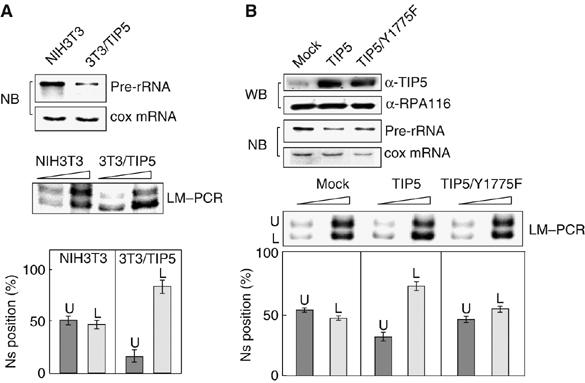
NoRC alters the ratio of ‘active' and ‘silent' nucleosomes. (A) Overexpression of TIP5 represses Pol I transcription and shifts nucleosomes into the ‘silent' position. Mononucleosomal DNA from NIH3T3 and 3T3/TIP5 cells was subjected to LM-PCR to determine specific nucleosome positions. The bar diagram below shows the percentage of the upper (dark bar) and lower PCR fragment (light bar) estimated in three independent experiments. A Northern blot (NB) monitoring the level of pre-RNA synthesis in NIH3T3 and 3T3/TIP5 cells is shown above. (B) Functional NoRC is required for shifting nucleosomes into the ‘silent' position. Flag-tagged TIP5 or TIP5/Y1775F was expressed in NIH3T3 cells by retroviral infection using pBABE, pBABE-flTIP5 or pBABE-flTIP5/Y1775F. The Western blot (upper panel, WB) shows the amount of overexpressed wild-type and mutant TIP5 (flTIP5 and flTIP5/Y1775F) as well as endogenous Pol I (α-RPA116). The Northern blot (middle panel, NB) shows the level of pre-rRNA and cytochrome c oxidase (cox) mRNA. The nucleosome positions as determined in three independent experiments are shown below (LM-PCR). The dark bars (labeled U) indicate the relative amount of nucleosomes in the ‘active' position; the light bars (labeled L) mark nucleosomes in the ‘silent' position.
To provide further evidence that NoRC is the molecular machine that moves the nucleosome into the inactive position, we sought to analyze the ratio of the two nucleosome positions after overexpressing a mutant form of TIP5 that does not trigger silencing of Pol I transcription. We used TIP5/Y1175F, a point mutant that does not associate with chromatin and is not capable of establishing heterochromatic features and transcriptional silencing (Zhou and Grummt, 2005). As shown in Figure 3B, overexpression of wild-type TIP5, but not TIP5/Y1175F, repressed Pol I transcription. Comparison of nucleosome positions by LM-PCR after overexpression of wild-type or mutant TIP5 revealed that TIP5/Y1175F did not alter the ratio of ‘active' versus ‘silent' nucleosome positions, indicating that TIP5/Y1175F was not capable of shifting ‘active' nucleosomes into the ‘inactive' position. This result demonstrates that active NoRC is required for shifting the promoter-bound nucleosome into the downstream ‘inactive' position that is associated with transcriptional silencing.
Epigenetic changes at the rDNA promoter during adipocyte differentiation
To evaluate the biological significance of NoRC-mediated chromatin remodeling, we have examined epigenetic changes at the rDNA promoter during adipocyte differentiation. For this, confluent cultures of 3T3-L1 cells were induced to differentiate into adipocytes and differentiation was monitored after 10 days by staining with the lipophilic dye Oil Red O (Figure 4A). In differentiated cells, pre-rRNA synthesis was reduced by more than 80% (Figure 4B). This decrease in Pol I transcription was accompanied by decreased levels of Pol I and UBF and an increase in TIP5 (Figure 4C). Transcriptional repression and enhanced NoRC levels were accompanied by an increase in heterochromatic histone modifications, such as hypoacetylation of histone H4 and dimethylation of histone H3 at lysine 9 (Figure 4D).
Figure 4.
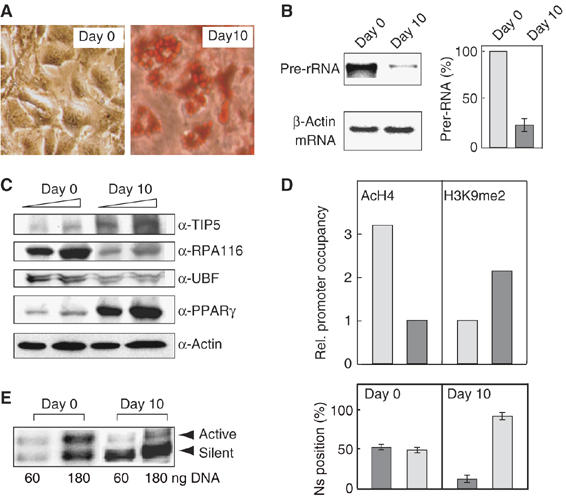
Repression of rDNA transcription in differentiated cells is accompanied by changes in nucleosome positions at the rDNA promoter. (A) Differentiation of 3T3-L1 cells into adipocytes. Phase-contrast micrographs of uninduced 3T3-L1 cells (day 0), and 10 days after induction of differentiation. The images show cells stained with Oil Red-O. (B) Pre-RNA synthesis is decreased in differentiated adipocytes. RNA from undifferentiated (day 0) and differentiated (day 10) 3T3-L1 cells was subjected to Northern blot analysis (left panel) and quantitative RT–PCR (bars) to compare the relative level of 45S pre-rRNA. The percentage of pre-rRNA levels in undifferentiated (light bar) and differentiated 3T3-L1 cells (dark bar) as determined in three independent experiments is shown. (C) Western blots showing the level of cellular Pol I (RPA116), UBF, TIP5, SIRT1, PPARγ and β-actin in undifferentiated (day 0) and differentiated (day 10) 3T3-L1 cells. (D) Heterochromatic histone modifications at the rDNA promoter are enhanced in differentiated adipocytes. The bars show the relative levels of histone H4 acetylation (AcH4) and H3K9 dimethylation (H3K9me2) in undifferentiated cells (day 0, light bars) and in differentiated adipocytes (day 10, dark bar). Values represent the average of two independent experiments. (E) Nucleosome positions are altered during adipocyte differentiation. Nucleosome positioning was analyzed in undifferentiated and differentiated 3T3-L1 cells by LM-PCR. The bar diagram shows the ratio of nucleosomes in the ‘active' (dark bars) and ‘silent' position (light bars) as determined in three independent experiments. Error bars indicate standard deviations.
To examine whether the ratio of nucleosomes in the ‘active' versus the ‘silent' position was changed in differentiated cells, we compared the nucleosome positions at the rDNA promoter in 3T3-L1 cells and adipocytes. The results in Figure 4E demonstrate that differentiation resulted in repositioning of nucleosomes, the majority of nucleosomes being shifted from the ‘active' into the ‘silent' position. The 50:50% ratio of nucleosomes in the ‘active' and ‘silent' position in undifferentiated 3T3-L1 cells (day 0) was changed to 10:90% after differentiation into adipocytes (day 10). This result demonstrates that cessation of rDNA transcription after exit from the cell cycle is caused or accompanied by an increase of NoRC, which induces a switch of nucleosome positions and mediates epigenetic changes at the rDNA promoter.
Discussion
Previous studies have demonstrated the importance of nucleosome positioning in the organization of nucleoprotein complexes at promoters and regulatory elements. Positioned nucleosomes may either occlude or facilitate binding of basal transcription factors to chromatin, thereby repressing or activating transcription. In some cases, nucleosomes are positioned as a consequence of specific factor binding, whereas in other cases certain DNA sequences are able to position nucleosomes in vitro (Grunstein, 1990; Simpson, 1991). However, most of the sequences identified in vitro fail to precisely position nucleosomes in vivo. This suggests that in addition to DNA structure and flexibility, other mechanisms define nucleosome positioning in cellular chromatin. DNA binding factors, such as the α2-MCM1 complex, actively position nucleosomes at repressed genes in yeast α-cells. This process requires the intact histone H4 tail, indicating the involvement of ISWI-like chromatin remodeling activities (Shimizu et al, 1991; Clapier et al, 2001). Likewise, in budding yeast, the global corepressor Ssn6–Tup1 is responsible for nucleosome positioning both at specific genes and the recombination enhancer at silent mating-type loci (Cooper et al, 1994; Weiss and Simpson, 1997; Kastaniotis et al, 2000; Fleming and Pennings, 2001; Li and Reese, 2001). These studies indicated that Tup1 and the Isw2 chromatin remodeling complex collaborate in nucleosome positioning and gene repression (Zhang and Reese, 2004). Although Ssn6–Tup1 is necessary for nucleosome positioning at the RNR3 gene, it requires the ISW2 remodeling complex to precisely position nucleosomes. This suggests an intimate link between mechanisms that regulate the structure and dynamics of chromatin with those that are involved in nucleosome positioning. The results of the present study support this view, demonstrating that chromatin remodeling complexes trigger changes in nucleosome positions, which in turn define the transcriptional readout.
We have identified two distinct nucleosome positions at the murine rDNA promoter that mark active and silent gene copies, respectively. At potentially active genes, a nucleosome occupies sequences from −157 to the transcription start site, whereas at silent genes, the nucleosome covers sequences from −132 to +22. The key player that shifts the nucleosome downstream of the transcription start site at silent gene copies is NoRC, the chromatin remodeling complex that recruits histone-modifying enzymes and DNA methyltransferase(s), thereby triggering heterochromatin formation and silencing of a fraction of rDNA (Strohner et al, 2001). Methylation of the rDNA promoter at CpG at −133 prevents binding of the basal transcription factor UBF to the UCE, which in turn leads to impaired preinitiation complex formation and repression of Pol I transcription (Santoro and Grummt, 2001; Santoro et al, 2002).
Previous studies have revealed that the overall organization of rDNA reconstituted into chromatin depends on binding of TTF-I to the promoter-proximal terminator T0 (Längst et al, 1997). This suggested that TTF-I might serve as a boundary factor that positions nucleosomes next to its cognate site. However, subsequent studies revealed that binding of TTF-I to T0 is required for the establishment of both the active and silent state of rDNA in vivo and in vitro, a finding that suggested a dynamic and active role of TTF-I in modulating the chromatin structure and activity state of rRNA genes. The current view is that TTF-I interacts with different chromatin remodeling complexes that recruit either specific co-activators or co-repressors to the rDNA promoter that establish the euchromatic or heterochromatic rDNA conformation. Our present data extend this view, showing that active and silent rDNA copies are not only characterized by distinct epigenetic marks but also by different nucleosome positions. Apparently, different chromatin remodeling complexes differ in their capability to move or position nucleosomes at specific DNA sequences. In support of this, targeting of NoRC to the rDNA promoter alters nucleosome positions, placing the 3′ border of the nucleosome 22 bp downstream of the transcription initiation site, that is, a position that marks silent rDNA copies.
On active genes, UBF has been shown to bind as a dimer to both the UCE and the core promoter. The tandem HMG boxes enable a UBF dimer to wrap the DNA in a right-handed direction, forming a loop of 360° once every ∼140 bp (Bazett-Jones et al, 1994; Stefanovsky et al, 2001). As a consequence, the UCE and the core promoter are brought into close proximity, providing the correct scaffolding for productive interactions between UBF and TIF-IB/SL1 bound to the two promoter elements. Wrapping the rDNA promoter by either UBF or a positioned nucleosome might be instrumental for PIC formation and transcription activation. The core and UCE elements are located within the realm of the nucleosome but are located at the opposite DNA exit sites of the nucleosome. The specific nucleosomal architecture of active genes places the DNA exit sites into close proximity, potentially allowing cooperative binding of UBF and TIF-IB/SL1 to the positioned nucleosome. Consistent with this, changing either the distance between the two promoter elements or increasing the distance of the TTF-I binding site relative to the transcription start site inactivated Pol I transcription (Clos et al, 1986; Längst et al, 1998), indicating that a proper promoter architecture is a prerequisite for the exact positioning of the promoter-bound nucleosome, which in turn is required for cooperative binding of UBF and TIF-IB/SL1.
On silent genes, a nucleosome is positioned downstream of the transcription start site and both the UBF binding site and the functionally important CpG residue at nucleotide −133 are placed into the nucleosomal linker region that is occupied by nucleosome-bound NoRC (G Längst et al, unpublished). The core element, on the other hand, has been moved inside the nucleosome and the relative alignment of the DNA element with respect to the histone octamer surface has been changed. DNA elements being accessible at the ‘active' nucleosome position are facing towards the histone octamer surface in the ‘silent' nucleosome position. As a consequence, the core promoter should be less accessible for TIF-IB/SL1 binding. Thus, while at active genes the nucleosome juxtaposes the core and UCE sequences, both sequence elements are separated at silent genes, not allowing cooperative binding of UBF and TIF-IB/SL1. This result supports the view that specific nucleosome positions affect transcription factor binding and gene expression. The identification of NoRC as the major determinant of the ‘silent' nucleosome position suggests that remodeling complexes are not only the major determinants of chromatin dynamics but are also capable of defining a specific chromatin structure. This dual function may also explain why chromatin remodeling complexes are so diverse and abundant in the cell. Differential gene regulation by specifically positioned nucleosomes is an attractive mechanism that would allow the cell to keep a high signal to noise ratio of DNA-dependent processes and reduce the complexity of regulation by establishing chromatin structures that allow or prevent binding of transcription factors to regulatory elements.
Materials and methods
Cell lines
NIH3T3 and BD EcoPack-293 Packaging cells (cat. no. 631507) were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% FCS, and antibiotics. Retroviral infection and selection with puromycin (5 μg/ml, 10 days) was performed as described (Picard et al, 2004). 3T3-L1 cells were cultured in DMEM containing 25 mM glucose, 10% FCS, 1 mM glutamine and 1 mM sodium pyruvate. At confluence, preadipocytes were cultured for 2 days in culture medium supplemented with 0.5 mM 3-isobutyl-1-methylxanthine, 1 μM dexamethasone and 2 μM insulin. Cells were cultured for 2 days in the presence of 2 μM insulin and for another 4–8 days in the absence of insulin. After this treatment, more than 90% of the cells were differentiated into adipocytes as revealed by droplet accumulation and Oil Red-O staining.
RNA analysis
Cellular RNA was isolated with TRIzol®Reagent (Invitrogen) and 45S pre-rRNA levels were monitored on Northern blots by hybridization to antisense RNA complementary to the first 155 nucleotides of unprocessed 45S pre-RNA (Voit et al, 1999). To normalize for differences in RNA loading, the filter was also hybridized with a probe that is complementary to β-actin mRNA. Alternatively, pre-rRNA was analyzed by reverse transcription followed by quantitative real-time PCR (Li et al, 2005).
Analysis of nucleosome positions by LM-PCR
LM-PCR was performed as described (McPherson et al, 1993; Soutoglou and Talianidis, 2002). 2 × 106 NIH3T3 cells were crosslinked with 1% formaldehyde for 10 min, permeabilized with 0.05% lysolecitin (Sigma) for 1 min at 37°C in 150 mM sucrose, 80 mM KCl, 35 mM HEPES (pH 7.4), 5 mM K2HPO4, 5 mM MgCl2, 0.5 mM CaCl2 and treated with 50 U of MNase (Roche) in 150 mM sucrose, 50 mM Tris–HCl (pH 7.4), 50 mM NaCl, 2 mM CaCl2 for 10 min at room temperature. Mononucleosome-sized DNA was isolated from agarose gels, phosphorylated with T4 polynucleotide kinase (New England Biolabs), ligated to linker oligonucleotides and purified DNA was amplified by PCR using the linker oligonucleotide and 5′-labeled rDNA-specific primers. PCR products were analyzed on 8% polyacrylamide gels.
Bisulfite modification of genomic DNA
Mononucleosomal DNA was ligated to linker oligonucleotides and cytosines were modified with the EpiTect Bisulfite kit (Qiagen). After washing and desulfonation, DNA was eluted and subjected to LM-PCR using primers that are specific for bisulfite-treated DNA. Purified PCR products were ligated into pCR2.1pTOPO-TA (Invitrogen) and introduced into competent cells (Dh5α). A minimum of 10 clones selected at random from each DNA were sequenced.
Chromatin immunoprecipitation and DNA methylation assays
Antibodies were incubated with crosslinked chromatin overnight at 4°C and collected with protein A agarose for 2 h. After reversal of the crosslink and digestion with proteinase K, DNA was extracted with phenol–chloroform and amplified by PCR (Santoro and Grummt, 2005). PCR products were visualized on ethidium bromide-stained 2% agarose gels or subjected to real-time PCR. The ratio of rDNA in the immunoprecipitates versus rDNA in the input chromatin was normalized to control reactions from undifferentiated cells. To monitor CpG methylation, mononucleosomal DNA was digested with 20 U HpaII before PCR amplification with the linker primer and the rDNA-specific primer A. For quantification, real-time PCR was performed with a LightCycler (Roche) using the SYBR Green detection system. Quantification of DNA methylation was performed as described (Li et al, 2005).
Antibodies
Antibodies against TIP5, RPA116 and UBF have been described (Voit et al, 1999; Strohner et al, 2001). Antibodies to modified histones were from Upstate Biotechnology.
Acknowledgments
We thank Yonggong Zhou for providing pBABE-TIP5 and pBABE-TIP5/Y1775F. This work was supported by the Deutsche Forschungsgemeinschaft (SFB/Transregio 5, SP ‘Epigenetics'), the EU-Network ‘Epigenome' and the Fonds der Chemischen Industrie.
References
- Bazett-Jones DP, Leblanc B, Herfort M, Moss T (1994) Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science 264: 1134–1137 [DOI] [PubMed] [Google Scholar]
- Clapier CR, Längst G, Corona DF, Becker PB, Nightingale KP (2001) Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol Cell Biol 21: 875–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clos J, Normann A, Öehrlein A, Grummt I (1986) The core promoter of mouse rDNA consists of two functionally distinct domains. Nucleic Acids Res 14: 7581–7595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JP, Roth SY, Simpson RT (1994) The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes Dev 8: 1400–1410 [DOI] [PubMed] [Google Scholar]
- Corona DF, Tamkun JW (2004) Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochim Biophys Acta 1677: 113–119 [DOI] [PubMed] [Google Scholar]
- Fleming AB, Pennings S (2001) Antagonistic remodelling by Swi–Snf and Tup1–Ssn6 of an extensive chromatin region forms the background for FLO1 gene regulation. EMBO J 20: 5219–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I, Pikaard CS (2003) Epigenetic silencing of RNA polymerase I transcription. Nat Rev Mol Cell Biol 4: 641–649 [DOI] [PubMed] [Google Scholar]
- Grunstein M (1990) Histone function in transcription. Annu Rev Cell Biol 6: 643–678 [DOI] [PubMed] [Google Scholar]
- Kastaniotis AJ, Mennella TA, Konrad C, Torres AM, Zitomer RS (2000) Roles of transcription factor Mot3 and chromatin in repression of the hypoxic gene ANB1 in yeast. Mol Cell Biol 20: 7088–7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Längst G, Becker PB, Grummt I (1998) TTF-I determines the chromatin architecture of the active rDNA promoter. EMBO J 17: 3135–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Längst G, Blank TA, Becker PB, Grummt I (1997) RNA polymerase I transcription on nucleosomal templates: the transcription termination factor TTF-I induces chromatin remodeling and relieves transcriptional repression. EMBO J 16: 760–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Reese JC (2001) Ssn6–Tup1 regulates RNR3 by positioning nucleosomes and affecting the chromatin structure at the upstream repression sequence. J Biol Chem 276: 33788–33797 [DOI] [PubMed] [Google Scholar]
- Li J, Santoro R, Koberna K, Grummt I (2005) The chromatin remodeling complex NoRC controls replication timing of rRNA genes. EMBO J 24: 120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson CE, Shim EY, Friedman DS, Zaret KS (1993) An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell 75: 387–398 [DOI] [PubMed] [Google Scholar]
- McStay B (2006) Nucleolar dominance: a model for rRNA gene silencing. Genes Dev 20: 1207–1214 [DOI] [PubMed] [Google Scholar]
- Nemeth A, Strohner R, Grummt I, Längst G (2004) The chromatin remodeling complex NoRC and TTF-I cooperate in the regulation of the mammalian rRNA genes in vivo. Nucleic Acids Res 32: 4091–4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Oliveira R, Leid M, McBurney M, Guarente L (2004) Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature 429: 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R, Grummt I (2001) Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol Cell 8: 719–725 [DOI] [PubMed] [Google Scholar]
- Santoro R, Grummt I (2005) Epigenetic mechanism of rRNA gene silencing: temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol Cell Biol 25: 2539–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R, Li J, Grummt I (2002) The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet 32: 393–396 [DOI] [PubMed] [Google Scholar]
- Shimizu M, Roth SY, Szent-Gyorgyi C, Simpson RT (1991) Nucleosomes are positioned with base pair precision adjacent to the alpha 2 operator in Saccharomyces cerevisiae. EMBO J 10: 3033–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson RT (1991) Nucleosome positioning: occurrence, mechanisms, and functional consequences. Prog Nucleic Acid Res Mol Biol 40: 143–184 [DOI] [PubMed] [Google Scholar]
- Soutoglou E, Talianidis I (2002) Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295: 1901–1904 [DOI] [PubMed] [Google Scholar]
- Stefanovsky VY, Pelletier G, Bazett-Jones DP, Crane-Robinson C, Moss T (2001) DNA looping in the RNA polymerase I enhancesome is the result of non-cooperative in-phase bending by two UBF molecules. Nucleic Acids Res 29: 3241–3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, Längst G, Grummt I (2001) NoRC—a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J 20: 4892–4900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohner R, Nemeth A, Nightingale KP, Grummt I, Becker PB, Längst G (2004) Recruitment of the nucleolar remodeling complex NoRC establishes ribosomal DNA silencing in chromatin. Mol Cell Biol 24: 1791–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voit R, Hoffmann M, Grummt I (1999) Phosphorylation by G1-specific cdk–cyclin complexes activates the nucleolar transcription factor UBF. EMBO J 18: 1891–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss K, Simpson RT (1997) Cell type-specific chromatin organization of the region that governs directionality of yeast mating type switching. EMBO J 16: 4352–4360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Reese JC (2004) Redundant mechanisms are used by Ssn6–Tup1 in repressing chromosomal gene transcription in Saccharomyces cerevisiae. J Biol Chem 279: 39240–39250 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Grummt I (2005) The PHD finger/bromodomaim of NoRC interacts with acetylated histone H4K16 and is sufficient for rDNA silencing. Curr Biol 15: 1434–1438 [DOI] [PubMed] [Google Scholar]


