Abstract
DNA polymerase mu (Pol µ) is a novel family X DNA polymerase that has been suggested to play a role in micro-homology mediated joining and repair of double strand breaks. We show here that human Pol µ is not able to discriminate against the 2′-OH group of the sugar moiety. It inserts rNTPs with an efficiency that is <10-fold lower than that of dNTPs, in sharp contrast with the >1000-fold discrimination characteristic of most DNA-dependent DNA polymerases. The lack of sugar discrimination by Pol µ is demonstrated by its ability to add rNTPs to both DNA and RNA primer strands, and to insert both deoxy- and ribonucleotides on growing nucleic acid chains. 3D-modelling of human Pol µ based on the available Pol β and TdT structural information allowed us to predict candidate residues involved in sugar discrimination. Thus, a single amino acid substitution in which Gly433 residue of Pol µ was mutated to the consensus tyrosine present in Pol β, produced a strong increase in the discrimination against ribonucleotides. The unusual capacity to insert both rNTPs and dNTPs will be discussed in the context of the predicted roles of Pol µ in DNA repair.
INTRODUCTION
Traditionally, polymerases have been classified into several classes on the basis of their substrate specificities: usage of DNA and/or RNA as a template and dNTPs or rNTPs as substrates. The two main classes, DNA and RNA polymerases, use DNA as template but differ in their ability to incorporate either dNTPs or rNTPs on nucleic acid chains. Natural DNA-dependent DNA polymerases exhibit high sugar discrimination, selecting dNTPs over rNTPs by several orders of magnitude (1–3). Based on existing evidence, the ability to discriminate between nucleotide substrates is a consequence of subtle structural differences among DNA polymerases. Thus, single amino acid substitutions introduced in the active site of different DNA polymerases had dramatic effects on the degree of discrimination between the 2′-H or -OH group of the sugar moiety, producing mutant DNA polymerases capable of incorporating rNTPs efficiently (2,4–7).
A striking exception to this rule is terminal deoxynucleotidyl transferase (TdT), a DNA polymerase which is not strictly DNA-dependent. TdT contributes to the diversification of antigen receptors by adding non-templated nucleotides to gene segment junctions of Ig and T-cell receptor genes during V(D)J recombination (reviewed in 8). Soon after its isolation, it was shown that TdT could use both rNTPs and dNTPs as nucleotide polymerization substrates (9). These results have been recently re-evaluated, confirming that murine TdT, although having a strong preference for deoxynucleotide-terminated over ribonucleotide-terminated primers, is almost incapable of discriminating between rNTPs and dNTPs (10).
TdT belongs to the Pol X DNA polymerase family, evolutionarily related to a larger group of nucleotidyltransferases (11,12). Four Pol X family members are expressed in vertebrates: TdT (9) and Pol β (13), both studied earlier, and the recently described Pol λ (14) and Pol µ (15). While Pol λ is structurally and functionally similar to Pol β, Pol µ is the closest relative to TdT, showing a 42% amino acid identity (16).
Pol µ is a recently described DNA polymerase with an intrinsic terminal transferase activity, error-proneness during DNA-dependent DNA synthesis, and specific expression pattern in secondary lymphoid organs in mammals. More over, Pol µ has the unusual ability to promote transient primer-template misalignments, which mainly result in frameshift (17) and base substitution (J.F. Ruiz, K. Bebenek, T. Kunkel and L. Blanco, unpublished results) mutations. These data, together with the recently demonstrated interaction with several components of the non-homologous end joining (NHEJ) machinery (18), suggest that Pol µ could have an important role in the end-joining pathway for repair of DNA double strand breaks. Moreover, the misalignment capacity of Pol µ appears to be crucial to carry out bypass synthesis of certain DNA lesions (19,20).
We provide here novel insights into the biochemical characterization of human Pol µ, demonstrating its striking ability to incorporate and elongate rNTPs to nucleic acid chains. This unusual capacity mainly relies on a single glycine residue that substitutes for a conserved aromatic residue present in Pol β and in most members of the Pol X family of DNA-dependent DNA polymerases.
MATERIALS AND METHODS
Materials
Unlabelled ultrapure dNTPs and rNTPs and [γ-32P]ATP (3000 Ci/mmol) were purchased from Amersham Pharmacia Biotech. Synthetic DNA oligonucleotides were obtained from Invitrogen (P15-DNA, 5′ TCTGTGCAGGTTCTT 3′; T32-DNA (A), 5′ TGAAGTCCCTCTCGACAAAGAACCTGCACAGA 3′; T32-DNA (C), 5′ TGAAGTCCCTCTCGACCAAGAACCTGCACAGA 3′; T32-DNA (G), 5′ TGA AGTCCCTCTCGACGAAGAACCTGCACAGA 3′; T32-DNA (T), 5′ TGAAGTCCCTCTCGACTAAGAACCTGCACAGA 3′; D16-DNA, 5′ GTCGAGAGGGACTTCA 3′) and RNA oligonucleotide from Genotek (P15-RNA, 5′ UCUGUGCAGGUUCUU 3′). All the oligonucleotides described above were purified by electrophoresis on 8 M urea, 20% polyacrylamide gels. T4 polynucleotide kinase and T4 DNA ligase were from New England Biolabs. TdT, Taq DNA polymerase and Pfu DNA polymerase were from Promega.
Construction and purification of wild-type and mutant forms of human Pol µ
Human Pol µ cDNA was obtained as previously described (15). Site-directed mutations were introduced into a human Pol µ overexpression plasmid (pRSETa-hPol µ) by a PCR-based method (QuikChange® Site-Directed Mutagenesis kit, Stratagene) with the following oligonucleotides: 5′ TGCTCGGTTTTACTGGCTCCAAGCT 3′ and its reverse complementary oligonucleotide for the W434F mutation; 5′ CTTTCGCCCTGCTCGGTGGGACTG 3′ and its reverse complementary oligonucleotide for the W434R mutation; 5′ TTCGCCCTGCTCTATTGGACTGGCTCC 3′ and its reverse complementary oligonucleotide for the G433Y mutation; 5′ CTTTCGCCCTGCTCTATTTTACTGGCTCCA 3′ and its reverse complementary oligonucleotide for the G433Y/W434F double mutation. Expression of Pol µ variants was carried out in the Escherichia coli strain BL21(DE3)pLysS under standard conditions. Pol µ proteins (either wild-type or mutant) were purified to homogeneity as previously described (15).
DNA polymerization assays on defined DNA molecules
Polymerase activity was evaluated by using synthetic double-stranded oligonucleotides as substrates. These substrates were prepared by annealing a 5′-32P-end-labelled primer (DNA or RNA) to different oligonucleotides to generate open (P15/T32) and gapped (P15/T32/D16) template/primer substrates. In polymerization reactions, the incubation mixture contained, in 12.5 µl, 50 mM Tris–HCl, pH 7.5, 2 mM MgCl2 or 1 mM MnCl2, 1 mM DTT, 4% glycerol, 0.1 mg/ml BSA, different concentrations of the indicated dNTPs or rNTPs, 4 nM of 5′-labelled substrate, and the indicated amount and concentration of either purified Pol µ (wild-type or mutants) or the indicated DNA polymerase. After incubation for 30 min at 30°C (conditions shown to be linear both in time and enzyme doses), reactions were stopped by adding gel loading buffer [95% (v/v) formamide, 10 mM EDTA, 0.1% (w/v) xylene cyanol and 0.1% (w/v) Bromophenol Blue]. Before loading the gel, samples were denatured by heating at 95°C for 5 min. Products were resolved and analysed by denaturing 8 M urea–20% PAGE and autoradiography. Quantitation of autoradiographs was done by densitometric analysis of the band(s) corresponding to primer extension products with the Bio-Rad Quantity One software.
Determination of Pol µ sugar selectivity
To analyse Pol µ sugar selectivity, DNA oligonucleotide P15 was hybridized to the four variants of the T32 template oligonucleotide (T32(A), T32(C), T32(G) and T32(T), differing in the first templating base) and to the D16 oligonucleotide. Then 4 nM of each DNA-gapped substrate was incubated with 100 nM Pol µ, 2 mM MgCl2 and the indicated amounts of each pair of nucleotides (dATP + rATP; dCTP + rCTP; dGTP + rGTP; dTTP + rUTP) in the presence of the same incubation mixture described above. After incubation for 30 min at 30°C (conditions shown to be linear both in time and enzyme amount), reactions were stopped by adding gel loading buffer and products were resolved and analysed by denaturing 8 M urea–20% PAGE and autoradiography. Calculation of the relative amount of primers extended either with dNTP or with rNTP was done by densitometric analysis of the band(s) corresponding to one-nucleotide primer extension products with the Bio-Rad Quantity One software. The sugar selectivity factor (S) is the mean ratio of the efficiencies for dNTP and rNTP additions (% primer extension with dNTP/% primer extension with rNTP) calculated for different nucleotide concentrations and three independent experiments.
Amino acid sequence comparisons
An initial alignment of several members of the Pol X family was done by using the MULTALIN program (http://www.toulouse.inra.fr/multalin.html). As a second step, the alignment obtained was adjusted manually and refined on the basis of the secondary elements of rat Pol β, as deduced from its crystal structure (21).
Three-dimensional structure extrapolations
The structural model for human Pol µ was automatically generated using the program Swiss Model (http://www.expasy.ch/swissmod/swiss-model.html) and the recently obtained three-dimensional structure of murine TdT (22) as a template. TdT PDB coordinates (1KEJ) were obtained from Protein Data Bank (http://www.rcsb.org/pdb/). Next, this Pol µ model was aligned with the structure of DNA Pol β complexed with gapped DNA and ddCTP (PDB coordinates 1BPY). 1BPY coordinates were selected as they correspond to the fully closed conformation of Pol β, shown to have the maximal structural similarity (overlapping) to the crystal structure of TdT (22). Putative amino acid residues of both Pol µ (modelled) and Pol β implicated in sugar discrimination were identified and compared by using the Swiss PDB Viewer program (23; http://www.expasy.ch/spdbv/). Single (G433Y, W434F, W434R) and double (G433Y/W434F) amino acid substitutions to be introduced in Pol µ were modelled by using the same program, and the side chain conformations with the lowest energy were selected.
RESULTS
Human Pol µ incorporates and elongates dNTPs and rNTPs on a DNA primer strand with a similar efficiency
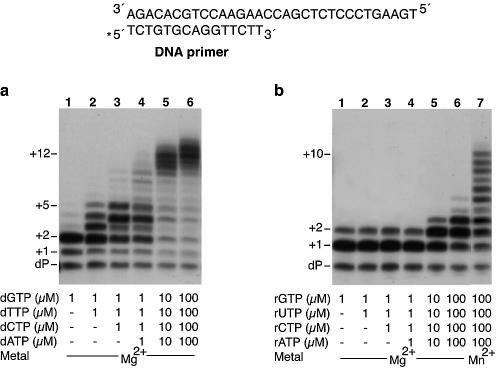
Although Pol µ has an intrinsic terminal transferase activity, its preferred substrates are template-primer structures (15). Thus, in the presence of a DNA template/primer and the four dNTPs as substrates, Pol µ catalyses efficient polymerization of dNTPs onto the DNA primer strand (Fig. 1a). Moreover, this polymerization occurs in a template-directed manner, as suggested by the change in the elongation pattern observed by providing subsets of the four dNTPs (Fig. 1a, lanes 1 to 4). However, although most Pol µ insertions are template-directed, it frequently catalyses misincorporation of dNTPs (Fig. 1a, lane 2), preferentially due to transient template misalignments (17).
Figure 1.
Pol µ efficiently polymerizes ribonucleotides on a DNA primer. The assay was carried out using 4 nM of a 5′-32P-labelled 15mer DNA primer (dP) hybridized to a DNA template oligonucleotide (a scheme is shown), as described in Materials and Methods. Polymerization was assayed in the presence of 2 mM MgCl2, 100 nM purified human Pol µ, and the indicated concentrations of either dNTPs (a) or rNTPs (b). Where indicated, MnCl2 (1 mM) was used instead of MgCl2. After incubation for 30 min at 30°C, extension of the 5′-labelled oligonucleotide was analysed by 8 M urea–20% PAGE and autoradiography. Relevant extension products are indicated.
We checked the possibility that rNTPs could be used as substrates by Pol µ. As shown in Figure 1b, human Pol µ efficiently incorporates rNTPs on a DNA primer strand, as judged by the proportion of extended primer strands (compare Fig. 1a and b). However, elongation of ribonucleotides or with ribonucleotides is less efficient, as the pattern observed is shorter than that obtained with identical concentrations of dNTPs (compare Fig. 1a and b, lanes 1 to 6). In fact, at a low ribonucleotide concentration (1 µM), the insertion/elongation pattern obtained in the presence of the four rNTPs is very similar to that obtained with rGTP alone (the first rNTP to be inserted according to the template). This limited pattern suggests that ribonucleotide incorporation by Pol µ might be compromised by the template sequence context or by a specific ribonucleotide preference (see below). However, this pattern could be further elongated either by increasing the rNTP concentration in the reaction (Fig. 1b, lanes 5 and 6), or by using Mn2+ instead of Mg2+ as metal cofactor in the reaction (lane 7). Insertions of rNTPs occurred mainly as template-directed events, even in the presence of Mn2+ ions, and resulted from template misalignments (results not shown).
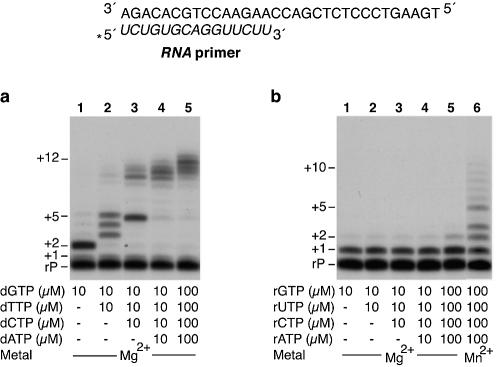
Pol µ is able to polymerize nucleotides on RNA primer strands
The strong accumulation of the +1 product shown in Figure 1b (lanes 1–4) could reflect a lower efficiency to recognize a primer-terminus having a 2′-OH group. To clarify this point, we directly tested whether Pol µ has the capacity to use a pre-existing RNA primer to incorporate ribo- or deoxynucleotides. For a more direct comparison with the results shown in Figure 1, we used a primer strand having the same nucleotide sequence, but made of rNTPs, hybridized to the same DNA template sequence. As shown in Figure 2, Pol µ is able to polymerize both dNTPs (Fig. 2a) and rNTPs (Fig. 2b) on a RNA primer hybridized to a DNA template strand. By providing dGTP alone, elongation is mainly restricted to the insertion of two residues (Fig. 2a), as expected for a faithful reaction. However, addition of different combinations of dNTPs allowed further extension, generating products longer than expected, consistent with Pol µ’s propensity to insert nucleotides in a variety of misalignment-mediated events. As shown in Figure 2, there is a significant amount of unextended RNA primers, higher than that of the unextended DNA primer molecules shown in Figure 1a. This was due to a 3-fold lesser hybridization efficiency of the RNA primers to the template DNA, as determined by polyacrylamide gel electrophoresis at low ionic strength (results not shown). On the other hand, the nucleotide extension pattern of these RNA and DNA primers is very similar. Again, the elongation is more efficient with dNTPs than with rNTPs substrates (Fig. 2a and b, respectively). RNA primer elongation could be strongly improved by using Mn2+ instead of Mg2+ as metal cofactor in the reaction (Fig. 2b, lane 6).
Figure 2.
Pol µ efficiently polymerizes both deoxy- and ribonucleotides on RNA primers. The assay was carried out using 4 nM of a 5′-32P-labelled 15mer RNA primer (rP) hybridized to a DNA template oligonucleotide (a scheme is shown), as described in Materials and Methods. Polymerization was assayed in the presence of 2 mM MgCl2, 100 nM purified human Pol µ, and the indicated concentrations of either dNTPs (a) or rNTPs (b). Where indicated, MnCl2 (1 mM) was used instead of MgCl2. After incubation for 30 min at 30°C, extension of the 5′-labelled oligonucleotide was analysed by 8 M urea–20% PAGE and autoradiography. Relevant extension products are indicated.
According to these results we can conclude that Pol µ is also able to use a DNA/RNA template/primer substrate to incorporate and elongate both ribo- and deoxynucleotides.
Ribonucleotide insertion/extension by other Pol X family members
Pol µ belongs to the Pol X family of DNA polymerases, together with TdT, Pol β, Pol λ and the most distantly related Pol σ (reviewed in 24). While Pol λ is more related to Pol β, Pol µ is closely related to TdT, an enzyme whose ability to insert rNTPs has been previously described (10). Unlike Pol β, Pol λ, Pol µ and TdT have a BRCT domain at their N-terminus. However, although all of them could share a similar three-dimensional structure, their biochemical activities appear to be quite diverse. Therefore, we compared the ability of DNA polymerases of the Pol X family to incorporate rNTPs on DNA primers in our experimental conditions. As shown in Figure 3, Pol β, Pol λ and Pol µ behaved as proficient DNA-dependent DNA polymerases, polymerizing dNTPs on a DNA template/primer substrate. TdT, as expected, was less efficient to polymerize dNTPs on this kind of substrate, since it displayed its optimal activity on non-templated single stranded DNA molecules (not shown; 25). On the other hand, the ability to incorporate rNTPs on these DNA template/primer molecules, which can be clearly seen with both TdT and Pol µ, was null in Pol λ and very low in Pol β (Fig. 3). The same results were obtained under different reaction conditions and other DNA substrates as gapped DNA (data not shown). Moreover, as judged by the extension pattern shown in Figure 3, ribonucleotide incorporation by Pol µ was template-directed, while TdT polymerized rNTPs in a template-independent manner.
Figure 3.
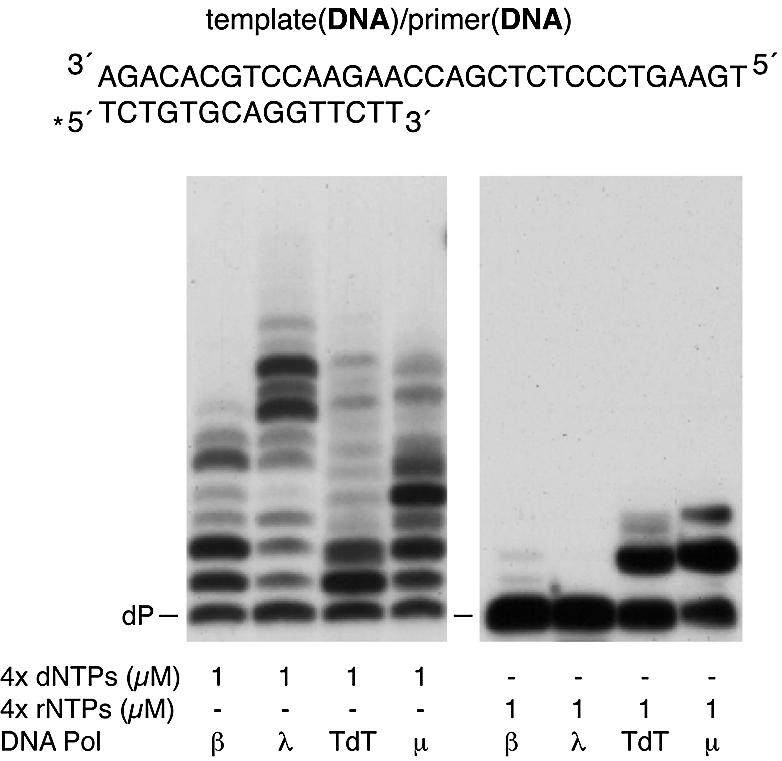
Comparative analysis of rNTP insertion by Pol X DNA polymerases. The assay was carried out using 4 nM of a 5′-labelled 15mer DNA primer (dP) hybridized to a DNA template oligonucleotide (see Materials and Methods). Polymerization was assayed in the presence of 2 mM MgCl2, 1 µM of either dNTPs or rNTPs, and 100 nM of either Pol β, Pol λ, TdT or Pol µ. After incubation for 30 min at 30°C, extension of the 5′-labelled oligonucleotide was analysed by 8 M urea–20% PAGE and autoradiography.
These results suggest that the ability to efficiently use rNTPs as substrates to elongate DNA primers appears to be specific to TdT-like DNA polymerases (TdT, Pol µ), while other members of the Pol X family, such as Pol β and Pol λ do not have this capacity.
Pol µ polymerize ribo- and deoxynucleotides distributively on template/primer substrates
DNA polymerases from the Pol X family display a strictly distributive polymerization mode, i.e. they dissociate from a template-primer molecule after each nucleotide addition cycle. This behaviour has been shown for Pol β (26), Pol λ (27) and TdT (10), but it has not been reported yet in the case of Pol µ. In this paper, we assessed Pol µ’s processivity when polymerizing either dNTPs or rNTPs on DNA template/primer substrates by analysing the chain length distribution at various enzyme/DNA substrate ratios. As shown in Figure 4, the length of the products synthesized by Pol µ when polymerizing either dNTPs or rNTPs decreased with the enzyme/substrate ratio (Fig. 4a and b, left panels). In both cases, the limit length is 1, in agreement with a fully distributive polymerization pattern. We also analysed a possible effect of the metal cofactor used in the polymerization reaction on this low Pol µ processivity. As shown in Figure 4a and b (right panels), the distributive behaviour of Pol µ is also maintained in the presence of Mn2+ activating ions.
Figure 4.
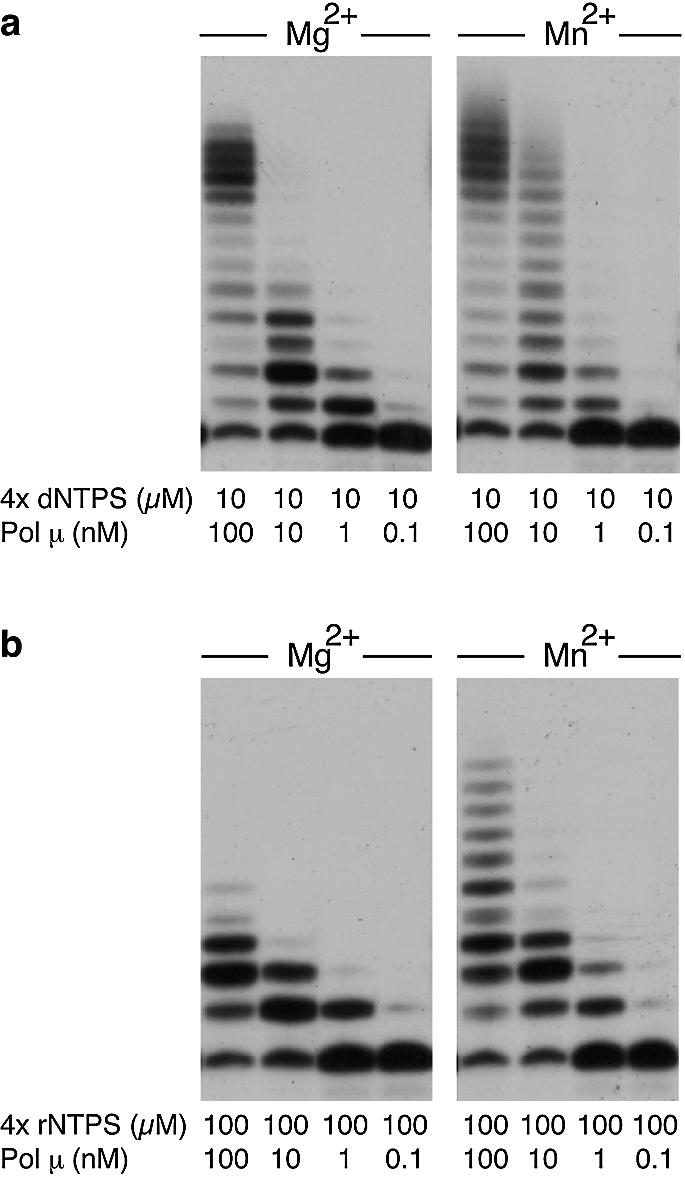
Distributive polymerization of both dNTPs and rNTPs by human Pol µ. Reactions were performed by using 4 nM of a 5′-labelled 15mer DNA primer hybridized to a 32mer template oligonucleotide, as described in Materials and Methods. Polymerization reactions were carried out in the presence of either 2 mM MgCl2 or 1 mM MnCl2, 10 µM dNTPs, and the indicated amounts (from 100 to 0.1 nM) of Pol µ. After incubation for 30 min at 30°C, extension of the 5′-labelled oligonucleotide was analysed by 8 M urea–20% PAGE and autoradiography.
Therefore, we can conclude that Pol µ polymerizes both rNTPs and dNTPs on a DNA primer in a distributive manner, independently of the metal activator present (Mg2+ or Mn2+).
Sugar discrimination by Pol µ depends on each particular nucleotide base pair
To analyse the discrimination ratio for each rNTP/dNTP pair by Pol µ, we designed a set of 1-nucleotide gapped DNA substrates with each of the four (A, C, G or T) bases as templates. Since Pol µ is not able to carry out strand displacement (our unpublished data) such DNA substrates will restrict polymerization to a single nucleotide incorporation, and eliminate the additional complexity resulting from the frequency of promiscuous insertion events catalysed by Pol µ. Thus, we experimentally determined the value of the Pol µ sugar selectivity factor (S) for each deoxy/ribonucleotide pair (dATP/rATP, dCTP/rCTP, dGTP/rGTP and dTTP/rUTP) using a competition assay (10) where both sugar variants (ribose and deoxyribose) of each nucleotide are simultaneously provided. Since the rNTP and dNTP have different molecular weights, the +1 extended primers can be easily separated by gel electrophoresis and then quantified. The S factor is given by the ratio between the amount of primers extended with either dNTPs or rNTPs (% primer extension with dNTP/% primer extension with rNTP).
Figure 5 shows a representative assay for an S factor determination, and Table 1 summarizes the values obtained for all four dNTP/rNTP pairs. The S values range between 1.34 for dGTP over rGTP and 11.04 for dATP over rATP. These values are comparable to those obtained for TdT in untemplated reactions (10), and several orders of magnitude smaller than the sugar selectivity factors reported for other DNA polymerases (1–3). Interestingly, the selectivity factors for Pol µ vary depending on the nature of the base. The lack of discrimination between ribo- and deoxynucleotides is more pronounced for Gs and Cs than for Ts and As (Table 1).
Figure 5.
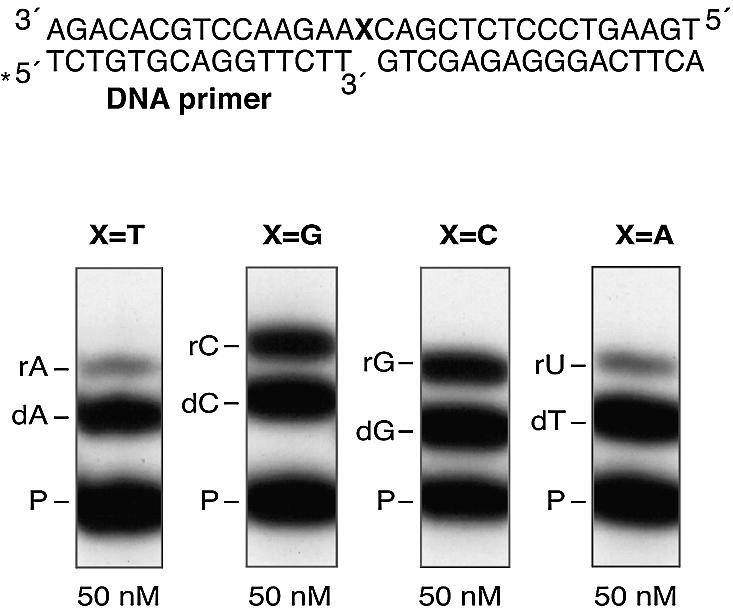
Sugar discrimination by Pol µ depends on the nature of the base. Single nucleotide gap-filling assays were performed using the four 1 nt-gapped DNA substrates (a scheme is shown) differing in the templating base (X). Reactions were performed by using 4 nM of each 5′-32P-labelled 1 nt-gapped DNA substrate, 100 nM Pol µ, 2 mM MgCl2, and the indicated concentration of each of the four pairs of deoxy- and ribonucleotide. After 30 min at 30°C, reactions were subjected to 8 M urea–20% PAGE, and the deoxy/ribonucleotide +1 extended primers were detected by autoradiography.
Table 1. Sugar selectivity factors for the insertion of deoxy/ribonucleotide pairs by Pol µ.
| Template | Nucleotide mix | WT | W434F | W434R | G433Y |
|---|---|---|---|---|---|
| dT | dA/rA | 11.04 ± 0.70a | 17.09 ± 0.59 | 17.87 ± 2.65 | >30 |
| dG | dC/rC | 2.70 ± 0.67 | 3.41 ± 0.74 | 7.54 ± 1.65 | >40 |
| dC | dG/rG | 1.34 ± 0.26 | 1.77 ± 0.20 | 7.67 ± 0.91 | >25 |
| dA | dT/rU | 8.64 ± 1.18 | 5.93 ± 1.42 | 7.65 ± 1.55 | >30 |
| dNb | dN/rN | 5.93 ± 0.70 | 7.05 ± 0.73 | 10.18 ± 1.69 | >31 |
aFigures indicate the mean values of independent experiments and their standard deviation.
bThe sugar selectivity factors obtained for each deoxy/ribonucleotide pair were used to calculate the mean value for the four bases (N).
Ribonucleotides inhibit DNA polymerization by Pol µ
As has been previously shown, cells have a ribo/deoxyribonucleotide pool imbalance, with an excess of rNTPs over dNTPs (28). Thus, once established that Pol µ is able to incorporate rNTPs on both DNA and RNA primers, we analysed the incorporation of both dNTPs and rNTPs on DNA strands in vitro under conditions that reproduce this pool imbalance. We carried out polymerization reactions in which each individual rNTP was added at a 10-fold excess over the four dNTPs. As shown in Figure 6, the effect of the rNTP varied depending on the nature of the base. Thus, while the addition of pyrimidines (rUTP or rCTP) had no apparent effect on the Pol µ elongation pattern (Fig. 6, lanes 3 and 4, respectively), the addition of purines (rATP and mainly rGTP) inhibited elongation (lanes 2 and 5, respectively). The stronger inhibition by rGTP could be explained either by the fact that the first two consecutive templating bases are dC, combined with the low discrimination of rGTP over dGTP (Table 1), or by a poorer extension of rGMP-terminated primers by Pol µ. To clearly demonstrate that rNTPs can be incorporated into DNA by Pol µ activity, we carried out in vitro polymerization reactions in which we added three dNTPs and the fourth was replaced by the rNTP counterpart in a 10-fold excess, confirming that, under these conditions, Pol µ activity is able to incorporate rNTPs into DNA polymerization products (data not shown).
Figure 6.
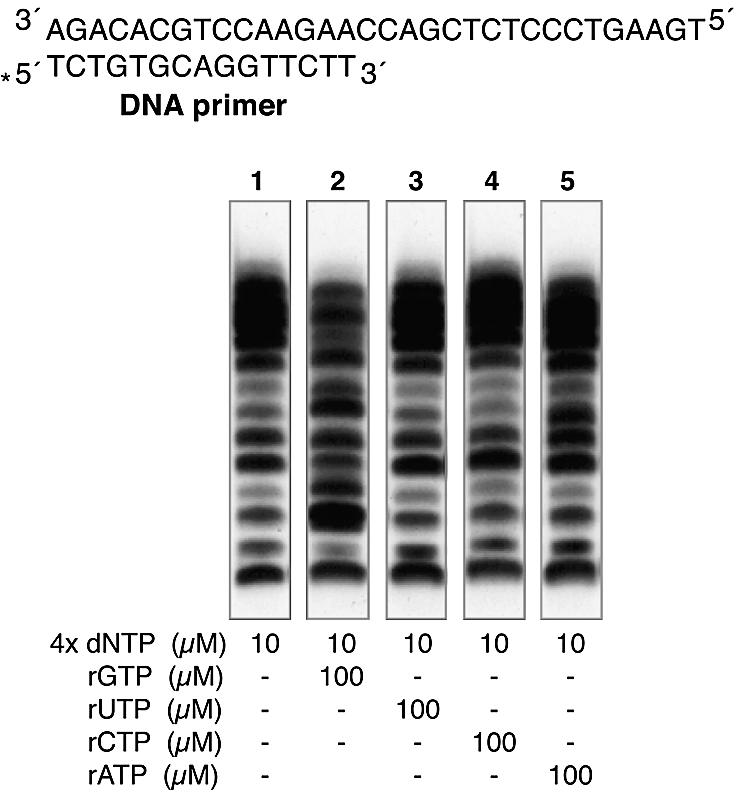
Insertion of rNTPs inhibit DNA polymerization by Pol µ. Primer extension assays were performed using 4 nM 5′-32P-labelled DNA primer/template, 100 nM Pol µ, 2 mM MgCl2, and the indicated concentration of either dNTPs and rNTPs. Different combinations of nucleotides were used to evaluate the interference of rNTPs on dNTP polymerization. After 30 min at 30°C, polymerization products were subjected to 8 M urea–20% PAGE and autoradiography.
Gly433 of Pol µ is essential to allow insertion of rNTPs
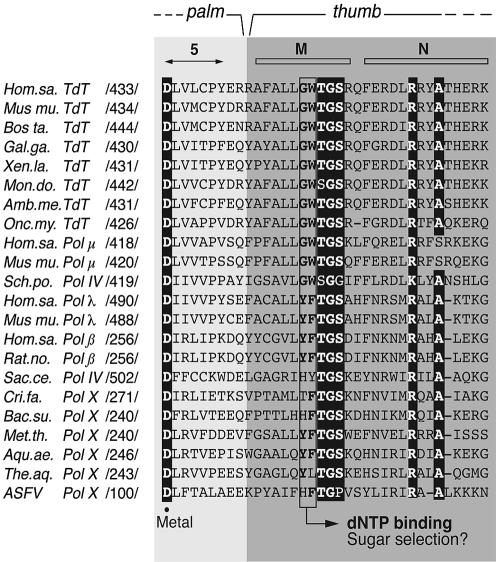
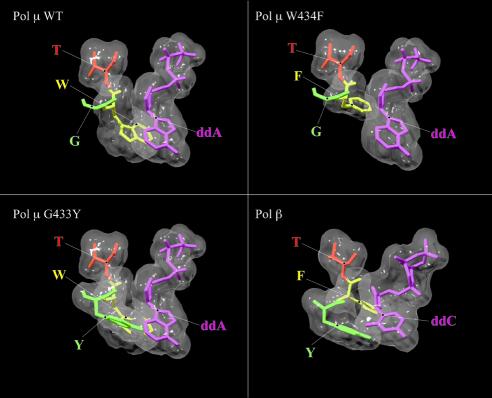
As previously described, there are three Pol β residues that interact by van der Waals contacts with the sugar moiety (C2′ and C3′ carbons) of the incoming nucleotide: Tyr271, Phe272 and Gly274, located at the end of α-helix M in the thumb subdomain, and predicted to participate in nucleotide selectivity of DNA over RNA (21). As shown in Figure 7, a counterpart of Pol β Gly274 is invariantly present in all members of the Pol X family, including TdT and Pol µ. Interestingly, Tyr271 and Phe272 residues of Pol β are substituted by invariant Gly and Trp residues, respectively, in TdT and Pol µ from different species, and in Pol IV from Schizosaccharomyces pombe. Therefore, according to this, it seemed very likely that residues Gly433 and Trp434 of Pol µ could be responsible for the lack of sugar discrimination between rNTPs and dNTPs.
Figure 7.
Multiple alignment of the amino acid region likely involved in sugar discrimination in the Pol X family. Numbers between slashes indicate the amino acid position relative to the N-terminus of each polymerase. According to Pol β structural data (21), this region connects subdomains palm and thumb, and contains one of the three aspartates acting as metal ligands (black dot above the sequences). Invariant residues among Pol X family members (29) are indicated in white letters over a black background. The two amino acid residues most likely implicated in sugar discrimination are indicated in bold, inside a box. Abbreviations used are: Hom.sa., Homo sapiens, Mus mu., Mus musculus, Bos ta., Bos taurus, Gal.ga., Gallus gallus, Xen.la., Xenopus laevis, Mon.do., Monodelphis domestica, Amb.me., Ambystoma mexicanum, Onc.my., Oncorrhynchus mykiss, Sch.po., Schizosaccharomyces pombe, Rat.no., Rattus norvergicus, Sac.ce., Saccharomyces cerevisiae, Cri.fa., Crithidia fasciculata, Bac.su., Bacillus subtilis, Met.th., Methanothermobacter thermautotrophicus, Aqu.ae., Aquifex aeolicus and The.aq., Thermus aquaticus.
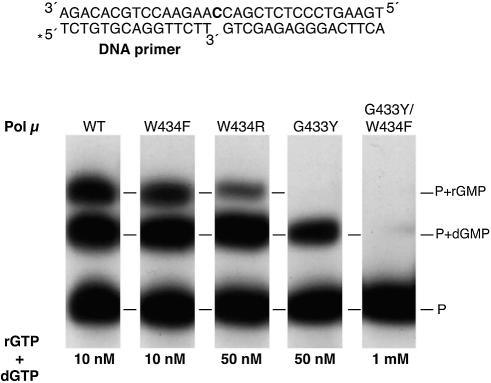
To test this prediction, we mutated the amino acid residues G433 and W434 of Pol µ by site-directed mutagenesis (see Materials and Methods), and checked how efficiently the mutant proteins discriminate between dNTPs and rNTPs. Single mutations selected were G433 to Tyr and W434 to Phe, to restore individual consensus residues of Pol β-like enzymes. Two additional mutants were obtained: W434 to Arg, to evaluate the effect of a non-conservative change at this position, and the double mutant G433Y/W434F, to fully restore the Pol β consensus.
Interestingly, the W434F mutant protein had a polymerization activity similar to that of the wild-type protein, as assessed by dNTP incorporation, and was still able to polymerize ribonucleotides (Fig. 8). As shown in Table 1, sugar selectivity factors for mutant W434F were similar to those obtained with the wild-type protein, with a maximal difference of 1.5-fold for dA/rA selectivity. In agreement with the conservation of an aromatic residue in most Pol X polymerases, the catalytic activity of mutant W434R decreased about 5-fold compared to the wild-type protein. This effect appears to be higher for rNTP than dNTP polymerization, as shown in Figure 8. However, as shown in Table 1, the change in discrimination between ribo- and deoxyribonucleotides by the W434R mutant protein seems to be dependent on the nature of the base, as selectivity factors for C and G were the most affected (about 3- and 6-fold, respectively) by the mutation (Table 1). No change in sugar discrimination was observed for the dA/rA and dT/rU pairs. Strikingly, the ability to incorporate rNTPs drastically dropped in the G433Y mutant protein, as shown in Figure 8. The S values obtained for this mutant (Table 1) reflect a large gain of sugar discrimination for all dNTP/rNTP pairs. On the other hand, as shown in Figure 8, this mutation results in a decrease in the dNTP polymerization efficiency in relation to wild-type Pol µ, which suggests that this amino acid residue is also important for the optimal catalytic activity of the enzyme.
Figure 8.
Lack of sugar discrimination by Pol µ resides in Gly433. Single nucleotide gap-filling assays were performed using 4 nM of a 5′-32P-labelled 1 nt-gapped DNA substrate having a dC as templating base (a scheme is shown), 2 mM MgCl2, the indicated concentration of both dGTP and rGTP, and 100 nM of either the wild-type Pol µ (WT), or each of the site-directed mutant proteins W434F, W434R, G433Y and G433/W434F. After 30 min at 30°C, the ratio of deoxy- versus ribonucleotide-extended primers was analysed by 8 M urea–20% PAGE and autoradiography.
The double mutant protein G433Y/W434F had a very low, almost null, polymerization activity with both dNTPs and rNTPs (Fig. 8), indicating the essential role of these amino acid residues for an optimal polymerization activity of Pol µ, in addition to their role in sugar discrimination.
DISCUSSION
Pol µ is a novel nucleotidyltransferase belonging to the Pol X family. Despite its previously described terminal transferase activity (15), Pol µ should be considered a DNA-dependent DNA polymerase versatile in its use of substrates. In fact, Pol µ was originally described as an extremely unfaithful DNA polymerase (15), and it was later shown that its infidelity is related to its extraordinary ability to accept misaligned template-primer structures as a substrate (17). As we have shown here, an additional feature that reflects the versatility of substrate usage by Pol µ is its ability to insert ribonucleotides to the 3′-hydroxyl primer ends of either DNA template/primer or DNA-gapped molecules, in sharp contrast with most DNA polymerases. Moreover, the sugar selectivity factors measured in single nucleotide addition assays (between 1 and 11, for dG/rG and dA/rA, respectively; see Table 1), indicate that Pol µ efficiently incorporates both ribo- and deoxyribonucleotides. This is a very rare behaviour for a DNA synthesizing enzyme, since most DNA polymerases strongly favour the insertion of deoxynucleotides to avoid the incidental insertion of ribonucleotides into the DNA. In fact, as shown here, other Pol X enzymes as Pol β or Pol λ, strongly discriminate against ribonucleotides. Exceptionally, TdT (the closest counterpart of Pol µ) is known to have a similar ribonucleotide insertion capacity (10). After our work was completed, Nick McElhinny and Ramsden reported a series of results very similar to those described here, demonstrating that Pol µ is a DNA-dependent DNA and RNA polymerase (30).
For several DNA polymerases, sugar discrimination has been shown to depend on a single residue in motif A that acts as a steric barrier for the 2′-hydroxyl of an incoming ribonucleotide (1,5). However, crystallographic analysis of Pol β suggested a different mechanism, in which exclusion of the ribose moiety is carried out by a three-residue peptide backbone (Tyr271–Phe272–Thr273) located in α-helix M of the thumb subdomain (21). More recently, the crystal structure of murine TdT has been resolved (22). According to this structure, the sugar moiety of the incoming nucleotide is located in the neighbourhood of a number of α-helix M residues (Fig. 7): Trp450 on one side, and the cis-peptide bond between Gly452 and Ser453 on the other side. However, none of these residues are close enough to the 2′ position of the sugar moiety of an incoming nucleotide, thus accounting for the ability of TdT to efficiently incorporate ribonucleotides. The extensive amino acid sequence similarity between Pol µ and TdT allowed us to generate a model of Pol µ’s three-dimensional structure based on the crystal structure of murine TdT (22). This model indicates a similar organization of the active site of both enzymes. It suggests that the Pol µ residues closest to the 2′ position of the ribose of an incoming nucleotide would be Gly433 and Trp434, located in α-helix M (Fig. 9, top left), whose counterparts in Pol β are Tyr271 and Phe272 (Fig. 9, bottom right). Our results demonstrate that, while ribonucleotide insertion is not greatly affected when Trp434 is substituted, the small size of the Gly433 side chain is essential to allow the polymerization of ribonucleotides by Pol µ. In fact, when Gly433 is changed to a residue with a bulkier side chain (Tyr) as that present in Pol β (Fig. 9, bottom left), a dramatic increase in the sugar selectivity factors between deoxy- and ribonucleotides is observed. Thus, the size of the side chain present at this position probably regulates ribonucleotide insertion in Pol X family enzymes. Based on this mutational analysis and the amino acid sequence alignment shown in Figure 7, a similar efficiency of rNTP insertion, as that shown by TdT and Pol µ, is also predicted for Pol IV from S.pombe.
Figure 9.
Structural basis for the lack of sugar discrimination by Pol µ. Generation of a structural model of Pol µ based on TdT structure (22), and comparison with that of a Pol β ternary complex (21) were performed as indicated in Materials and Methods. The figure shows the spatial position of the wild-type Pol µ amino acid residues (Gly433, Trp434 and Thr435) that are closest to the sugar moiety of an incoming nucleotide (left-top panel). Site directed substitution of either Gly433 to Tyr (G433Y; left-bottom panel) or Trp434 to Phe (W434F; right-top panel) was modelled. A structural comparison of the Pol β counterpart residues (Tyr271, Phe272 and Thr273) is also shown (right-bottom panel). Figure was created using the Swiss PDB Viewer program (23; http://www.expasy.ch/spdbv/), and rendered with POV Ray (www.povray.org).
The highly efficient insertion of rNTPs by both TdT and Pol µ is extremely intriguing taking into account that incorporation of ribonucleotides in the DNA creates a lesion that has to be repaired at a risk of recruiting various error-free and error-prone DNA repair systems. However, the biological consequences could differ as TdT synthesis is mostly template-independent, while the ribonucleotide insertion capacity described here for Pol µ is directed by the template. Furthermore, whereas TdT activity is restricted to a very specific stage of lymphocyte differentiation, Pol µ seems to be expressed in most cell types, which points to the potential relevance of rNTP insertion into DNA by Pol µ in a/some widely occurring process.
As shown here, Pol µ efficiently inserts rNTPs into DNA, but polymerization efficiency decreases after the incorporation of a limited number of rNMPs. When both rNTPs and dNTPs are present, Pol µ synthesizes mixed products. However, since Pol µ extends ribonucleotide-terminated primers with a lower catalytic rate than deoxyribonucleotide-terminated ones, mixed products are shorter in length. A similar effect has been described recently for murine TdT (10), leading to the proposal that rNTP insertion could serve to limit non-templated nucleotide additions (N-additions) by TdT to the junctions of rearranging V, D and J gene segments. The terminal transferase activity of Pol µ, together with its lack of sugar discrimination for nucleotides suggest that Pol µ could have a role in V(D)J recombination, perhaps complementing TdT additions at V(D)J recombinational joints. Interestingly, second V(D)J recombination waves (receptor editing) have been described to occur in a subset of germinal centre B lymphocytes (31), just the place where higher Pol µ levels have been found (15,16).
Recent data demonstrated that Pol µ forms a complex with the end-joining factors Ku and XRCC4-Ligase IV in a manner very similar to that of TdT (18,32). Moreover, this complex facilitates the ability of XRCC4-Lig IV and Ku to join ends with partially complementary overhangs in vitro, supporting the proposal of a participation of Pol µ in the repair of DNA double strand breaks (16–18). It is not obvious how insertion of rNTPs by Pol µ could have a relevant role in NHEJ and perhaps also in lesion bypass of DNA damage, but a tempting speculation is that insertion of rNTPs could stabilize template misalignment intermediates during these processes. An interesting proposal, recently made by Nick McElhinny and Ramsden (30), is that Pol µ may have evolved rNTP polymerization activity to be effective when nuclear dNTP pools are lowest, since rNTP pools remain high throughout the cell cycle and in non-cycling cells.
It is worth noting that the unusual capacity of Pol µ to accept misaligned template-primer molecules as a substrate, a convenient feature for microhomology-mediated NHEJ, is not affected in the G433Y mutant (our unpublished data), suggesting that the ability to insert ribonucleotides is not a side-effect (i.e. as a consequence of a less tightened active site) and therefore is likely to have a biological meaning. The identification of the critical residue involved in the lack of sugar discrimination by human Pol µ will guide future studies to demonstrate the relevance of rNTP incorporation for the biological function of this enzyme.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Kasia Bebenek (NIEHS, NC, USA) for critical reading of the manuscript. This work was supported by Ministerio de Ciencia y Tecnología Grant BMC2000–1138, and by an institutional grant from Fundación Ramón Areces. J.F.R, R.J., M.G.-D. and A.J.P. were recipients of a fellowship from the Ministerio de Educación y Ciencia. S.G.-B. was postdoctoral fellow from the Comunidad de Madrid and A.R.F.d.H. was a fellow from the Ramón y Cajal Research Program.
REFERENCES
- 1.Joyce C.M. (1997) Choosing the right sugar: How polymerases select a nucleotide substrate. Proc. Natl Acad. Sci. USA, 94, 1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Astatke M., Ng,K., Grindley,N.D.F. and Joyce,C.M. (1998) A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proc. Natl Acad. Sci. USA, 95, 3402–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cases-González C.E., Gutiérrez-Rivas,M. and Menéndez-Arias,L. (2000) Coupling ribose selection to fidelity of DNA synthesis. The role of Tyr-115 of human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem., 275, 19759–19767. [DOI] [PubMed] [Google Scholar]
- 4.Gao G., Orlova,M., Georgiadis,M.M., Hendrickson,W.A. and Goff,S.P. (1997) Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. Proc. Natl Acad. Sci. USA, 94, 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnin A., Lázaro,J.M., Blanco,L. and Salas,M. (1999) A single tyrosine prevents insertion of ribonucleotides in the eukaryotic-type phi29 DNA polymerase. J. Mol. Biol., 290, 241–251. [DOI] [PubMed] [Google Scholar]
- 6.Patel P.H. and Loeb,L.A. (2000) Multiple amino acid substitutions allow DNA polymerases to synthesize RNA. J. Biol. Chem., 275, 40266–40272. [DOI] [PubMed] [Google Scholar]
- 7.Ogawa M., Tosaka,A., Ito,Y., Yoshida,S. and Suzuki,M. (2001) Enhanced ribonucleotide incorporation by an O-helix mutant of Thermus aquaticus DNA polymerase I. Mutat. Res., 485, 197–207. [DOI] [PubMed] [Google Scholar]
- 8.Bassing C.H., Swat,W. and Alt,F.W. (2002) The mechanism and regulation of chromosomal V(D)J recombination. Cell, 109, S45–S55. [DOI] [PubMed] [Google Scholar]
- 9.Kato K.I., Goncalves,J.M., Houts,G.E. and Bollum,F.J. (1967) Deoxynucleotide-polymerizing enzymes of calf thymus gland. II. Properties of the terminal deoxynucleotidyltransferase. J. Biol. Chem., 242, 2780–2789. [PubMed] [Google Scholar]
- 10.Boulé J.B., Rougeon,F. and Papanicolaou,C. (2001) Terminal deoxynucleotidyl transferase indiscriminately incorporates ribonucleotides and deoxyribonucleotides. J. Biol. Chem., 276, 31388–31393. [DOI] [PubMed] [Google Scholar]
- 11.Holm L. and Sander,C. (1995) DNA polymerase β belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem. Sci., 20, 345–347. [DOI] [PubMed] [Google Scholar]
- 12.Aravind L. and Koonin,E.V. (1999) DNA polymerase β-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res., 27, 1609–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.SenGupta D.N., Zmudzka,B.Z., Kumar,P., Cobianchi,F., Skowronski,J. and Wilson,S.H. (1986) Biochem. Biophys. Res. Commun. 136, 341–347. [DOI] [PubMed] [Google Scholar]
- 14.García-Díaz M., Domínguez,O., López-Fernández,L.A., Laín de Lera,T., Saníger,M.L., Ruiz,J.F., Párraga,M., García-Ortiz,M.J., Kirchhoff,T., del Mazo,J., Bernad,A. and Blanco,L. (2001) DNA polymerase lambda (Pol λ), a novel eukaryotic DNA polymerase with a potential role in meiosis. J. Mol. Biol., 301, 851–867. [DOI] [PubMed] [Google Scholar]
- 15.Domínguez O., Ruiz,J.F., Laín de Lera,T., García-Díaz,M., González,M.A., Kirchhoff,T., Martínez,A.C., Bernad,A. and Blanco,L. (2000) DNA polymerase mu (Pol µ), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J., 19, 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruiz J.F., Domínguez,O., Laín de Lera,T., García-Díaz,M., Bernad,A. and Blanco,L. (2001) DNA polymerase mu, a candidate hypermutase? Phil. Trans. R. Soc. Lond. B, 356, 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y., Wu,X., Yuan,F., Xie,Z. and Wang,Z. (2001) DNA polymerase µ. Mol. Cell. Biol., 21, 7995–8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahajan K.N., Nick McElhinny,S.A., Mitchell,B.S. and Ramsden,D.A. (2002) Association of DNA polymerase µ (pol µ) with ku and ligase IV: role for pol µ in end-joining double-strand break repair. Mol. Cell. Biol., 22, 5194–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y., Wu,X., Guo,D., Rechkoblit,O., Taylor,J.S., Geacintov,N.E. and Wang,Z. (2002) Lesion bypass activities of human DNA polymerase µ. J. Biol. Chem., 277, 44582–44587. [DOI] [PubMed] [Google Scholar]
- 20.Havener J.M., Nick McElhinny,S.A., Bassett,E., Gauger,M., Ramsden,D.A. and Chaney,S.G. (2003) Translesion synthesis past platinum DNA adducts by human DNA polymerase µ. Biochemistry, 42, 1777–1788. [DOI] [PubMed] [Google Scholar]
- 21.Pelletier H., Sawaya,M.R., Kumar,A., Wilson,S.H. and Kraut,J. (1994) Structures of ternary complexes of rat DNA polymerase β, a DNA template-primer, and ddCTP. Science, 264, 1891–1903. [PubMed] [Google Scholar]
- 22.Delarue M., Boulé,J.M., Lescar,J., Expert-Bezançon,N., Jourdan,N., Sukumar,N., Rougeon,F. and Papanicolaou,C. (2002) Crystal structure of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase. EMBO J., 21, 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guex N. and Peitsch,M.C. (1997) SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- 24.Burgers P.M.J., Koonin,E.V., Bruford,E., Blanco,L., Burtis,K.C., Christman,M.F., Copeland,W.C., Friedberg,E.C., Hanaoka,F., Hinkle,D.C. et al. (2001) Eukaryotic DNA polymerases: proposal for a revised nomenclature. J. Biol. Chem., 276, 43487–43490. [DOI] [PubMed] [Google Scholar]
- 25.Bollum F.J. (1962) Oligodeoxyribonucleotide-primed reactions catalyzed by calf thymus polymerase. J. Biol. Chem., 237, 1945–1949. [PubMed] [Google Scholar]
- 26.Kunkel T.A. (1985) The mutational specificity of DNA polymerases-alpha and gamma during in vitro DNA synthesis. J. Biol. Chem., 260, 12866–12874. [PubMed] [Google Scholar]
- 27.García-Díaz M., Bebenek,K., Sabariegos,R., Domínguez,O., Rodríguez,J., Kirchhoff,T., García-Palomero,E., Picher,A.J., Juárez,R., Ruiz,J.F., Kunkel,T.A. and Blanco,L. (2002) DNA polymerase λ, a novel DNA repair enzyme in human cells. J. Biol. Chem., 277, 13184–13191. [DOI] [PubMed] [Google Scholar]
- 28.Meuth M. (1984) The genetic consequences of nucleotide precursor pool imbalance in mammalian cells. Mutat. Res., 126, 107–112. [DOI] [PubMed] [Google Scholar]
- 29.Oliveros M., Yáñez,R.J., Salas,M.L., Viñuela,E. and Blanco,L. (1997) Characterization of an African swine fever virus 20-kDa DNA polymerase involved in DNA repair. J. Biol. Chem., 272, 30899–30910. [DOI] [PubMed] [Google Scholar]
- 30.Nick McElhinny S.A. and Ramsden,D.A. (2003) Polymerase mu is a DNA-directed DNA/RNA polymerase. Mol. Cell. Biol., 23, 2309–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han S., Dillon,S.R., Zheng,B., Shimoda,M., Schlissel,M.S. and Kelsoe,G. (1997) V(D)J recombinase activity in a subset of germinal center B lymphocytes. Science, 278, 301–305. [DOI] [PubMed] [Google Scholar]
- 32.Mahajan K.N., Gangi-Peterson,L., Sorscher,D.H., Wang,J., Gathy,K.N., Mahajan,N.P., Reeves,W.H. and Mitchell,B.S. (1999) Association of terminal deoxynucleotidyl transferase with Ku. Proc. Natl Acad. Sci. USA, 96, 13926–13931. [DOI] [PMC free article] [PubMed] [Google Scholar]