Abstract
Regulation of CLB2 is important both for completion of the normal vegetative cell cycle in Saccharo myces cerevisiae and for departure from the vegetative cell cycle upon nitrogen deprivation. Cell cycle-regulated transcription of CLB2 in the G2/M phase is known to be brought about by a set of proteins including Mcm1p, Fkh2/1p and Ndd1p that associate with a 35 bp G2/M-specific sequence common to a set of co-regulated genes. CLB2 transcription is regulated by additional signals, including by nitrogen levels, by positive feedback from the Clb2–Cdc28 kinase, and by osmotic stress, but the corresponding regulatory sequences and proteins have not been identified. We have found that the essential Reb1 transcription factor binds with high affinity to a sequence upstream of CLB2, within a region implicated previously by others in regulated expression, but upstream of the known G2/M-specific site. CLB2 sequence from the region around the Reb1p site blocks activation by the Gal4 protein when positioned downstream of the Gal4-binding site. Since a mutation in the Reb1p site abrogates this effect, we suggest that Reb1p is likely to occupy this site in vivo.
INTRODUCTION
Progress through the cell cycle is mediated by the ordered appearance and disappearance of particular cyclins, a process controlled in turn by regulated transcription of their respective genes and regulated proteolysis of the cyclin proteins. In Saccharomyces cerevisiae, the mitotic cyclin Clb2p, which is coordinately expressed with Clb1p during the G2/M phase of the cell cycle (1–3), associates with the cyclin-dependent kinase Cdc28p and acts to bring about mitosis (4) and isotropic bud growth (5). The central distinct role of CLB2 among the cyclins is suggested by the observation that expression of CLB2 alone, in the absence of the other three mitotic cyclins CLB1, CLB3 and CLB4, is sufficient for cell cycle progression, while deletion of CLB2 (even in the presence of the other three mitotic cyclins) causes morphological changes associated with delayed mitosis (1–3). Clb2p function is controlled in several ways: by regulated expression of the CLB2 gene, by regulated localization and degradation of Clb2 protein, and by regulation of the activity of the Clb2–Cdc28 kinase (6–10).
Cell cycle regulation of CLB2 transcription plays an important part in regulation of Clb2 activity. Transcription of CLB2 during G2/M is brought about by at least three proteins: the essential Mcm1 protein (11,12), Fkh1 and Fkh2 proteins (6,13–17), and Ndd1 protein (18). Mcm1, Fkh2/1 and Ndd1 proteins associate with an essential regulatory site in the CLB2 promoter, referred to as the G2/M-specific site (see Fig. 1), both in vitro and in vivo (11,12,19). Current evidence suggests that Fkh2p plays a more important role in CLB2 regulation than Fkh1p, but the specific role(s) of the Fkh proteins remains to be defined (14,17,19). Moreover, sequences similar to the Mcm1p/Fkh2/1p/Ndd1p-binding site in CLB2 are found in the upstream regions of a set of genes transcribed during the G2/M phase (20,21).
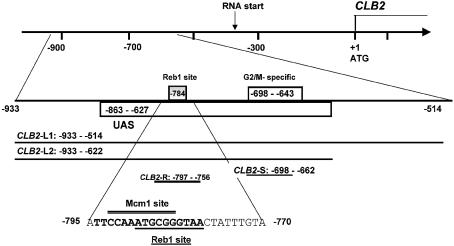
Figure 1.
Diagram of the regulatory sequences of the CLB2 gene. Sequences from –863 to –627, called UAS, are sufficient to confer full expression on a reporter gene (18). The G2/M-specific site from –698 to –643 is bound by Mcm1p, Fkh2p and Ndd1p and is found in a set of G2/M-regulated genes. The Reb1p site at –784 was identified in the course of these experiments; the sequence protected from DNase I digestion (–795 to –770) as well as the Reb1p-binding site (underlined) and putative Mcm1p-binding site (overlined) are shown. DNA fragments used in the binding experiments in this study are illustrated here: CLB2-L1 (–933 to –514), CLB2-L2 (–933 to –622), CLB2-S (–698 to –662) and CLB2-R (–797 to –756).
Among these regulators, Ndd1 protein is the best candidate to be directly responsible for G2/M-regulated expression of CLB2. The NDD1 gene, which was first identified as a high copy suppressor of a cdc28-1N mutant strain (18), is perfectly poised for direct regulation of CLB2: NDD1 is transcribed during S phase; Ndd1 protein is phosphorylated in G2/M (14,16); and Ndd1p is recruited to CLB2 in a cell cycle-specific manner (14,19). Neither Mcm1p nor Fkh2p is as likely to be directly responsible for cell cycle regulation. Both Mcm1 protein, a MADS box protein with homology to the serum response factor, and Fkh2p, one of four proteins in yeast containing a forkhead domain, are involved in regulation of many genes with different patterns of expression (19,22). Furthermore, both Mcm1p and Fkh2p are found upstream of the CLB2 gene throughout the cell cycle (19). However, Fkh2p may play a role in cell cycle regulation, since cell cycle-regulated phosphorylation of Fkh2p has been observed (16).
In addition to its role in the normal cell cycle, Clb2p is implicated in a second role in an environmentally triggered departure from vegetative growth at G2. In low nitrogen, diploid dimorphic yeast strains undergo a transition at G2 from vegetative growth to pseudohyphal development, in which cells become elongated and buds do not septate (23,24). At least two signal transduction pathways mediate this process, one of the end results of which is expression of the MUC1/FLO11 gene (24–28). While there is much still to decipher between the signal and the end result, Rua et al. (24) suggest that CLB2 may be the crucial target of the signal transduction pathways. This is based on two observations. First, deletion of CLB2 triggers pseudohyphal development in the absence of environmental signals (29). Secondly, overproduction of CLB2 is refractory to pseudohyphal development in the presence of a constitutively active signaling pathway (29). Moreover, CLB2 transcription in a diploid yeast cell is regulated by nitrogen availability (30): limitation of nitrogen results in a 5-fold decrease in CLB2 transcript in diploids. Since either low nitrogen availability or deletion of CLB2 can produce the transition from diploid vegetative growth to pseudohyphal development (24), nitrogen regulation of CLB2 transcription is likely to be important in this decision. It is unknown how nitrogen-dependent regulation is mediated.
There is evidence that regulation of CLB2 transcription may involve sequences outside the known G2/M-specific regulatory sequences (see Fig. 1) and/or proteins in addition to Mcm1p, Fkh2/1p and Ndd1p. First, Loy et al. (18) found that a 240 bp CLB2 upstream regulatory sequence (UAS) from –863 to –627 (relative to the ATG) was sufficient to confer full G2/M-regulated expression on a reporter gene, whereas the 55 bp G2/M-specific sequence from –698 to –643, which is bound by Mcm1p, Fkh2p, and Ndd1p, was not (see Fig. 1). We refer to the sequences from –863 to –627 as the CLB2 UAS. One interpretation of these results is that factors in addition to Mcm1p/Fkh2p/Ndd1p aid in expression/regulation of CLB2 and bind outside the G2/M-specific site. Secondly, Loy et al. (18) identified an activity associated with CLB2 UAS sequences that exhibited properties expected of a repressor, since the activity was reduced in extracts from cells arrested in M phase, when CLB2 is transcribed (18). The protein responsible for this binding activity was not identified, although some experiments indicated that it might be Mcm1p. Thirdly, there is evidence that CLB2 transcription may also be regulated by other signals outside of the cell cycle, such as by nitrogen, as discussed above (30), by hypertonic stress (31), and by active Cdc28p–Clb2p in a positive feedback loop (32).
Since CLB2 expression is central to growth and cell cycle progression and is subject to multiple other regulatory circuits, identification of the proteins involved in its regulation is crucial. We present evidence here that the essential Reb1 protein is responsible for a major CLB2 UAS DNA-binding activity, found in extracts. We also show that CLB2 sequences containing the Reb1p site block activation by Gal4 protein, an effect relieved by a point mutation in the Reb1p site. Thus, we suggest that Reb1p binding or its binding site is functional in vivo.
MATERIALS AND METHODS
Strains, plasmids and media
Yeast proteins were purified from EJG573, derived from JHRY20-2Ca [see Martzen et al. (33)], which is MATa, his3Δ200, leu2-3,112, ura3-52, pep4Δ::URA3, cyhR, MCM1-(HA)3. Both MHK7.2.4 and YT1091, which are derived from EG123 [see Kuo et al. (34)], are MATa, ura3-52, leu2-3, 112, trp1-1, with mcm1Δ::TRP1; MHK7.2.4 carries YCplac33 bearing URA3 and MCM1(1–98) (HA)3, while YT1091 carries YCplac33, URA3 and MCM1 (1–286) (HA)3. Yeast strains bearing a deletion of the chromosomal reb1 gene were obtained from John Warner (35,36). Both J342 and J343 are derived from W303 (J47A) of genotype Mata ade2-1, his3-11,15, leu2-3,112, trp1-1, ura3-1, can1-100. J342 is reb1Δ::LEU2 and carries plasmid pBM272-41 with PGAL REB1, HIS3; J343 is derived from J342 by addition of plasmid pAB43 with URA3 and Kluyveromyces lactis REB1.
The PGAL CYC-lacZ reporter constructs were derived from pASGint (37), a gift from A. Johnson. The a-specific genes operator, called asg, which is a binding site for the Mcm1 and α2 proteins, was removed by digestion with SalI and SphI, and religation in the presence of 5′ TCGACCCGGGTACCTGTACAGCATG 3′. CLB2 sequences from –942 to –721 relative to the ATG (PCR amplified with appropriate restriction ends) were inserted between the SalI and SphI sites. This Reb1p site, AATGCGGgTAACTATTTGTA, which matches the consensus site defined by Liaw et al. (38), was mutated to AATGCGGTTAACTATTTGTA, called Reb1p site-m1 below. Plasmids were integrated into the ura3-52 allele in EMP20 MATa/MAtα, ura3-52/ura3-52, his3Δ200/his3Δ200, ade2-101ochre/ade2-101ochre. Four strains derived from EMP20 are reported here: CVS1055 carries the original pASG plasmid; CVS1056 carries the pASG Δasg site; CVS1060 has pASG Δasg CLB2 –942 to –731; and CVS1067 carries pASG Δasg CLB2 –942 to –731 Reb1p site-m1. Integration at the URA3 locus was checked by PCR amplification using one oligonucleotide complementary to chromosomal URA3 sequences and the other complementary to plasmid-specific lacZ sequences.. The resulting PCR products were subjected to sequence analysis to confirm their identity.
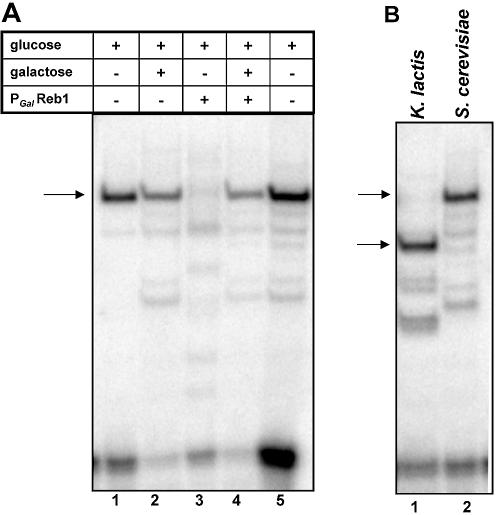
Medium was made as described by Sherman (39) with either 2% dextrose or 3% galactose as the carbon source. For the experiment described in Figure 5, cells were grown in medium containing 2% galactose plus 1% dextrose and shifted to medium with only 2% dextrose.
Figure 5.
Alteration of either the amount or size of Reb1 protein produced in yeast alters the EMSA complex. (A) EMSA with labeled CLB2-R DNA and crude extracts made from a wild-type yeast strain (J47A) and a conditional reb1 mutant (J342) (deleted for the chromosomal reb1 gene and expressing REB1 under control of the Gal promoter) (36). Yeast were grown in galactose + glucose, then shifted to glucose for 4–4.5 generations to repress REB1. Growth in galactose alone is toxic to strain J342, presumably because overproduction of Reb1 protein is toxic (36). Lanes 1 and 2, extracts from the wild-type parent J47A grown in glucose (lane 1) or glucose + galactose (lane 2); lanes 3 and 4, extracts from J342, carrying the conditional PGAL REB1 strain after the switch to glucose (lane 3) or grown in glucose + galactose (lane 4); lane 5, extracts from the wild-type EG573 yeast strain. (B) EMSA of complexes formed with labeled CLB2-R DNA using extracts derived from the J343 yeast strain expressing the 573 amino acid Reb1 protein homolog from K.lactis (lane 1) or from the J47A yeast strain expressing the 802 amino acid S.cerevisiae Reb1 protein (lane 2) (36).
DNAs
CLB2 sequences in the experiments shown in Figures 2, 3 and 4 came from CHR XVI, nucleotides 770 727–771 028, which is –933 to –514 relative to the CLB2 ATG and includes the UAS sequence shown by Loy et al. (18) to confer G2/M-regulated expression on a reporter gene. The CLB2, CLN3 and STE2 DNAs were previously described (22). Plasmid pFBW32 with CLB2 sequences –933 to –514, cloned into Bluescript KS+ (Stratagene), was described previously (22); CLB2 DNA fragments derived from this clone are labeled by the base pairs relative to the CLB2 ATG (CLB2-L1 –933 to –514; CLB2-L2 –933 to –622; CLB2-S –698 to –662; CLB2-R –797 to –756). The 200 bp competitor used in Figure 2B, obtained by PCR amplification from pFBW32, contained –825 to –622. Oligonucleotides used in the binding and competition experiments reported in Figure 3B were 62 bp; the wild-type oligonucleotide contains sequences corresponding to CLB2 –797 to –756 (agacagatcttattccaaatgcgggtaactatttgtataatatgtttacatggatcctgatc); other competitors were based on the same sequence with the mutations shown in Figure 3D. A competitor with a 26 bp CLB2 sequence from –795 to –770 (attccaaatgcgggtaactatttgta) was used in Figure 3C.
Figure 2.
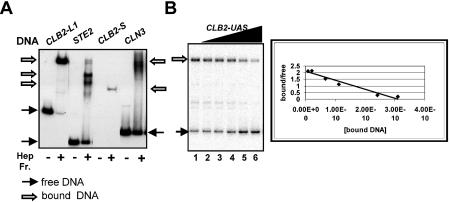
DNA binding of the heparin–agarose-purified yeast proteins assessed by EMSA. (A) The binding activity of heparin–agarose-purified yeast proteins for CLB2-L1 DNA (–933 to –514), which contains the UAS sequences, and CLB2-S DNA (–698 to – 662) containing the Mcm1p/Fkh2p/Ndd1p-binding sites, as well as STE2 and CLN3 UAS sequences, containing known Mcm1p-binding sites. The CLB2 DNAs are illustrated in Figure 1. Bound and free DNAs are indicated by arrows; the free CLB2-S DNA is not shown. (B) Binding of yeast heparin–agarose-purified proteins to 5 × 10–11 M labeled CLB2-L2 DNA (–933 to –622) was competed with unlabeled CLB2 DNA sequences from –825 to –622. Binding affinity was assessed by Scatchard analysis.
Figure 3.
Identification of DNA sequences important for binding. (A) The binding site of the heparin–agarose-purified protein fraction was mapped by DNase I protection on each strand of the CLB2-L2 DNA fragment. The protected region and sequence on each strand are illustrated, as well as the position of the known G2/M-specific sequences. (B) Binding and competition of binding with unlabeled oligonucleotides bearing the protected DNA sequence. DNA-binding activity was examined using EMSA with labeled CLB2-R containing 42 bp of CLB2 (–797 to –756). Oligonucleotides with either the wild-type sequence or mutations in the putative Mcm1p- and Fkhp-binding sites (shown in D) were also used as competitors. In each set, unlabeled competitors were at the following concentrations: 0, 2.5 × 10–10, 5 × 10–10, 7.5 × 10–10 and 10 × 10–10 M. (C) DNA binding to full-length CLB2-L2 (–933 to –622), assessed by EMSA, is competed by an oligonucleotide bearing only 26 bp of CLB2 (corresponding to the protected sequence –795 to –770). This oligonucleotide with a single mutation in the Reb1p (Reb1 site-m1) site fails to compete DNA binding. Competitor DNAs were present at 0, 0, 0.5 × 10–10, 1 × 10–10, 2.5 × 10–10, 5 × 10–10, 25 × 10–10 and 50 × 10–10 M. (D) DNA sequences of the protected region, the potential binding sites for Mcm1p, Fkh2p and Reb1p, and the mutations introduced into the oligonucleotides used in (B) and (C). For each putative binding site, the sequences that constitute a binding site are shown, with the most conserved bases in bold; the mutations introduced to impair binding are underlined.
Figure 4.

Reb1 protein, translated in vitro, binds CLB2 upstream sequences. Reb1 protein (±[35S]methionine) was made in a coupled in vitro transcription–translation system from a plasmid bearing the REB1 gene under control of the T7 promoter. Lanes 1–3: labeled CLB2-L2 DNA in EMSA using yeast heparin–agarose-purified proteins (lane 1), in vitro translated Reb1 protein (lane 2) and in vitro translated luciferase (lane 3). Lanes 4 and 5: 35S-labeled Reb1 protein in EMSA using unlabeled CLB2-L2 DNA (lane 4) or no added CLB2 DNA (lane 5).
DNA binding assays
DNA binding assays contained 24 mM Tris–HCl pH 7.5, 5 mM Tris–HCl pH 8, 160 mM NaCl, 3.8 mM MgCl2, 1.3 mM dithiothreitol (DTT), 1.6 mM (NH4)2SO4, 5 mM spermidine, 50 µg/ml bovine serum albumin (BSA), 20.5 mM EDTA [similar to Lydall et al. (40)], 0.25 mg/ml poly (dIdC:dIdC), 16% glycerol, 0.05 mg/ml calf thymus or salmon sperm DNA, 30 mM β-glycerophosphate, 1 mM Na orthovanadate, 20 nM okadaic acid, 1 mM NaF and 0.2–2 × 10–10 M labeled CLB2 DNA. [The labeled CLB2 DNA was at <0.5 × 10–10 M for experiments involving competition by unlabeled DNA fragments (Figs 2B, and 3B and C).] Binding reactions were incubated at room temperature for 15 min and subjected to electrophoresis in 5% acrylamide, 0.2% bisacrylamide gels containing 0.5× TBE and 10% glycerol; gels were dried and exposed on a phosphoimager. Quantitation of bands was done using the Image-Quant program.
Yeast protein preparation
Yeast EJG573 strain (3.2 l) was grown in YPD medium to OD600 = 3, harvested, washed twice with cold H2O, frozen at –70° C, washed with 10 ml of buffer ZA and resuspended in 10 ml of buffer ZA [0.2 M Tris–HCl pH 8.0, 0.8 M (NH4)2SO4, 10 mM DTT, 10 mM MgCl2, 2 mM CaCl2, 1.4 mM β-mercaptoethanol, 0.1% Triton X-100, 13% glycerol, 1 mM EDTA (41)] with protease and phosphatase inhibitors (1 mM NaF, 50 mM β-glycerophosphate, 0.4 mM Na-orthovanadate, 20 nM okadaic acid, 10 µg/ml leupeptin, 10 µg/ml pepstatin). Cells were lysed with glass beads as described (42), followed by centrifugation at 9300 g for 25 min to remove insoluble material from the crude extract (supernatant). (NH4)2SO4 was added slowly to the crude extract to a final concentration of 50% saturation, followed by stirring for 20 min, and centrifugation for 45 min to obtain the pellet of precipitated proteins which was stored at –70°C. The (NH4)2SO4 pellet was resuspended in a minimal volume (2.2 ml) of buffer C [20 mM HEPES pH 8.0, 5 mM EDTA, 7 mM β-mercaptoethanol, 20% glycerol, 5 mM DTT with 1 mM phenylmethylsulfonyl fluoride (PMSF) and additional protease and phosphatase inhibitors described above] and partially desalted by centrifugation through 2 ml of Sephadex G25 in a disposable polyprep column (Isolab). Material that flowed through the column was diluted 10-fold with buffer A-50 [50 mM Tris–HCl pH 8, 1 mM EDTA, 10% glycerol, 1 mM DTT, 50 mM (NH4)2SO4 with protease and phosphatase inhibitors], nutated with heparin–agarose for 90 min in the cold, after which the heparin–agarose was allowed to settle, and the supernatant was decanted. The heparin–agarose was poured into a column and eluted with a gradient in buffer A from 0 M NaCl, 50 mM (NH4)2SO4 to 2 M NaCl, 0 mM (NH4)2SO4. CLB2 DNA-binding activity eluted at 0.95 M NaCl, 25 mM (NH4)2SO4.
In vitro translation
Reb1p protein was produced in the coupled T7 transcription–rabbit reticulocyte translation system from Promega. Reactions (200 µl) contained 4 µg of plasmid DNA with REB1 under control of the T7 promoter and either [35S]methionine (Amersham-AG1594, 15 mCi/ml >1000Ci/mmol) or unlabeled methionine, and additional components as per Promega Technical Bulletin 126. After incubation at 30°C for 90 min, proteins were precipitated by slow addition of an equal volume of buffer CB (20 mM HEPES pH 8.0, 0.15 M K+ glutamate, 1 mM EDTA, 15% glycerol, 1 mM DTT) saturated with (NH4)2SO4 (70 g added per 100 ml), incubated on ice for an additional 45 min and centrifuged in a microfuge for 30 min. The pellet containing Reb1 protein was resuspended in buffer CB (100 µl) and desalted by passage through 200–250 µl of Sephadex G25 resin equilibrated with buffer CB. The flow through was divided into 11 µl aliquots, frozen under liquid nitrogen and stored at –70°C.
β-Galactosidase assays
The LacZ overlay assay was done according to the protocol by Greg Petsko (http://www.sacs.ucsf.edu/home/HerskowitzLab/protocols/xgalagar.html) with 0.25 mg/ml 5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside (X-Gal).
RESULTS
A CLB2 UAS-binding activity that requires sequences outside the G2/M region and that does not involve Mcm1 protein
In the course of examining crude extracts for activities that bind to DNA from the region upstream of CLB2, we observed strong binding to a large fragment (CLB2-L1) containing the entire UAS defined by Loy et al. (18) and partially purified this activity by heparin–agarose chromatography. As shown in Figure 2, CLB2-L1 (–933 to –514) DNA is bound by these heparin–agarose-purified yeast proteins as well as or better than the regulatory sequences of STE2 and CLN3, which are known to be bound by Mcm1p (Fig. 2A) (11,12,43–45). Most of the CLB2-L1 DNA is bound by these proteins under these conditions. Surprisingly, only a weak DNA–protein complex is observed with the smaller CLB2-S (–698 to –662) probe containing the known Mcm1p/Fkh2p-binding site, although similar amounts of radioactivity and DNA were present in each reaction. Since this heparin–agarose-purified protein fraction also bound to regulatory sequences of STE2 and CLN3, we inferred that Mcm1p was present in this preparation. We confirmed this directly by showing that equivalent fractions from a strain expressing hemagglutinin (HA) epitope-tagged Mcm1 protein reacted with anti-HA antibody (data not shown), as observed earlier [see Kuo et al. (34)]. The binding to CLB2-L1 but not to CLB2-S in fractions that have sufficient Mcm1p to bind other Mcm1p control regions efficiently suggested that a prominent CLB2-binding activity required sequences outside the cell cycle regulatory region of CLB2. Therefore, we continued to investigate this binding activity.
This CLB2 DNA-binding activity was of sufficient affinity and specificity that it was likely to occur in vivo. Moreover, sequences sufficient for UAS activity compete efficiently for this binding, suggesting that it occurs in a biologically important region. To show this, we measured the KD for the binding reaction with CLB2-L2 DNA, which includes sequences from –933 to –622, and competed binding with sequences from –825 to –622 upstream of CLB2, which are a subset of the sequences implicated in UAS function by Loy et al. (18). Binding to radioactive CLB2-L2 DNA, monitored by electrophoretic mobility shift assay (EMSA), was competed with increasing concentrations of unlabeled DNA (Fig. 2B). Based on Scatchard analysis of the data, the KD is ∼2 × 10–10 M and requires only sequences within the UAS region. Furthermore, the binding is not competed by a >100-fold mass excess of salmon sperm DNA, from which we infer that binding is specific enough to be relevant in vivo.
This initial observation suggests that sequences outside the G2/M regulatory region are important for the observed DNA-binding activity, but does not provide information on whether or not proteins previously implicated in CLB2 regulation are involved in the binding activity. For example, there are at least four sites outside of the G2/M control region that could be bound by Mcm1 protein, including two with sequences TTTCCN6GG that match well with conserved bases in known Mcm1p-binding sites and two others that are slightly poorer matches [(12,18,22), see Mai et al. (46)]. These sites could mediate cooperative interactions of Mcm1p binding, or Mcm1p alone could recognize one of these sequences better than the binding site in the G2/M-specific site. Alternatively, a completely different protein could be responsible for the observed CLB2 UAS binding.
Although Mcm1p was known to be present in the heparin–agarose-purified preparation, and is known to bind tightly to DNA alone, we conclude that Mcm1 protein is not present in the CLB2-L2 complex based on three lines of evidence. First, an antibody to an HA epitope tag on Mcm1 protein does not alter the CLB2-L2 DNA–protein complex mobility in EMSA. While the HA antibody quantitatively shifted the Mcm1p–CLN3 DNA complex as expected (43), it had little, if any, effect on the protein–CLB2-L2 complex (see Supplementary material). Secondly, a change in the size of the Mcm1 protein does not alter the mobility of the complex in EMSA. The mobility of the CLB2 DNA–protein complexes made with extracts from two strains that differ only in the size of the Mcm1p is similar (see Supplementary material). Thirdly, the proteins that bind to the CLN3 DNA and to the CLB2 DNA differ from each other: the CLB2-binding activity is not competed by CLN3 DNA with known Mcm1p-binding sites. Both CLB2 and CLN3 compete in the 10–10–10–9 M range for binding to themselves (data not shown).
The essential Reb1 protein binds the CLB2 UAS upstream of the G2/M-specific regulatory region
To further characterize the DNA-binding activity, the binding site was mapped using DNase I protection analysis. As shown in Figure 3A, the observed protected region is ∼26 bp long and covers similar regions of each strand. It is located between –795 and –770, ∼95 bp upstream of the known G2/M-specific sequences (see Fig. 1). Thus, this binding activity differs in both location and protein content from the previously described binding of Mcm1p/Fkh2p/Ndd1p to the G2/M cell cycle regulatory region (11,12).
We suggest that the activity responsible for DNase I protection is also responsible for the complex detected by EMSA, based on two observations. First, a strong binding activity is found using a labeled 57 bp fragment which includes 42 bp of the protected sequences (Fig. 3B) (CLB2-R, from –797 to –756) and 15 flanking base pairs, and binding to this probe is self-competed by DNA at <5 × 10–10 M, a concentration similar to that required for competition of binding by the 200 bp UAS fragment (Fig. 2B). Secondly, the protein–DNA complex formed with the 300 bp CLB2-L2 (–933 to –622) fragment is competed by an oligonucleotide in which the only CLB2 sequences are the 26 bp protected from DNase I digestion (see Fig. 3C). Thus the EMSA-detected binding activity and binding detected by DNase I protection are probably due to the same binding activity, and sequences outside the protected region are not required for high affinity binding.
Within and surrounding the protected region, there are potential binding sites for Mcm1p, Reb1p and possibly Fkh-like proteins (see Figs 1 and 4D). Thus it is feasible that any of these proteins or proteins with related DNA-binding domains could interact with this sequence. For example, in yeast, in addition to Mcm1p, there are three proteins, Arg80p, Rlm1p and Smp1p (47–49), that contain MADS box DNA-binding domains. Since MADS box proteins recognize related DNA sequences (50) and in some cases act cooperatively (51), any of these proteins could recognize this site. Likewise, in addition to Fkh2p, there are three additional forkhead-containing proteins, Hcm1p, Fkh1p and Fhl1p (13,17,52,53). Reb1p, the product of an essential gene, is implicated in transcription regulation at many sites (54–56). One yeast protein, the product of YDR026, has significant homology to the myb-like Reb1p DNA-binding domains, although this protein has not been previously characterized.
To learn which (if any) of the consensus binding sites was important for the binding activity, oligonucleotides with and without mutations in the putative Mcm1p-, Fkhp- and Reb1p-binding sites were used as competitors of the DNA-binding activity (Fig. 3B and C). In the experiment shown in Figure 3B, complexes were formed with the 57 bp labeled DNA CLB2-R, with CLB2 sequence from –797 to –756, and formation of the labeled complex was competed with unlabeled DNA containing the same CLB2 sequences bearing the mutations indicated in Figure 3D. Competitor DNAs with mutations in either the Mcm1p-binding site or the putative Fkhp-binding site were as effective competitors as DNA with the wild-type sequence (Fig. 3B and D). Since the mutations in the Mcm1p-binding site alter highly conserved bases implicated in MADs box binding (47,57–60), it is unlikely that any MADS box protein is required for binding. Since introduction of three mutations into the putative Fkhp-binding site did not impair binding, it is unlikely that Fkh-like proteins are involved.
In the experiment shown in Figure 3C, complexes were formed with labeled full-length CLB2-L2 DNA (–933 to –622), and binding was competed with DNAs containing only 26 bp homologous to CLB2 (–795 to –770), corresponding to the protected region shown in its entirety in Figure 3D. DNA with the wild-type sequence is an effective competitor (Fig. 3C). In contrast, an oligonucleotide with a mutation in a conserved base in the putative Reb1p-binding site is severely impaired as a competitor (Fig. 3C and D). Since the sequence GCGGGT was obtained by selection of Reb1p-binding sites from random sequences (38) and is highly conserved at known Reb1p-binding sites, the specific mutation site was chosen to alter the Reb1p consensus sequence GCGGGT without affecting the Mcm1p-binding site (Fig. 3D). We infer that this part of the sequence is important for specific binding and that Reb1p (or its homologs) may bind CLB2-L fragments. Moreover, the apparent KD calculated from oligonucleotide competition of CLB2-L2 binding (from data in Fig. 3C, see Supplementary material) is 3 × 10–10 M, very similar to that for CLB2 UAS competition of CLB2-L2 binding (2 × 10–10 M, Fig. 2B). This suggests that the oligonucleotide contains the sequences important for CLB2-L2 binding.
To provide support for this proposal, we show directly that Reb1 protein, made in vitro in a rabbit reticulocyte extract, is able to bind the CLB2-L2 fragment containing the CLB2 UAS (Fig. 4). As shown in Figure 4, the Reb1p–CLB2 DNA complex observed with Reb1 protein translated in vitro (lanes 2 and 4) migrates to the same position as the CLB2 complex with yeast heparin–agarose-purified proteins (lane1). Reb1p binding to CLB2-L2 can be observed either with labeled CLB2-L2 DNA and unlabeled in vitro translated Reb1 protein (lane 2) or with radioactively labeled Reb1 protein and unlabeled CLB2-L2 DNA (lane 4). Both Reb1 protein and CLB2 DNA are required for complex formation since no complex is observed either when labeled CLB2-L2 DNA is incubated with a lysate programmed to produce luciferase (lane 3) or when radioactive Reb1 protein is examined in the absence of CLB2 DNA (lane 5). In addition, two faster migrating complexes are seen with in vitro transcribed/translated Reb1 protein; these are most probably due to truncated Reb1 protein, since their formation requires both CLB2 DNA and Reb1 protein. Since all reactions contain a 45-fold mass excess of salmon sperm DNA over CLB2 DNA and the salmon sperm DNA is present in the no CLB2 control in lane 5, the Reb1p DNA-binding activity appears to be specific for CLB2 DNA.
Reb1p is responsible for the binding activity observed in crude extracts
Two lines of evidence indicate that Reb1 protein is responsible for the DNA-binding activity that protects –795 to –770 upstream of CLB2 and is the most prominent activity observed with yeast crude extracts (data not shown). First, if REB1 expression is altered, the amount of CLB2–protein complex is altered. We demonstrated this using a strain in which expression of the S.cerevisiae REB1 gene is under galactose control. As shown in Figure 5A, DNA binding to CLB2-R (–797 to –756) is severely depleted in extracts from the PGAL-REB1 conditional mutant made 3–4 generations after shifting the strains to glucose medium (in which expression of REB1 is nearly shut off) (lane 3) in comparison with extracts made from the same strain grown in glucose plus galactose (lane 4) (in which some expression of REB1 occurs). This is not observed in the parental strains (lanes 1 and 2). We infer either that Reb1 protein is required for the CLB2-binding activity or that a fairly labile gene product whose expression is under Reb1p control is required for the activity. Furthermore, we note that the major CLB2–protein complex from these extracts co-migrates with the major CLB2-binding activity found in our pep4Δ strain (lane 5).
Secondly, if the size of the Reb1 protein is altered, the mobility of the protein–DNA complex is also changed. We showed this by using a strain that expresses the K.lactis homolog of REB1, which encodes a 573 amino acid protein instead of the 802 amino acid protein encoded by the S.cerevisiae REB1 gene (36). As can be seen in Figure 5B, the CLB2-R–protein complex observed with extracts from a strain in which K.lactis Reb1p is the primary source of Reb1 protein (Fig. 5B, lane 1) migrates significantly faster than the complex with the S.cerevisae Reb1p (Fig. 5B, lane 2). This result provides strong evidence that Reb1 protein is part of the major CLB2–protein complex in vitro.
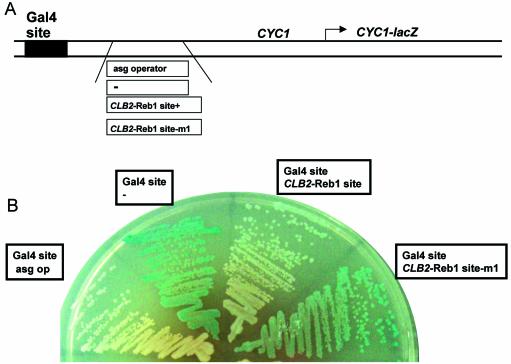
CLB2 sequences with the Reb1p site block Gal4 transcription activation
To find out if the Reb1p site is occupied in vivo, we inserted CLB2 sequences containing the Reb1p site downstream of a binding site for the Gal4 transcription activator and upstream of the TATA box adjacent to a CYC1-lacZ reporter gene (37). Use of this construct allows identification of sequences that either activate transcription in glucose or interfere with Gal4 activation in galactose. CLB2 sequences containing the Reb1p site but lacking the G2/M-specific activation region (see Figs 1 and 6A) were introduced into the reporter gene, which was then integrated into the chromosome at the URA3 locus. Both of the control constructs, with either the Gal4 site alone or the Gal4 site and the asg operator, behave as expected. Expression of CYC1-lacZ on galactose is observed in constructs bearing the Gal4-binding site alone (Fig. 6B); little/no expression of this construct is seen on glucose (data not shown). The presence of a binding site for a known repressor [the a-specific genes (asg) operator] reduces galactose-dependent expression of CYC1-lacZ (Fig. 6B) (37); the α2 protein, a strong repressor, binds the asg operator with Mcm1 protein (61,62).
Figure 6.
The CLB2 Reb1 site modulates Gal4p activation of gene expression. (A) Diagram of the four reporter constructs that were integrated at the URA3 locus and assayed in (B). The Gal4p DNA-binding site and the a-specific genes (asg) operator are present in the original construct (37). The asg operator was replaced by CLB2 sequences from –942 to –721 (either wild type or bearing the Reb1p site m1 mutation) or by a linker sequence as shown. (B) The expression of the CYC1-lacZ gene from yeast strains bearing each plasmid was examined on plates containing X-gal and galactose.
We find that the CLB2 sequences interfere with Gal4-dependent expression of CYC1-lacZ, and that interference activity is dependent upon an intact Reb1p-binding site. CYC1-lacZ expression on galactose is substantially reduced when the CLB2 sequences –942 to –721 are inserted downstream of the Gal4-binding site. Moreover, a single mutation in a conserved base in the Reb1p-binding site (38) substantially restores galactose-dependent expression of CYC1-lacZ, i.e. the mutation reduces interference by these CLB2 sequences. We have shown that this base change, a G→T mutation (seen in Fig. 3C and D), almost completely abolished Reb1p binding in vitro based on the inability of this DNA to compete specific binding. Thus, the Reb1p site is likely to be occupied in vivo. We note that the CLB2 sequences do not cause activation in glucose, suggesting that there is no strong activation function associated with these CLB2 sequences (data not shown).
DISCUSSION
We have provided evidence that the essential Reb1 transcription factor binds a sequence upstream of the CLB2 gene in a region implicated in transcription regulatory function (18). A tight, specific DNA-binding activity protects a region of CLB2 95 bp upstream of the previously identified G2/M-specific site, which is bound by Mcm1p/Fkh2p/Ndd1p. Although a consensus Mcm1p-binding site is present in the protected region, neither the sequence nor the protein requirements for binding are consistent with a role for Mcm1p in this complex. In support of this, we find that (i) competition for binding to CLB2 sequences is not impaired by mutations in the conserved bases of the Mcm1p-binding site at the protected region; (ii) the CLB2-binding activity is not competed by DNAs containing known Mcm1p-binding sites; and (iii) the mobility of the CLB2–protein complex is not affected by antibodies directed at an HA epitope tag on Mcm1p, or by the size of the Mcm1 protein present in the yeast extract. Reb1 protein is responsible for DNA binding based on the following evidence: (i) mutations in the putative Reb1p-binding site found at the protected region do impair binding, since an oligonucleotide with a mutation in the Reb1p site is unable to competitively inhibit specific DNA binding; (ii) Reb1 protein made in vitro or in Escherichia coli (data not shown) is able to bind these CLB2 sequences and generate a protein–DNA complex of mobility similar to the yeast complex; (iii) the size of the Reb1 protein in a yeast extract affects the mobility of the protein–CLB2 complex; and (iv) depletion of Reb1 protein from yeast cells eliminates the CLB2–protein complex.
It is likely that this upstream sequence of CLB2 is bound by Reb1p in vivo. First, Gal4-mediated transcription activation is blocked by upstream CLB2 sequences that contain the Reb1p site; this effect is significantly diminished by the introduction of a single base change in the Reb1p-binding site. The ability of this site to affect gene expression in vivo suggests that it is normally bound in vivo. Secondly, while this work was in progress, Lee et al. (63) mapped the location of 106 transcription factors in the intergenic regions of the S.cerevisiae genome by tagging the regulators and performing chromatin immunoprecipitation (ChIP) analysis. Their data provide independent support for the presence of Reb1p at CLB2 in vivo; they report a high probability that Reb1 protein is associated with CLB2 intergenic sequences (ρ = 2.4 × 10–5, with ρ = 0.001 an acceptable score) (63).
Reb1 protein is an essential, auto-regulated DNA-binding protein that binds to many sites in the S.cerevisiae genome (35,64); upstream regions from 146–250 yeast genes are enriched in genomic ChIP analysis with Reb1p (63). Reb1p sites, which can have either positive or negative effects on expression, are found upstream of a number of RNA polymerase II-dependent genes, including many important metabolic genes (56,65–69), as well as upstream and downstream of rRNA genes (55,65,70,71). Reb1p, as well as its homologs in Scizosaccharomyces pombe and mice (72–74), is implicated in termination of rRNA transcription (55,73,75).
Reb1p may exert many of its effects on gene expression by modulating chromatin structure, based on three observations. First, Reb1 protein is implicated in the separation of silent chromatin domains from expressed domains. Reb1p-binding sites are present in insulator elements that have this effect, and a synthetic multimer of Reb1p-binding sites exhibits insulator activity (76). Secondly, the Reb1 protein in K.lactis, which is highly homologous to and can functionally substitute for Reb1p in S.cerevisiae protein, is responsible for transcriptional silencing at the mating type loci (77); this activity is primarily associated with chromatin domains. Thirdly, Reb1 protein is found associated with several proteins implicated in chromatin structure or remodeling, including Rsc2 and Rsc3, components of a histone deacetylase complex, and Eaf1, part of a complex with the histone acetyltransferase Esa1 (78,79). Interestingly, yeast with mutations in RSC2 arrest in the G2/M phase of the cell cycle (80–82), although the basis for this phenotype is unknown.
We can envisage several possible roles for Reb1p and this regulatory site in CLB2 control, most of which are not necessarily mutually exclusive. Since there are overlapping Mcm1p and Reb1p consensus sites at this position, Mcm1p and Reb1p may compete for binding to this site in CLB2. Regulation might be mediated by competition, in which the important role of Reb1p is primarily occlusion of Mcm1 protein, or vice versa. If so, deletion of the site could easily fail to reveal its potential regulatory effects since binding of both proteins would be lost simultaneously; mutation of individual sites will be required. Moreover, the situation may be complicated by the presence of additional Mcm1p-binding sites upstream of CLB2.
Alternatively, the Reb1p site might be required for differential regulation such as repression in low nitrogen, osmotic regulation, activation by Cdc28–Clb2 or repression during sporulation. Reb1p might act alone or in concert with other regulators. For example, Miled et al. (30) found that Xbp1 (a stress-inducible transcription factor) was required for repression of CLB2 transcription in low nitrogen. It is unknown if Xbp1 binds directly to the CLB2 regulatory region, although Mai and Breeden (83,84) noted a consensus binding site for Xbp1 in the CLB2 upstream region.
Another alternative is that at CLB2, Reb1p may work primarily by recruiting specific chromatin-remodeling agents; we can envisage three different effects these might have on CLB2 expression. First, the Reb1p–CLB2 recruited complex could potentiate activation by the Mcm1p–Fkh2p–Ndd1p complex. If this is correct, then Reb1p binding is the explanation for the observation by Loy et al. (18) that the 250 bp UAS was required to activate a reporter gene rather than simply the 55 bp G2/M regulatory sequence. Secondly, Reb1p-recruited agents could facilitate repression of CLB2 during the S and G1 phases of the cell cycle. Evidence that such repression exists is based on the observation that deletion of FKH1 and FKH2 has three consequences: loss of cell cycle control of CLB2 transcription, an increase in basal transcription of CLB2 throughout the cell cycle, and suppression of the requirement for NDD1 for viability (14). Thirdly, Reb1p-recruited complexes could insulate the CLB2 gene from external signals commencing at adjacent genes that could result in inappropriate expression since either too much transcript, too little transcript or expression at the wrong time is likely to have consequences. Fourel et al. (85) propose that Reb1 protein, with a set of general regulatory factors, may act to partition the genome, in particular insulating one chromosomal domain from another.
Reb1p is likely to play an important role in the complex regulation of CLB2 which will in turn influence the timing or progression of the cell cycle or the switch to pseudohyphal development. Identification of other proteins that affect this promoter and their interactions will allow us to understand not only the regulatory circuitry of the CLB2 gene but also the mechanisms by which the cell cycle is regulated.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Andrei Alexandrov, Feng Xing, Mark Dumont, Jane Jackman, and in particular Eric Phizicky for helpful comments on the manuscript, J. Warner for strains, and A. Johnson for plasmids. This work was supported by grant HG02311 from the National Institutes of Health to Eric M. Phizicky.
REFERENCES
- 1.Surana U., Robitsch,H., Price,C., Schuster,T., Fitch,I., Futcher,A.B. and Nasmyth,K. (1991) The role of CDC28 and cyclins during mitosis in the budding yeast S.cerevisiae. Cell, 65, 145–161. [DOI] [PubMed] [Google Scholar]
- 2.Richardson H., Lew,D.J., Henze,M., Sugimoto,K. and Reed,S.I. (1992) Cyclin-B homologs in Saccharomyces cerevisiae function in S phase and in G2. Genes Dev., 6, 2021–2034. [DOI] [PubMed] [Google Scholar]
- 3.Fitch I., Dahmann,C., Surana,U., Amon,A., Nasmyth,K., Goetsch,L., Byers,B. and Futcher,B. (1992) Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell, 3, 805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nasmyth K. (1996) At the heart of the budding yeast cell cycle. Trends Genet., 12, 405–412. [DOI] [PubMed] [Google Scholar]
- 5.Lew D.J. and Reed,S.I. (1993) Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol., 120, 1305–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hollenhorst P.C., Bose,M.E., Mielke,M.R., Muller,U. and Fox,C.A. (2000) Forkhead genes in transcriptional silencing, cell morphology and the cell cycle. Overlapping and distinct functions for FKH1 and FKH2 in Saccharomyces cerevisiae. Genetics, 154, 1533–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheu Y.J., Barral,Y. and Snyder,M. (2000) Polarized growth controls cell shape and bipolar bud site selection in Saccharomyces cerevisiae. Mol. Cell. Biol., 20, 5235–5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeong F.M., Lim,H.H., Padmashree,C.G. and Surana,U. (2000) Exit from mitosis in budding yeast: biphasic inactivation of the Cdc28–Clb2 mitotic kinase and the role of Cdc20. Mol. Cell, 5, 501–511. [DOI] [PubMed] [Google Scholar]
- 9.Hood J.K., Hwang,W.W. and Silver,P.A. (2001) The Saccharomyces cerevisiae cyclin Clb2p is targeted to multiple subcellular locations by cis- and trans-acting determinants. J. Cell Sci., 114, 589–597. [DOI] [PubMed] [Google Scholar]
- 10.Schwab M., Lutum,A.S. and Seufert,W. (1997) Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell, 90, 683–693. [DOI] [PubMed] [Google Scholar]
- 11.Althoefer H., Schleiffer,A., Wassmann,K., Nordheim,A. and Ammerer,G. (1995) Mcm1 is required to coordinate G2-specific transcription in Saccharomyces cerevisiae. Mol. Cell. Biol., 15, 5917–5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maher M., Cong,F., Kindelberger,D., Nasmyth,K. and Dalton,S. (1995) Cell cycle-regulated transcription of the CLB2 gene is dependent on Mcm1 and a ternary complex factor. Mol. Cell. Biol., 15, 3129–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hollenhorst P.C., Pietz,G. and Fox,C.A. (2001) Mechanisms controlling differential promoter-occupancy by the yeast forkhead proteins Fkh1p and Fkh2p: implications for regulating the cell cycle and differentiation. Genes Dev., 15, 2445–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koranda M., Schleiffer,A., Endler,L. and Ammerer,G. (2000) Forkhead-like transcription factors recruit Ndd1 to the chromatin of G2/M-specific promoters. Nature, 406, 94–98. [DOI] [PubMed] [Google Scholar]
- 15.Kumar R., Reynolds,D.M., Shevchenko,A., Goldstone,S.D. and Dalton,S. (2000) Forkhead transcription factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control transcription required for M-phase. Curr. Biol., 10, 896–906. [DOI] [PubMed] [Google Scholar]
- 16.Pic A., Lim,F.L., Ross,S.J., Veal,E.A., Johnson,A.L., Sultan,M.R., West,A.G., Johnston,L.H., Sharrocks,A.D. and Morgan,B.A. (2000) The forkhead protein Fkh2 is a component of the yeast cell cycle transcription factor SFF. EMBO J., 19, 3750–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu G., Spellman,P.T., Volpe,T., Brown,P.O., Botstein,D., Davis,T.N. and Futcher,B. (2000) Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature, 406, 90–94. [DOI] [PubMed] [Google Scholar]
- 18.Loy C.J., Lydall,D. and Surana,U. (1999) NDD1, a high-dosage suppressor of cdc28–1N, is essential for expression of a subset of late-S-phase-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 3312–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simon I., Barnett,J., Hannett,N., Harbison,C.T., Rinaldi,N.J., Volkert,T.L., Wyrick,J.J., Zeitlinger,J., Gifford,D.K., Jaakkola,T.S. and Young,R.A. (2001) Serial regulation of transcriptional regulators in the yeast cell cycle. Cell, 106, 697–708. [DOI] [PubMed] [Google Scholar]
- 20.Spellman P.T., Sherlock,G., Zhang,M.Q., Iyer,V.R., Anders,K., Eisen,M.B., Brown,P.O., Botstein,D. and Futcher,B. (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell, 9, 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho R.J., Campbell,M.J., Winzeler,E.A., Steinmetz,L., Conway,A., Wodicka,L., Wolfsberg,T.G., Gabrielian,A.E., Landsman,D., Lockhart,D.J. and Davis,R.W. (1998) A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell, 2, 65–73. [DOI] [PubMed] [Google Scholar]
- 22.Kuo M.H. and Grayhack,E. (1994) A library of yeast genomic MCM1 binding sites contains genes involved in cell cycle control, cell wall and membrane structure and metabolism. Mol. Cell. Biol., 14, 348–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gancedo J.M. (2001) Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev., 25, 107–123. [DOI] [PubMed] [Google Scholar]
- 24.Rua D., Tobe,B.T. and Kron,S.J. (2001) Cell cycle control of yeast filamentous growth. Curr. Opin. Microbiol., 4, 720–727. [DOI] [PubMed] [Google Scholar]
- 25.Gimeno C.J., Ljungdahl,P.O., Styles,C.A. and Fink,G.R. (1992) Unipolar cell divisions in the yeast S.cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell, 68, 1077–1090. [DOI] [PubMed] [Google Scholar]
- 26.Lorenz M.C. and Heitman,J. (1997) Yeast pseudohyphal growth is regulated by GPA2, a G protein α homolog. EMBO J., 16, 7008–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan X. and Heitman,J. (1999) Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 4874–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rupp S., Summers,E., Lo,H.J., Madhani,H. and Fink,G. (1999) MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J., 18, 1257–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahn S.H., Acurio,A. and Kron,S.J. (1999) Regulation of G2/M progression by the STE mitogen-activated protein kinase pathway in budding yeast filamentous growth. Mol. Biol. Cell, 10, 3301–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miled C., Mann,C. and Faye,G. (2001) Xbp1-mediated repression of CLB gene expression contributes to the modifications of yeast cell morphology and cell cycle seen during nitrogen-limited growth. Mol. Cell. Biol., 21, 3714–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexander M.R., Tyers,M., Perret,M., Craig,B.M., Fang,K.S. and Gustin,M.C. (2001) Regulation of cell cycle progression by Swe1p and Hog1p following hypertonic stress. Mol. Biol. Cell, 12, 53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amon A., Tyers,M., Futcher,B. and Nasmyth,K. (1993) Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell, 74, 993–1007. [DOI] [PubMed] [Google Scholar]
- 33.Martzen M.R., McCraith,S.M., Spinelli,S.L., Torres,F.M., Fields,S., Grayhack,E.J. and Phizicky,E.M. (1999) A biochemical genomics approach for identifying genes by the activity of their products. Science, 286, 1153–1155. [DOI] [PubMed] [Google Scholar]
- 34.Kuo M.H., Nadeau,E.T. and Grayhack,E.J. (1997) Multiple phosphorylated forms of the Saccharomyces cerevisiae Mcm1 protein include an isoform induced in response to high salt concentrations. Mol. Cell. Biol., 17, 819–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ju Q.D., Morrow,B.E. and Warner,J.R. (1990) REB1, a yeast DNA-binding protein with many targets, is essential for growth and bears some resemblance to the oncogene myb. Mol. Cell. Biol., 10, 5226–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrow B.E., Ju,Q. and Warner,J.R. (1993) A bipartite DNA-binding domain in yeast Reb1p. Mol. Cell. Biol., 13, 1173–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Redd M.J., Stark,M.R. and Johnson,A.D. (1996) Accessibility of alpha 2-repressed promoters to the activator Gal4. Mol. Cell. Biol., 16, 2865–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liaw P.C. and Brandl,C.J. (1994) Defining the sequence specificity of the Saccharomyces cerevisiae DNA binding protein REB1p by selecting binding sites from random-sequence oligonucleotides. Yeast, 10, 771–787. [DOI] [PubMed] [Google Scholar]
- 39.Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194, 3–21. [DOI] [PubMed] [Google Scholar]
- 40.Lydall D., Ammerer,G. and Nasmyth,K. (1991) A new role for MCM1 in yeast: cell cycle regulation of SW15 transcription. Genes Dev., 5, 2405–2419. [DOI] [PubMed] [Google Scholar]
- 41.Company M., Adler,C. and Errede,B. (1988) Identification of a Ty1 regulatory sequence responsive to STE7 and STE12. Mol. Cell. Biol., 8, 2545–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phizicky E.M., Martzen,M.R., McCraith,S.M., Spinelli,S.L., Xing,F., Shull,N.P., Van Slyke,C., Montagne,R.K., Torres,F.M., Fields,S. and Grayhack,E.J. (2002) Biochemical genomics approach to map activities to genes. Methods Enzymol., 350, 546–559. [DOI] [PubMed] [Google Scholar]
- 43.McInerny C.J., Partridge,J.F., Mikesell,G.E., Creemer,D.P. and Breeden,L.L. (1997) A novel Mcm1-dependent element in the SWI4, CLN3, CDC6 and CDC47 promoters activates M/G1-specific transcription. Genes Dev., 11, 1277–1288. [DOI] [PubMed] [Google Scholar]
- 44.Passmore S., Elble,R. and Tye,B.K. (1989) A protein involved in minichromosome maintenance in yeast binds a transcriptional enhancer conserved in eukaryotes. Genes Dev., 3, 921–935. [DOI] [PubMed] [Google Scholar]
- 45.Jarvis E.E., Clark,K.L. and Sprague,G.F.,Jr (1989) The yeast transcription activator PRTF, a homolog of the mammalian serum response factor, is encoded by the MCM1 gene. Genes Dev., 3, 936–945. [DOI] [PubMed] [Google Scholar]
- 46.Mai B., Miles,S. and Breeden,L.L. (2002) Characterization of the ECB binding complex responsible for the M/G(1)-specific transcription of CLN3 and SWI4. Mol. Cell. Biol., 22, 430–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jamai A., Dubois,E., Vershon,A.K. and Messenguy,F. (2002) Swapping functional specificity of a MADS box protein: residues required for Arg80 regulation of arginine metabolism. Mol. Cell. Biol., 22, 5741–5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watanabe Y., Irie,K. and Matsumoto,K. (1995) Yeast RLM1 encodes a serum response factor-like protein that may function downstream of the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell. Biol., 15, 5740–5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.West A.G. and Sharrocks,A.D. (1999) MADS-box transcription factors adopt alternative mechanisms for bending DNA. J. Mol. Biol., 286, 1311–1323. [DOI] [PubMed] [Google Scholar]
- 50.Shore P. and Sharrocks,A.D. (1995) The MADS-box family of transcription factors. Eur. J. Biochem., 229, 1–13. [DOI] [PubMed] [Google Scholar]
- 51.Amar N., Messenguy,F., El Bakkoury,M. and Dubois,E. (2000) ArgRII, a component of the ArgR–Mcm1 complex involved in the control of arginine metabolism in Saccharomyces cerevisiae, is the sensor of arginine. Mol. Cell. Biol., 20, 2087–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu G. and Davis,T.N. (1998) The fork head transcription factor Hcm1p participates in the regulation of SPC110, which encodes the calmodulin-binding protein in the yeast spindle pole body. Biochim. Biophys Acta, 1448, 236–244. [DOI] [PubMed] [Google Scholar]
- 53.Hermann-LeDenmat S., Werner,M., Sentenac,A. and Thuriaux,P. (1994) Suppression of yeast RNA polymerase III mutations by FHL1, a gene coding for a fork head protein involved in rRNA processing. Mol. Cell. Biol., 14, 2905–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y., Moir,R.D., Sethy-Coraci,I.K., Warner,J.R. and Willis,I.M. (2000) Repression of ribosome and tRNA synthesis in secretion-defective cells is signaled by a novel branch of the cell integrity pathway. Mol. Cell. Biol., 20, 3843–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson S.P. and Warner,J.R. (1991) Termination of transcription of ribosomal RNA in Saccharomyces cerevisiae. Mol. Cell. Biochem., 104, 163–168. [DOI] [PubMed] [Google Scholar]
- 56.Chasman D.I., Lue,N.F., Buchman,A.R., LaPointe,J.W., Lorch,Y. and Kornberg,R.D. (1990) A yeast protein that influences the chromatin structure of UASG and functions as a powerful auxiliary gene activator. Genes Dev., 4, 503–514. [DOI] [PubMed] [Google Scholar]
- 57.Pellegrini L., Tan,S. and Richmond,T.J. (1995) Structure of serum response factor core bound to DNA. Nature, 376, 490–498. [DOI] [PubMed] [Google Scholar]
- 58.Tan S. and Richmond,T.J. (1998) Crystal structure of the yeast MATα2/MCM1/DNA ternary complex. Nature, 391, 660–666. [DOI] [PubMed] [Google Scholar]
- 59.Tan S., Hunziker,Y., Pellegrini,L. and Richmond,T.J. (2000) Crystallization of the yeast MATα2/MCM1/DNA ternary complex: general methods and principles for protein–DNA cocrystallization. J. Mol. Biol., 297, 947–959. [DOI] [PubMed] [Google Scholar]
- 60.Acton T.B., Zhong,H. and Vershon,A.K. (1997) DNA-binding specificity of Mcm1: operator mutations that alter DNA-bending and transcriptional activities by a MADS box protein. Mol. Cell. Biol., 17, 1881–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keleher C.A., Redd,M.J., Schultz,J., Carlson,M. and Johnson,A.D. (1992) Ssn6–Tup1 is a general repressor of transcription in yeast. Cell, 68, 709–719. [DOI] [PubMed] [Google Scholar]
- 62.Smith R.L. and Johnson,A.D. (2000) Turning genes off by Ssn6–Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci., 25, 325–330. [DOI] [PubMed] [Google Scholar]
- 63.Lee T.I., Rinaldi,N.J., Robert,F., Odom,D.T., Bar-Joseph,Z., Gerber,G.K., Hannett,N.M., Harbison,C.T., Thompson,C.M., Simon,I., Zeitlinger,J., Jennings,E.G., Murray,H.L., Gordon,D.B., Ren,B., Wyrick,J.J., Tagne,J.B., Volkert,T.L., Fraenkel,E., Gifford,D.K. and Young,R.A. (2002) Transcriptional regulatory networks in Saccharomyces cerevisiae. Science, 298, 799–804. [DOI] [PubMed] [Google Scholar]
- 64.Wang K.L. and Warner,J.R. (1998) Positive and negative autoregulation of REB1 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol., 18, 4368–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang H., Nicholson,P.R. and Stillman,D.J. (1990) Identification of a Saccharomyces cerevisiae DNA-binding protein involved in transcriptional regulation. Mol. Cell. Biol., 10, 1743–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Packham E.A., Graham,I.R. and Chambers,A. (1996) The multifunctional transcription factors Abf1p, Rap1p and Reb1p are required for full transcriptional activation of the chromosomal PGK gene in Saccharomyces cerevisiae. Mol. Gen. Genet., 250, 348–356. [DOI] [PubMed] [Google Scholar]
- 67.Schuller H.J., Schutz,A., Knab,S., Hoffmann,B. and Schweizer,E. (1994) Importance of general regulatory factors Rap1p, Abf1p and Reb1p for the activation of yeast fatty acid synthase genes FAS1 and FAS2. Eur. J. Biochem., 225, 213–222. [DOI] [PubMed] [Google Scholar]
- 68.Moreira J.M., Horz,W. and Holmberg,S. (2002) Neither Reb1p nor poly(dA*T) elements are responsible for the highly specific chromatin organization at the ILV1 promoter. J. Biol. Chem., 277, 3202–3209. [DOI] [PubMed] [Google Scholar]
- 69.Carmen A.A. and Holland,M.J. (1994) The upstream repression sequence from the yeast enolase gene ENO1 is a complex regulatory element that binds multiple trans-acting factors including REB1. J. Biol. Chem., 269, 9790–9797. [PubMed] [Google Scholar]
- 70.Kang J.J., Yokoi,T.J. and Holland,M.J. (1995) Binding sites for abundant nuclear factors modulate RNA polymerase I-dependent enhancer function in Saccharomyces cerevisiae. J. Biol. Chem., 270, 28723–28732. [DOI] [PubMed] [Google Scholar]
- 71.Kulkens T., van der Sande,C.A., Dekker,A.F., van Heerikhuizen,H. and Planta,R.J. (1992) A system to study transcription by yeast RNA polymerase I within the chromosomal context: functional analysis of the ribosomal DNA enhancer and the RBP1/REB1 binding sites. EMBO J., 11, 4665–4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao A., Guo,A., Liu,Z. and Pape,L. (1997) Molecular cloning and analysis of Schizosaccharomyces pombe Reb1p: sequence-specific recognition of two sites in the far upstream rDNA intergenic spacer. Nucleic Acids Res., 25, 904–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jansa P. and Grummt,I. (1999) Mechanism of transcription termination: PTRF interacts with the largest subunit of RNA polymerase I and dissociates paused transcription complexes from yeast and mouse. Mol. Gen. Genet., 262, 508–514. [DOI] [PubMed] [Google Scholar]
- 74.Bartsch I., Schoneberg,C. and Grummt,I. (1988) Purification and characterization of TTFI, a factor that mediates termination of mouse ribosomal DNA transcription. Mol. Cell. Biol., 8, 3891–3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reeder R.H., Guevara,P. and Roan,J.G. (1999) Saccharomyces cerevisiae RNA polymerase I terminates transcription at the Reb1 terminator in vivo. Mol. Cell. Biol., 19, 7369–7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fourel G., Revardel,E., Koering,C.E. and Gilson,E. (1999) Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J., 18, 2522–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sjostrand J.O., Kegel,A. and Astrom,S.U. (2002) Functional diversity of silencers in budding yeasts. Eukaryot. Cell, 1, 548–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gavin A.C., Bosche,M., Krause,R., Grandi,P., Marzioch,M., Bauer,A., Schultz,J., Rick,J.M., Michon,A.M., Cruciat,C.M., Remor,M., Hofert,C., Schelder,M., Brajenovic,M., Ruffner,H., Merino,A., Klein,K., Hudak,M., Dickson,D., Rudi,T., Gnau,V., Bauch,A., Bastuck,S., Huhse,B., Leutwein,C., Heurtier,M.A., Copley,R.R., Edelmann,A., Querfurth,E., Rybin,V., Drewes,G., Raida,M., Bouwmeester,T., Bork,P., Seraphin,B., Kuster,B., Neubauer,G. and Superti-Furga,G. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- 79.Bader G.D. and Hogue,C.W. (2002) Analyzing yeast protein–protein interaction data obtained from different sources. Nat. Biotechnol., 20, 991–997. [DOI] [PubMed] [Google Scholar]
- 80.Cairns B.R., Schlichter,A., Erdjument-Bromage,H., Tempst,P., Kornberg,R.D. and Winston,F. (1999) Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH and bromodomains. Mol. Cell, 4, 715–723. [DOI] [PubMed] [Google Scholar]
- 81.Goodwin G.H. and Nicolas,R.H. (2001) The BAH domain, polybromo and the RSC chromatin remodelling complex. Gene, 268, 1–7. [DOI] [PubMed] [Google Scholar]
- 82.Ng H.H., Robert,F., Young,R.A. and Struhl,K. (2002) Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev., 16, 806–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mai B. and Breeden,L. (1997) Xbp1, a stress-induced transcriptional repressor of the Saccharomyces cerevisiae Swi4/Mbp1 family. Mol. Cell. Biol., 17, 6491–6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mai B. and Breeden,L. (2000) CLN1 and its repression by Xbp1 are important for efficient sporulation in budding yeast. Mol. Cell. Biol., 20, 478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fourel G., Miyake,T., Defossez,P.A., Li,R. and Gilson,E. (2002) General regulatory factors (GRFs) as genome partitioners. J. Biol. Chem., 277, 41736–41743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.