Abstract
A solitary long terminal repeat (LTR) of ERV-9 human endogenous retrovirus is located upstream of the HS5 site in the human β-globin locus control region and possesses unique enhancer activity in erythroid K562 cells. In cells transfected with plasmid LTR-HS5-εp-GFP, the LTR enhancer activates the GFP reporter gene and is not blocked by the interposed HS5 site, which has been reported to have insulator function. The LTR enhancer initiates synthesis of long RNAs from the LTR promoter through the intervening HS5 site into the ε-globin promoter and the GFP gene. Synthesis of the sense, long LTR RNAs is correlated with high level synthesis of GFP mRNA from the ε-globin promoter. Mutations of the LTR promoter and/or the ε-globin promoter show that (i) the LTR enhancer can autonomously initiate synthesis of LTR RNAs independent of the promoters and (ii) the LTR RNAs are not processed into GFP mRNA or translated into GFP. However, reversing the orientation of the LTR in plasmid (LTR)rev-HS5-εp-GFP, thus reversing the direction of synthesis of LTR RNAs in the antisense direction away from the ε-globin promoter and GFP gene drastically reduces the level of GFP mRNA and thus LTR enhancer function. The results suggest that the LTR-assembled transcription machinery in synthesizing non-coding, LTR RNAs can reach the downstream ε-globin promoter to activate transcription of the GFP gene.
INTRODUCTION
The solitary long terminal repeats (LTRs) of human endogenous retroviruses are middle repetitive DNAs characterized as retrotransposons (1–4). They contain the U3 enhancer and promoter region, the transcribed R region, whose 5′ end marks the initiation site of retroviral RNA synthesis, and the U5 region (5,6), but no internal gag, pol or env genes. In the human genome, retroelements, including the Alu (SINE) and L1 (LINE) elements and the solitary LTRs of endogenous retroviruses, are present at thousands to hundreds of thousands of copies and comprise up to 45% of the total chromosomal DNA (7). The functional roles in the human genome of these transposed, repetitive DNAs are not clear. The retroelements have been proposed to be selfish DNAs serving no relevant host function (8). However, recent findings show that they contain regulatory sequences including enhancers and promoters (9–14) and can serve a relevant cellular function in modulating the transcription of cis-linked host genes, thus providing plasticity and adaptive advantages to the host.
The human genome contains ∼50 copies of the ERV-9 endogenous retrovirus and an additional 3000–4000 copies of solitary ERV-9 LTRs (1,3, 15–17). Compared with the LTRs of other families of endogenous retroviruses, the ERV-9 LTRs exhibit an unusual sequence feature: the U3 regions contain from 5 to 17 tandem repeats of 37–41 bases (16,18,19) with recurrent GATA (20), CCAAT (21) and CACCC (22) motifs potentially capable of binding to cognate transcription factors abundantly expressed in blood cells.
The human β-globin gene locus spans the embryonic ε-, the fetal Gγ- and Aγ- and the adult δ- and β-globin genes. The β-globin locus control region (β-LCR), located 10–60 kb upstream of the globin genes and defined by DNase I hypersensitive sites HS5, HS4, HS3, HS2 and HS1 (23–25), plays a pivotal role in transcriptional activation of the far downstream β-like globin genes. It is not clearly understood whether the β-LCR acts over distance by a looping or a tracking mechanism (for reviews, see 26–29). In an effort to define the 5′ border of the β-LCR with the hope of gaining insight into the mechanism of β-LCR function, we previously cloned and sequenced the DNA further upstream of the HS5 site (19) and discovered a solitary ERV-9 LTR located 1.5 kb upstream of the HS5 site in the 5′ boundary area of the human β-LCR. The 5′ HS5 LTR possesses prominent enhancer activity in erythroid cells (19,30). Here, functional dissection of the 9 kb boundary area of the β-LCR shows that the LTR U3 region spanning 660 bases of DNA possessed unique enhancer and promoter activities not shared by the other DNA in the boundary area. However, the 5′ HS5 LTR is not linked to immediately downstream retroviral or cellular genes (GenBank accession no. AF064190) that could be activated by the LTR enhancer and promoter. This suggests that the ERV-9 LTR enhancer may interact with the proximal HS5 site in modulating transcription of the β-LCR in erythroid cells.
However, like the chicken HS4 insulator in the 5′ boundary of the chicken β-globin gene locus, the HS5 site in the 5′ boundary of the human β-LCR has been shown to possess insulator function (31–33) and bind to CTCF protein that is essential for insulator activity (34). In recombinant plasmids in which the HS5 site is placed between the HS3 or HS2 enhancer of the β-LCR and a promoter, the interposed HS5 site blocks enhancer–promoter communication and drastically diminishes enhancer function in activating mRNA synthesis from the cis-linked reporter gene. It is thus possible that the HS5 site located naturally downstream of the ERV-9 LTR (see Fig. 1a) could similarly block the LTR enhancer, thus precluding any functional interactions of the LTR enhancer with the further downstream HS4, HS3, HS2 and HS1 sites in the β-LCR and the globin genes.
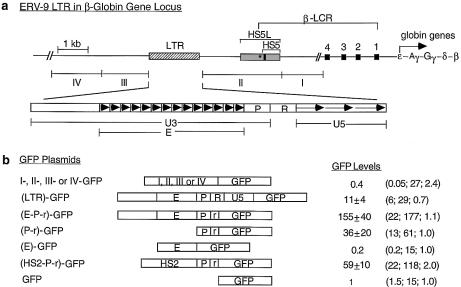
Figure 1.
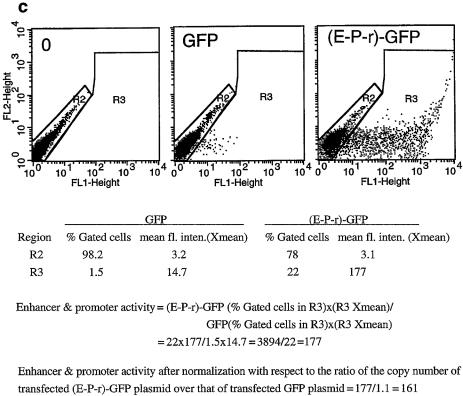
(Opposite) Structural and functional analyses of the 5′ boundary area of the human β-LCR. (a) Map of the ERV-9 LTR and the 5′ boundary area of the human β-LCR. Hatched box, LTR; stippled box, HS5L, the 1.2 kb HS5 site; vertical bar, CTCF binding site; *, NF-E2 binding site; HS5, the 0.5 kb HS5 site (see Table 1 for constructs containing HS5L or HS5); solid boxes, DNase I hypersensitive sites HS4, HS3, HS2 and HS1 of the β-LCR. The 9 kb DNA between // signs is drawn to scale; the 65 kb DNA to the right of // spans the β-LCR and the human β-like globin genes and is not drawn to scale. DNA fragments I, II, III and IV, DNAs in the boundary area that flank the LTR. The LTR is enlarged to show the U3, R and U5 regions. E, the U3 enhancer spanning 14 tandem repeats of 40 bp each denoted by the arrowheads; P, the 90 bp U3 promoter; open box to the left of E, the 590 bp of U3 GC-rich DNA 5′ of the enhancer; horizontal arrows, the three tandem repeats of ∼80 bp each in U5. (b) Maps of the recombinant GFP plasmids containing subfragments of the 5′ boundary area, I-, II-, III- and IV-, the LTR and components of the LTR. (HS2-P-r)-GFP contained the HS2 enhancer, the U3 promoter and the 5′ half of the R region of the 5′HS5 LTR coupled to the GFP gene. GFP, the reference enhancerless and promoterless GFP plasmid. The third column gives the GFP levels of the test plasmids relative to that of the reference GFP plasmid in transfected cells; values are averages of two determinations. For the I-, II-, III- and IV-GFP plasmids, the representative values of the III-GFP plasmid are presented; the GFP levels of the I-, II- and IV-GFP plasmids were even lower, in the 0.2–0.4 range, and are not shown. The three numbers in parentheses are, respectively, the percentage of fluorescent cells, the mean fluorescence intensities of the fluorescent cells [see (c)] and the ratio of the plasmid copy number of the transfected test plasmid to that of the reference GFP plasmid. (c) Sample calculation of the enhancer/promoter activity of the transfected (E-P-r)-GFP plasmid. (Left, middle and right) Dot plots by FACS analyses of K562 cells transfected with Tris buffer and GFP and (E-P-r)-GFP plasmids, respectively. x-axis, GFP fluorescence intensities of the transfected cells; y-axis, FL2 channel. The dot plots are the same whether FL2 or side scatter was used as the y-axis in the Cellquest program. However, using FL2 as the y-axis produced more compact and thus more easily gated fluorescent and non-fluorescent cell populations. R2 region, non-fluorescent cells; R3 region, fluorescent cells. The table below the dot plots gives quantitative analysis by the Cellquest program of the dot plot data. Xmean, mean fluorescence intensities of the gated cells. (Bottom) Calculation of the enhancer/promoter activity of (E-P-r)-GFP in K562 cells.
To investigate this possibility, we constructed recombinant plasmid LTR-HS5-εp-GFP, in which the ERV-9 LTR and the HS5 site in their natural genomic order were coupled to the ε-globin promoter, the nearest downstream globin promoter and the GFP reporter gene. Transfection experiments show that contrary to the HS2 and HS3 enhancers, the ERV-9 LTR enhancer synergized with and was not blocked by the HS5 site. This indicates that manifestation of HS5 insulator activity is not universal and depends on specific interactions of the HS5 site with the individual enhancer. To investigate the molecular basis of ERV-9 LTR enhancer function and its interaction with the HS5 site, we used 5′-RACE (35) to analyze the RNAs transcribed from the transfected plasmid. The results show that the LTR enhancer initiated synthesis of long LTR RNAs from multiple sites in the LTR through the intervening HS5 site into the ε-globin promoter and the GFP gene. Disabling the LTR promoter and/or the ε-globin promoter in plasmid LTR-HS5-εp-GFP demonstrates that (i) synthesis of the long LTR RNAs was initiated autonomously by the LTR enhancer independent of the LTR and ε-globin promoters and (ii) the long LTR RNAs were associated with enhanced synthesis of GFP mRNA initiated from the ε-globin promoter but were themselves not processed into GFP mRNA. In plasmid (LTR)rev-HS5-εp-GFP, reversing the orientation of the LTR with respect to the ε-globin promoter and GFP gene, thus reversing the direction of synthesis of the long LTR RNAs from the sense to the antisense direction away from the ε-globin promoter and GFP gene, caused a drastic drop in the level of GFP mRNA and thus in LTR enhancer activity. We discuss the possibility that the LTR-assembled transcription machinery may mediate enhancer function over a long distance by a tracking and transcription mechanism.
MATERIALS AND METHODS
Construction of recombinant GFP/CAT plasmids
The GFP plasmids (Fig. 1) were made from pEGFP-C1 (Clontech) which was digested with AseI and NheI to generate the vector backbone containing the GFP reporter gene and the SV40 poly(A) signal downstream of the GFP gene. The inserts were generated by PCR with forward and reverse primers containing corresponding AseI and NheI ends from either a phage template spanning the 5′ boundary area of the human β-LCR (19) or K562 genomic DNA. The positions in GenBank accession no. AF064190 of the forward and reverse PCR primers to generate the following PCR DNAs are as follows. For the complete LTR, 2650–2672 and 4325–4350; for (E-P-r), 2650–2672 and 3965–3987; for (P-r), 3849–3867 and 3965–3987; for (E), 3248–3263 and 3849–3867; 5′ boundary fragment I, 6695–6717 and 8253–8278 (the same as 65–89, GenBank accession no. U01317); II, 4482–4502 and 6695–6717; III, 1021–1045 and 2650–2672; and for border fragment IV, 128–150 (GenBank accession no. AF149710) and 1157–1176 (GenBank accession no. AF064190). The plasmid (HS2-P-r)-GFP contained the 740 bp HS2 sequence between BamHI and BglII sites (36) spliced into a BglII site at the 5′ end of the P-r sequence in the (P-r)-GFP plasmid. In the plasmids (E-P-r)-, (P-r)- and HS2-P-r-GFP (Fig. 1), the 3′ end 40 bases of the R region including the AATAAA motif (see 19) were not included. The authenticities of the PCR fragments were confirmed by DNA sequencing. The reference GFP plasmid was made by re-circularizing the AseI + NheI cleaved vector with an AseI-NheI adapter.
The construction of the plasmids LTR-CAT, LTR-HS5L-εp-CAT, HS5L-εp-CAT and εp-CAT, shown in Figure 2 and Table 1, have been described (19). To make plasmids LTR-HS5-εp-CAT and HS5-εp-CAT, the 0.5 kb HS5, made by PCR from the 1.2 kb HS5L with primers containing a BamHI cloning site, was used to replace HS5L in the corresponding LTR-HS5L-εp-CAT and HS5L-εp-CAT plasmids. The PCR primers for making HS5 were: forward, bases 6115–6139; reverse, bases 6576–6600 (GenBank accession no. AF064190). To make HS4-εp-CAT, a HS4 fragment of 0.9 kb spanning the NF-E2, Ap-1 and GATA-1 sites in the HS4 core (37) was made by PCR from K562 DNA with PCR primers: forward, 1020–1050; reverse, 1888–1910 (GenBank accession no. U01317). The forward primer contained a SalI cloning site and the reverse primer a BamHI cloning site. The HS4 fragment was inserted into the corresponding sites upstream of the ε-globin promoter in plasmid εp-CAT. To make HS5L-HS4-εp-CAT, HS5L containing SalI cloning sites made by PCR was inserted into the SalI site upstream of HS4 in HS4-εp-CAT. Construction of HS2-εp-CAT containing a 0.74 kb HS2 DNA fragment has been described (36). Plasmid HS2-HS5L-εp-CAT was made by inserting HS5L with BamHI cloning sites into the BamHI site between HS2 and εp in plasmid HS2εp-CAT.
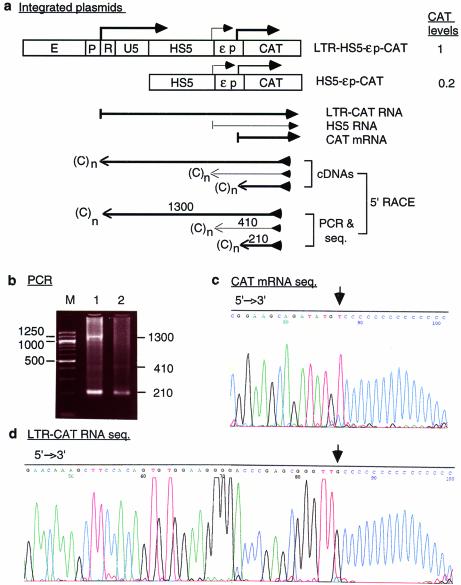
Figure 2.
(Opposite) Transcription of integrated plasmid LTR-HS5-εp-CAT in K562 cells. (a) Maps of the integrated LTR-HS5-εp-CAT and HS5-εp-CAT plasmids and transcription of the plasmids analyzed by 5′-RACE. Angled arrows, locations of transcriptional initiation sites in the LTR, HS5 site and the ε-globin promoter. Left to right arrows, RNAs initiated from within the LTR, HS5 and ε-globin promoter, with the thickness of the arrows showing the relative abundance of the RNAs. Right to left arrows, cDNAs reverse transcribed from the respective RNAs and PCR fragments subsequently amplified from the cDNAs by the 5′-RACE protocol. The PCR fragments depicted were the sequenced DNA strand; the arrows are aligned with the plasmid map on top. (C)n, poly(dC) tails added to the 3′ end of the cDNAs by the TdT enzyme. The arrowheads at the 5′ ends of the cDNA or the PCR fragment indicate the reverse primers used for cDNA synthesis, PCR amplification and DNA sequencing. Numbers are the sizes in nucleotides of the PCR fragments determined by gel electrophoresis and DNA sequencing [see (b), (c) and (d)]. CAT levels are relative levels of CAT enzyme produced by the LTR-HS5-εp-CAT plasmid (at 8 copies/cell) and HS5-εp-CAT (at 14 copies/cell) determined previously by CAT assay (19). (b) PCR products of 5′-RACE. Lanes 1 and 2, PCR bands generated by RNAs transcribed, respectively, from the LTR-HS5-εp-CAT and HS5-εp-CAT plasmids; M, 100 bp size markers. Numbers on the right and left margins are sizes in bp of the PCR fragments and the size markers. (c) DNA seqence of the 210 bp PCR fragment generated from CAT mRNA. The base marked by the arrow is the 5′ end/initiation site of CAT mRNA. (d) DNA sequence of the 1300 bp PCR fragment generated from the long LTR RNA; only the 5′ end of the sequence is shown. Arrow, 5′ end/initiation site of the LTR RNA. (e) DNA sequences near the transcriptional initiation sites of the LTR RNA and CAT mRNA. AATAAA, TATA box in the U3 or the ε-globin promoter; angled arrows, locations of transcriptional initiation sites; bold bases, transcribed bases.
Table 1. ERV-9 LTR enhancer activity is not blocked by the HS5 site in plasmids transiently transfected or stably integrated into K562 cells.
| Plasmid | CAT/GFP level | |
|---|---|---|
| 1. | LTR-CATa | 8 ± 1.6 |
| 2. | HS5L-εp-CATa | 5 ± 2.3 |
| 3. | LTR-HS5L-εp-CATa | 27 ± 4.5 |
| 4. | HS5-εp-CAT | 2 ± 0.5 |
| 5. | LTR-HS5-εp-CAT | 12 ± 2 |
| 6. | HS4-εp-CAT | 4 ± 1.5 |
| 7. | HS5L-HS4-εp-CAT | 28 ± 10 |
| 8. | εp-CAT | 1 |
| 9. | HS2-εp-CAT | 22 ± 5 |
| 10. | HS2-HS5L-εp-CAT | 8 ± 4 |
| 11. | LTR-GFP | 9 ± 3.3 (11± 4) |
| 12. | HS5-εp-GFP | 1.2 ± 0.4 (2 ± 0.3) |
| 13. | LTR-HS5-εp-GFP | 18 ± 5 (12 ± 0.7) |
| 14. | εp-GFP | 1 (1) |
HS5L, the 1.2 kb HS5 DNA; HS5, the truncated 0.5 kb HS5 DNA that contained the binding site for CTCF but not for NF-E2 (see Fig. 1a).
CAT levels were averages of three determinations with standard deviations. GFP levels were averages of two determinations. Numbers in parentheses are GFP levels of transiently transfected plasmids.
aCAT levels were determined in a previous study (19).
To construct HS5-εp-GFP containing 0.5 kb of HS5, shown in Table 1 and Figure 3, a PCR fragment spanning HS5 and the ε-globin promoter between 5′ AseI and 3′ AgeI cloning sites was made by PCR from plasmid HS5-εp-CAT (19) and inserted in clone II (Fig. 1a) in which the 1.2 kb HS5L and the genomic DNA 5′ of HS5L in clone II were deleted by AseI and AgeI digestion. To create the plasmid LTR-HS5-εp-GFP (construct 1, Fig. 3; construct 13, Table 1), plasmid HS5-εp-GFP, serving as the vector, was cleaved with AseI and SacI to remove the 5′-half of the HS5 site. The LTR between the AseI and AgeI sites excised from the (LTR)-GFP plasmid (Fig. 1b) and the 5′-half of HS5 between the AgeI and SacI sites generated by PCR were inserted into the cleaved vector. This plasmid contained the ERV-9 LTR of 1.7 kb spanning the U3 GC-rich region of 590 bp, U3 enhancer of 570 bp, U3 promoter of 90 bp, the R region of 96 bp and U5 of 344 bp (19), HS5 of 0.5 kb (bases 6100–6600, GenBank accession no. AF064190) containing a CTCF binding site, CCACTAGAG GGAAGAA, 50 bases from its 5′ end, the 0.2 kb ε-globin promoter (bases 19307–19505, GenBank accession no. U01317) and the 0.7 kb GFP gene in pEGFP-C1 (Clontech). In this plasmid, the SV40 splice site (Promega Bulletin 80) was inserted between the EcoRI and SalI sites in the polycloning region at the 3′ end of the GFP gene; the SV40 enhancer and promoter 5′ of the neomycin/kanamycin resistance gene were deleted by SspI and StuI double digestion.
Figure 3.
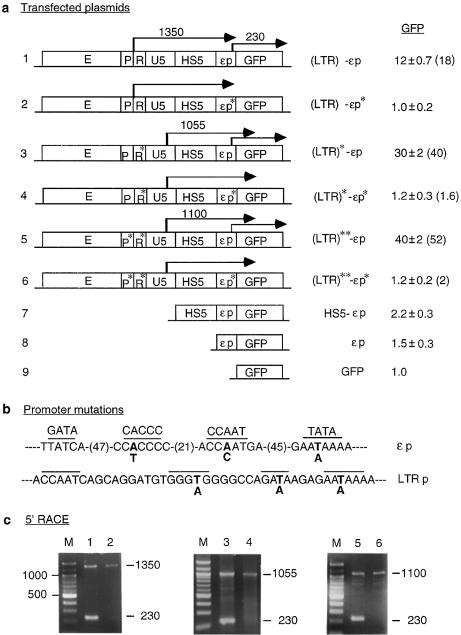
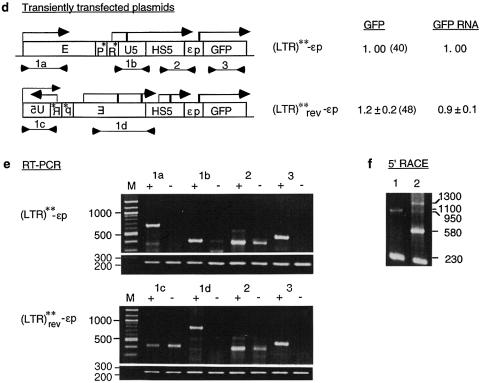
Effects of mutations in the LTR and the ε-globin promoters on transcription of LTR RNAs and GFP expression. (a) Maps of plasmids containing promoter mutations. P*, R* and εp*, mutated LTR promoter, R region and ε-globin promoter (see Materials and Methods and Fig. 3b). (LTR)* and (LTR)**, the LTR containing mutations in only the R region and in both the R region and LTR promoter, respectively. Angled arrows, LTR initiated long RNAs and GFP mRNA, with locations of the angled arrows marking transcription initiation sites mapped by 5′-RACE; the numbers above the arrows are the sizes in nucleotides of the 5′-RACE bands. Numbers in the column under GFP are GFP levels of transiently transfected plasmids expressed with respect to the GFP level of the enhancerless and promoterless GFP plasmid; the values are averages of two transfection experiments. Numbers in parentheses are GFP levels of integrated plasmids. Other designations are as in Figure 2a. (b) Mutations in the ε-globin and LTR promoters. Overlined bases, sequence motifs important to the function of the respective promoters. Bold letters, DNA bases in the promoters that were mutated to the bases below. In the ε-globin promoter, numbers in parentheses are the number of bases between the overlined motifs. (c) 5′-RACE. Lanes 1–6, 5′-RACE bands generated by RNAs transcribed from the transfected constructs 1–6; numbers on the right margins are sizes in bp of the 5′-RACE bands.
To make constructs 2 and 5, shown in Figure 3, containing, respectively, the mutated ε-globin and LTR promoters, multiple base substitutions in the promoter were made simultaneously with the following multiple sites mutagenesis protocol (H.Jin and A.Seyfang, in preparation). Briefly, to mutate each promoter using construct 1 as the template, a set of three reverse primers was synthesized. The middle primer, the mutagenesis primer, was complementary to the region in the promoter to be mutated and contained the desired base substitutions. The 5′ and 3′ primers were complementary to appropriate plasmid DNAs flanking the promoter and contained, respectively, a 5′ or a 3′ tail of a unique DNA sequence not found in the plasmid. The primers were annealed to denatured plasmid DNA and the gaps between them were synthesized with T4 DNA polymerase. The DNAs were then ligated with T4 ligase to generate the mutagenized DNA strand. Subsequently, the DNA was amplified by PCR using the unique 5′ and 3′ tails as primers, the amplified DNA was cleaved at unique restriction enzyme sites present in the flanking DNA and ligated with similarly cleaved construct 1. The mutagenesis primer for the LTR promoter was 5′-GGGCAGCCTGCTTTTTTTCTCTTTTCTGGCCCCTCCCACCATCCTGCTG-3′; for the ε-globin promoter to mutate the AATAAA box, 5′-GTCTGGCCTTTTTTTCTTTACTGCC-3′, and to mutate the CCAAT and CACCC motifs, 5′-CTTAAAAGTCATGGGTCAAGGCTGACCTGTGTCCTCAGGGGAGGAGTCAGGTCC-3′ (the bold letters represent mutated bases in the respective underlined motifs) (see Fig. 3b). Correct base substitutions were confirmed by DNA sequencing of the final plasmids. Constructs 4 and 6, (LTR)*-εp* and (LTR)**-εp*, were made by replacing the wild-type ε-globin promoter in constructs 3 and 5 with the base substituted ε-globin promoter excised from construct 2 by SacI cleavage in HS5 and EcoRI cleavage in the multi-cloning sites downstream of the GFP gene. In constructs 3–6 (Fig. 3) the R region mutations (AATAAA to AGTAAG) were made from construct 1 with a Quikchange site-directed mutagenesis kit (Stratagene).
To create the (LTR)rev-HS5-εp-GFP plasmid, shown in Figure 4, the LTR was generated from plasmid LTR-HS5-εp-GFP by PCR to contain an AgeI site at the 5′ end and AseI site at the 3′ end and together with the 5′-half of HS5 between the AgeI and SacI sites was inserted into the AseI and AgeI cleaved vector for construct 1 (Fig. 3) as described above. The (LTR)**rev-HS5-εp-GFP plasmid was made from (LTR)**-HS5-εp-GFP (construct 5, Fig. 3) by the same cloning strategy as plasmid (LTR)rev-HS5-εp-GFP.
Figure 4.
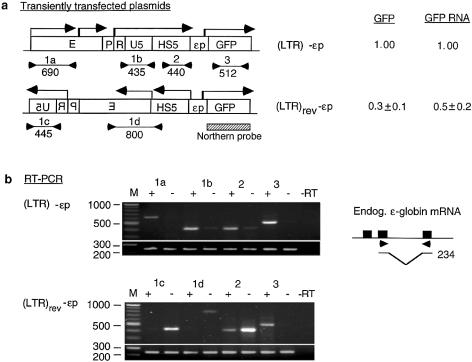
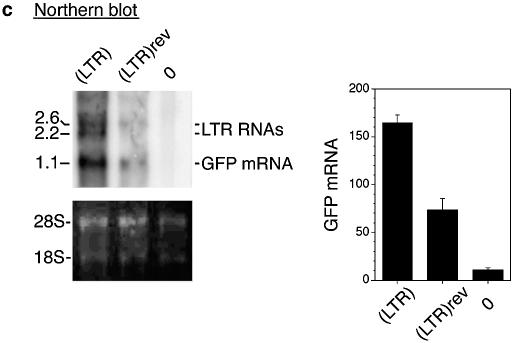
(Opposite and above) Effects of LTR orientation on GFP mRNA synthesis and GFP expression. (a) Maps of the reference (LTR)-εp and test (LTR)rev-εp plasmids. Locations and direction of angled arrows show the locations of RNA initiation sites and direction of RNA synthesis. The locations marked by heavy stems are initiation sites mapped by 5′-RACE and northern blot; locations marked by thin stems are initiation sites postulated to exist from the RT–PCR bands. Horizontal lines marked 1a, 1b, 1c, 1d, 2 and 3 are RT–PCR products generated by the respective primer pairs 1a–1d, 2 and 3; the lines are aligned with the plasmid maps on top; numbers underneath the lines are sizes (bp) of the RT–PCR bands. Hatched box, northern blot probe. GFP and GFP RNA, levels of GFP expression and GFP RNA of the test relative to those of the reference plasmids. Numbers in parentheses are GFP levels of the plasmids relative to that of the enhancerless and promoterless GFP plasmid. (b) Directional RT–PCR. Lanes marked 1a–1d, 2 and 3 are RT–PCR bands generated by the respective primer pairs; + and – lanes, RT–PCR bands generated by each primer pair from the sense and the antisense RNAs; –RT lane, no reverse transcriptase in the RT step to show absence of DNA contamination in the RNA samples; bottom rows of RT–PCR bands were generated from the endogenous ε-globin mRNA with a primer pair as depicted in the inset to its right and served as internal controls. (Inset) Solid boxes, the three exons of the ε-globin gene; arrowheads, the location of the primer pair spanning the second intron; horizontal lines, the spliced ε-globin mRNA amplified by the primer pair; the bent line, spliced out second intron; number to the right, size in base pairs of the RT–PCR band amplified by the primer pair from the ε-globin mRNA. (c) Northern blot of total cellular RNAs isolated from K562 cells transfected with (LTR)-εp plasmid [lane (LTR)], (LTR)rev-εp plasmid [lane (LTR)rev] and a non-transfected K562 control (lane 0). (Top left) Northern blot hybridized to the GFP gene probe. Numbers in the left margin are sizes in kilobases of the LTR RNAs and GFP mRNA. (Bottom left) Ethidium bromide stained bands of 28S and 18S RNAs to show equal loading of the RNA samples in different lanes in the agarose gel. Bar graphs are mean values of the radioactive counts, measured in arbitrary pixel units, of GFP mRNA from blots of three separate RNA preparations. (d and e) Maps of reference (LTR)**-εp and test (LTR)**rev-εp plasmids. Designations as in (a) and (b). (f) 5′-RACE of LTR RNAs and GFP mRNA transcribed from (LTR)**-εp (lane 1) and (LTR)**rev-εp (lane 2).
Transfection assays and fluorescent flow cytometry analyses
For transient transfections (Fig. 1), 10 µg of circular plasmids in duplicate were transfected by electroporation into 4 × 106 host cells in 400 µl of medium without fetal calf serum at 240 V, 960 µF in a Gene Pulser II (Bio-Rad). All other transient transfections (Figs 3 and 4) were carried out with linearized plasmids. Enhancer activities of the transfected plasmids were analyzed 48 h later in a FACScalibur with Cellquest software (Becton-Dickinson). Samples of 2 × 104 live cells were analyzed for each sample; necrotic cells were excluded from the FACS analyses by propidium iodide staining. The expression levels of the GFP gene in the transfected plasmids were calculated as shown in Figure 1c. The calculated GFP levels were then corrected with respect to the copy numbers of the transfected GFP gene as described (30). For transient transfections (Figs 3 and 4), 20 µg each of the plasmids linearized at a unique ApaLI site in the vector upstream of the LTR were electroporated and analyzed as described above. To screen for K562 cells harboring integrated plasmids (Fig. 3 and Table 1), the linearized plasmids were co-transfected with an expression plasmid for the neomycin resistance gene as described (36). The cells harboring integrated plasmids were harvested for FACS and RNA analyses after 3 weeks of G418 selection. The protocols for creating K562 cells harboring integrated CAT plasmids linearized at a unique AlwNI or NdeI site in the vector (Fig. 2 and Table 1) and CAT assays were previously described (19).
RNA isolation and 5′-RACE
Total cellular RNAs were isolated with a Totally RNA kit (Ambion). Before use, the RNAs were treated with RNase-free DNase I to eliminate possible DNA contamination. The 5′-RACE kit (Gibco BRL) was used according to the vendor protocol as described (30). Total cellular RNA, 5 µg/sample, was used in cDNA synthesis. After purification, 1 µg of cDNA per sample was used for oligo(dC) tailing. Finally, 0.2 µg of C-tailed cDNA per sample was used in nested PCR. After the second round of nested PCR, 10–15 µl samples from 50 µl were resolved in agarose gels. The amplicons were purified from agarose gels and sequenced by the Molecular Biology Core Laboratory. Alternatively, the authenticity of the 5′-RACE bands was confirmed with restriction enzyme digestions. The positions of the gene-specific, nested reverse primers used for (i) cDNA synthesis, (ii) and (iii) two rounds of PCR amplifications and (iv) DNA sequencing are as follows. In the CAT gene in plasmid pCAT-Basic (Fig. 2): (i) 2635–2655; (ii) 2575–2595; (iii) 2381–2401; (iv) 2328–2348 (Promega Manual). In the GFP gene in plasmid pEGFP-C1 (Fig. 3): (i) 826–850; (ii) 736–757; (iii) 617–640; (iv) 617–640 (Clontech).
RT–PCR and directional RT–PCR
Between 2 and 5 µg of total cellular RNAs isolated from transfected cells was used as the template for cDNA synthesis in each reverse transcription (RT) reaction. In directional RT–PCR (Fig. 4), to synthesize cDNAs from the sense RNAs, the reverse primers of the primer pairs spanning the regions of interest were used in RT. To synthesize cDNAs from the antisense RNAs, the forward primers of the primer pairs were used in RT. Aliquots of cDNAs transcribed from 400 ng of RNA were used in the subsequent PCRs with each of the forward and reverse primer pairs used in cDNA synthesis. The RT step was carried out with M-MLV reverse transcriptase (BRL) at 42°C for 60 min. The PCR con ditions were denaturation at 94°C for 1 min, annealing at 58°C for 1 min and extension at 72°C for 1 min, repeated for 32 cycles, which were chosen from pilot PCRs carried out for 28, 30, 32, 34 and 36 cycles. At 32 cycles, the PCR ingredients were not exhausted and the PCR product was within the linear range suitable for semi-quantitative analyses. Aliquots of the PCR products (5 µl of 50 µl) were analyzed in 2% agarose gels.
Note that in order to amplify only the RNAs transcribed from the transfected plasmids, the RT–PCR primer pairs were designed to span specific junction sequences not found in the K562 genome (see Fig. 4a and c for primer pair locations). In primer pair 1a, the forward primer spanned the junction between the LTR and the vector sequence; in primer pair 1b, the reverse primer spanned the junction between U5 and HS5, which in the genome were separated by >3 kb of DNA, a distance not efficiently amplified under the current RT–PCR conditions; the forward primer in primer pair 1c was located at the junction between U5 and the vector; primer pair 1d spanned the LTR enhancer in reverse orientation with respect to HS5, a configuration not found in the K562 genome; primer pair 2 spanned HS5 and the ε-globin promoter, which in the genome were separated by >20 kb of DNA; primer pair 3 amplified the GFP reporter gene which was a non-human sequence derived from jellyfish. The sequences of the primer pairs were as follows. 1a: forward, 4712–0008 (pEGFP-C1, Clontech); reverse, 3260–3281 (GenBank accession no. AF064190). 1b: forward, 4004–4028 (AF064190); reverse, 5′-TGACATACTGTGACCGGTAGCGCTAGCATT-3′ (the AgeI site at the junction between the LTR and HS5 is underlined). 1c: forward, 4712–0008 (pEGFP-C1, Clontech); reverse, 4004–4028 (GenBank accession no. AF064190). 1d: forward, 3260–3281; reverse, 6258–6280 (GenBank accession no. AF064190). 2: 6268–6292 (GenBank accession no. AF064190); reverse, 19338–19361 (U01317). 3: 616–631, 956–981 (pEGFP-C1).
An internal control was generated by a primer pair in the ε-globin mRNA (forward, 19957–19975; reverse, 21021–21045; GenBank accession no. U01317) as follows. The reverse primer in ε-globin mRNA was added together with each of the above test primers in the same tube as for RT to also synthesize cDNA from the endogenous ε-globin mRNA present in all the RNA samples. Equal aliquots of the cDNA stock were amplified in separate PCR tubes with the test primer pairs or the ε-globin primer pair for 32 and 30 cycles respectively. Samples (10–15 µl) from the 50 µl of PCR products were resolved in 2% agarose gels. The authenticity of the RT–PCR bands was confirmed either by DNA sequencing or restriction enzyme digestion. To quantify the RNA templates, the intensities of the RT–PCR bands were quantified with an IS1000 Image Analyzer (Alpha Innotech) and normalized with respect to the intensities of the band generated by endogenous ε-globin mRNA.
Northern blot
Aliquots of 15 µg of total cellular RNA isolated from the transfected cells (Fig. 4a) were resolved in 1.5% denaturing agarose gels, blotted and hybridized to 32P-labeled probe according to published protocols (38). Radioactive intensities of the RNA bands were quantified with a PhosphorImager.
RESULTS
Functional dissections of the 5′ boundary of the human β-LCR and the ERV-9 LTR
Using a newly developed transfection assay with the GFP gene as the reporter followed by FACS analyses (30), we determined the enhancer and promoter functions of the DNA in the 5′ boundary area of the β-LCR. The 9 kb boundary DNA including 3 kb of DNA spanning the HS5 site was sub-cloned into five recombinant GFP plasmids, clones I, II, LTR, III and IV (Fig. 1a), which were transfected separately into K562 erythroid cells. In a previous report (30), we observed that the percentages of fluorescent cells (first numbers in parentheses, Fig. 1b) correlated positively with the enhancer and promoter strengths as measured by the mean fluorescence intensities of the transfected cells (second numbers in parentheses, Fig. 1b). Hence, we took the product of the percentage of fluorescent cells multiplied by the mean fluorescence intensity of the fluorescent cells as a quantitative measure of the combined enhancer and promoter strengths of the LTR (see Fig. 1c). Consistent with previous assays using the CAT reporter gene (19), the (LTR)-GFP plasmid exhibited enhancer/promoter activities ∼10-fold above the reference GFP plasmid (Fig. 1b). Clones I–IV, including clone II, which spans the HS5 site, produced very few fluorescent cells and possessed no detectable enhancer/promoter activities.
We next determined the function of the U3, R and U5 regions of the ERV-9 LTR as demarcated by DNA sequence analyses (19). The U3 region spanning the 14 tandem repeats of 40 bases/repeat exhibited enhancer activity: plasmid (E-P-r)-GFP, containing the U3 enhancer and promoter together with the 5′ end of the R region spanning the retroviral transcriptional initiation site, expressed GFP 150-fold higher than the GFP plasmid (Fig. 1b). However, in the absence of the U3 promoter, the 14 U3 repeats by themselves in plasmid (E)-GFP did not activate the GFP gene (Fig. 1b), indicating a requirement for the LTR promoter in LTR enhancer function.
Plasmid (LTR)-GFP, containing the entire LTR including the U5 region, showed 15-fold lower enhancer activity than plasmid (E-P-r)-GFP (Fig. 1b). The reduction in GFP expression was probably due to the extra 400 bases of the R and U5 DNA that may contain transcriptional or translational inhibitors. In comparison with the LTR enhancer, the strong HS2 enhancer of the β-LCR (36) in plasmid (HS2-P-r)-GFP activated the GFP gene to a level 40% that of the (E-P-r)-GFP construct (Fig. 1b). These results indicate that among the DNA fragments in the 5′ boundary area of the β-LCR, the ERV-9 LTR possessed unique enhancer/promoter activity that was 2- to 3-fold higher than that of the HS2 enhancer as assayed in these GFP constructs.
ERV-9 LTR enhancer activity is not blocked by the HS5 site
We previously reported that in recombinant plasmid LTR-HS5L-εp-CAT containing the 1.2 kb HS5 fragment, the ERV-9 LTR enhancer activity was not blocked by the interposed HS5L (see constructs 1 and 3, Table 1) (19). However, the 1.2 kb HS5L spanned not only the CTCF binding site essential to HS5 insulator function (34) but also multiple binding sites for positive transcription factors such as erythroid NF-E2 and GATA-1 (39) (see Fig. 1a), which exhibited weak enhancer activity in transfected plasmid HS5L-εp-CAT (construct 2, Table 1). Thus, it is possible that the failure of HS5L to block ERV-9 LTR enhancer activity was due to the presence of these positive regulatory elements and/or the special vector backbone of the CAT plasmid that combined to negate the insulation effect of CTCF bound at the HS5 site. To investigate these possibilities, we used the pEGFP plasmid (Clontech) with a different vector backbone to construct the plasmid LTR-HS5-εp-GFP, in which the HS5 site was truncated to 0.5 kb to delete the NF-E2 site and nine GATA sites present in the first 700 bases of the 1.2 kb HS5L. The truncated HS5 core contained the CTCF binding site at its 5′ end and only 4 of the 13 GATA sites in HS5L (see Fig. 1a). For comparison, recombinant CAT plasmids containing the truncated 0.5 kb HS5 or the 1.2 kb HS5L coupled to the β-LCR HS4 or HS2 enhancers were also constructed (constructs 4–10, Table 1).
The plasmids were linearized at a site in the vector upstream of the LTR enhancer so that the enhancer could not act on the linked reporter gene from a location downstream of the gene as in a circular plasmid, thus by-passing the potential blocking effect of the HS5 insulator. To ensure that the linearized plasmids were not integrated into K562 cells in long tandem arrays so that the LTR enhancer in the downstream plasmid could directly interact with and activate the reporter gene of the upstream plasmid without an interposed HS5 site, the linearized plasmids were transfected into K562 cells by electroporation. Southern blots of the integrated plasmids following digestion by a restriction enzyme with a single cleavage site in the plasmids showed that the majority of the plasmids were intergrated in single copies into multiple, separate host sites (blots not shown) (40).
In integrated plasmid HS5-εp-CAT, the 0.5 kb HS5 did not exhibit appreciable enhancer activity (construct 4, Table 1). However, it still did not block ERV-9 LTR enhancer activity in LTR-HS5-εp-CAT, nor did it block LTR enhancer activity in LTR-HS5-εp-GFP (see constructs 1 and 5 and 11 and 13, Table 1). Furthermore, in the plasmid HS5L-HS4-εp-CAT, in which HS5L was coupled to the natural downstream HS4 site in the β-LCR (see Fig. 1a), HS5 strongly stimulated HS4, with inherently weak enhancer activity, to exhibit prominent enhancer activity comparable to that exhibited by the strong HS2 enhancer (compare constructs 6, 7 and 9, Table 1).
The failure of the HS5 site to exhibit insulator activity in these plasmids did not appear to be an artifact of the particular experimental conditions. In the plasmid HS2-HS5L-εp-CAT, similarly integrated into K562 cells with an identical transfection protocol, the HS5 site inserted downstream of the strong HS2 enhancer exhibited insulator properties in blocking HS2 enhancer activity (see constructs 9 and 10, Table 1). Taken together, the results indicate that when coupled in the genomic order to its natural neighbors in the β-LCR, i.e. either downstream of the ERV-9 LTR or upstream of the HS4 site, the HS5 site did not exhibit insulator properties. This indicates that manifestation of HS5 insulator activity is not universal and appears to depend on specific interactions between HS5 and the individual LTR, HS2 or HS3 enhancers.
In the LTR-HS5-εp-CAT plasmid the LTR enhancer activates synthesis of long RNAs that are initiated from the LTR promoter and extended through the HS5 site and the ε-globin promoter into the CAT gene
To investigate the molecular basis of ERV-9 LTR enhancer activity and its synergistic interaction with HS5, we used 5′-RACE to analyze the RNAs transcribed from plasmid LTR-HS5-εp-CAT (Fig. 2a). We detected long LTR RNAs that produced a 5′-RACE band of 1300 nt (Fig. 2b, lane 1). DNA sequencing of the entire 1300 bp band showed that the long LTR RNA was initiated from the LTR at the C base located 25 bases downstream of the AATAAA box in the U3 promoter (Fig. 2d and e) and extended through HS5 and the ε-globin promoter into the CAT gene (Fig. 2a). Correlated with the presence of the long LTR RNA, CAT mRNA, producing a 5′-RACE band of 210 bp, was synthesized more abundantly from plasmid LTR-HS5-εp-CAT than from plasmid HS5-εp-CAT (compare the intensities of the bands of 210 nt in Fig. 2b, lanes 1 and 2; the intensities of the 410 nt bands in these lanes produced by RNA initiated from within HS5 in the respective plasmids were comparable and served as the internal reference for the quality of the RNA templates and sample loading). DNA sequencing showed that the 210 nt band was produced by CAT mRNA initiated at the A base 22 bases downstream of the AATAAA (TATA) box in the ε-globin promoter (Fig. 2c and e). The enhanced synthesis of CAT mRNA from the integrated plasmid LTR-HS5-εp-CAT was observed repeatedly in three 5′-RACE assays using two different RNA preparations. The correlation between transcription of the long LTR RNAs and enhanced synthesis of CAT mRNA suggests that the LTR-initiated long RNA could be processed into CAT mRNA. The enhanced level of CAT enzyme synthesized from the plasmid LTR-HS5-εp-CAT suggests that the long LTR RNA could also be directly translated into the CAT enzyme.
The LTR-initiated long RNAs are not processed into mRNA or translated into protein products
To investigate the possibilities that the long LTR RNAs might be processed into CAT mRNA or directly translated into the protein product, we created three pairs of reference and test plasmids (constructs 1 and 2, 3 and 4 and 5 and 6, Fig. 3a) to study the relationship between syntheses of long LTR RNAs and mRNA. In these plasmids, we attempted to abrogate syntheses of the long LTR RNA or mRNA through disabling the LTR or ε-globin promoter. The ε-globin promoter was disabled by base substitutions in the AATAAA (TATA), CACCC and CCAAT motifs previously shown to be important for globin promoter function (41); the LTR promoter was disabled by similar base substitutions (Fig. 3b). The GFP reporter gene replaced the CAT gene in these constructs for ease of carrying out reporter gene assays by FACS analysis. The linearized plasmids were transiently or stably transfected into K562 cells by electroporation. GFP protein and mRNA in the transfected cells were analyzed by FACS and 5′-RACE.
In constructs 1 and 2, the reference plasmid LTR-HS5-εp-GFP, (LTR)-εp, either transiently transfected or stably integrated into K562 cells, expressed GFP at a level 12- to 18-fold higher than the enhancerless and promoterless GFP plasmid (construct 1, Fig. 3a). As with the integrated plasmid LTR-HS5-εp-CAT (Fig. 2), (LTR)-εp synthesized both the long LTR RNA and the short GFP mRNA, which produced, respectively, the 1350 and 230 nt bands in 5′-RACE (Fig. 3c, lane 1). Restriction enzyme digestions of the 1350 nt band (not shown) confirmed that this band was generated by LTR-GFP RNA initiated from within the LTR and extended through the HS5 and ε-globin promoter into the GFP gene. Sequencing of the 230 nt band (not shown) indicated that this band was generated by GFP mRNA initiated from the same specific site in the ε-globin promoter as CAT mRNA (Fig. 2c and e).
In contrast, the test plasmid LTR-HS5-εp*-GFP, (LTR)-εp*, containing the mutated ε-globin promoter, expressed GFP at a background level similar to that of the enhancerless and promoterless GFP plasmid (construct 2, Fig. 3a). In 5′-RACE, the 230 nt band produced by the GFP mRNA was not detectable, indicating that the mutant ε-globin promoter did not initiate synthesis of GFP mRNA. On the other hand, the long LTR RNA initiated from the LTR and extended through HS5 into the GFP gene was still synthesized and produced the 1350 nt band (Fig. 3c, lane 2). The LTR RNA extended into the 3′ end of the GFP gene as indicated by RT–PCR with a primer pair spanning the entire GFP gene (not shown). The clear absence of GFP mRNA from the test plasmid (Fig. 3c, lane 2) could not be due to less RNA template from the test plasmid used in the RT step or the PCR conditions of the 5′-RACE protocol, since an equal amount of RNA template from the reference plasmid, processed under an identical experimental protocol, produced not only the GFP mRNA band but also the LTR RNA band at a similar intensity to the test plasmid (compare intensities of the LTR RNA bands in lanes 1 and 2 and also in lanes 3 and 4 and lanes 5 and 6, Fig. 3c). These results, reproducibly observed in two independent experiments, indicate that the long LTR RNA was not processed into GFP mRNA nor was it efficiently translated into GFP.
Constructs 3 and 4, (LTR)*-εp and (LTR)*-εp* (Fig. 3a), confirmed these observations. These two plasmids contained (LTR)* in which the R region AATAAA motif was mutated to AGTAAG. FACS analysis showed surprisingly that construct 3, (LTR)*-εp, as a result of the two A→G base mutations in the transcribed R region, expressed GFP at a level almost 3-fold that of construct 1, (LTR)-εp (Fig. 3a). 5′-RACE showed that (LTR)*-εp synthesized the long LTR* RNA and also GFP, which produced, respectively, the bands of 1055 and 230 nt (Fig. 3c, lane 3). In contrast, the test plasmid (LTR)*-εp* containing the disabled ε-globin promoter synthesized no GFP mRNA but only the long LTR* RNA that produced the 1055 nt band and expressed GFP at near background level, 1.2-fold that of the GFP plasmid (Fig. 3a). These analyses again indicate that the long LTR* RNA was not processed into GFP mRNA or translated into GFP.
The size of the 1055 nt band produced by LTR* RNA was nearly 300 bases shorter than the 1350 nt band produced by the LTR RNA transcribed from constructs 1 and 2. The LTR* RNA was thus initiated not from the 5′ border of the R region but from a cryptic promoter 300 bases downstream in the U5 region within the intervening DNA between the second and third U5 repeats (see Fig. 1a), which contained no recognizable TATA box (30). Apparently, the mutated AGTAAG motif in the transcribed R region interacted with the transcriptional machinery assembled at the AATAAA (TATA) box located 80 bp further upstream in the LTR promoter to shift the major transcription site of the LTR* RNA by 300 bases into the U5 region.
The LTR enhancer can autonomously initiate RNA synthesis independent of the LTR promoter
To determine whether synthesis of the long LTR RNAs could be abrogated by disabling the LTR promoter, constructs 5 and 6, (LTR)**-εp and (LTR)**-εp*, were made (Fig. 3a). In these plasmids the LTR promoter was disabled by base substitutions in the AATAAA, GATA and CACCC motifs (Fig. 3b). In addition, the AATAAA motif in the R region was mutated to AGTAAG to prevent it from serving as a surrogate TATA box after the AATAAA (TATA) box in the LTR promoter was mutated. In construct 5, (LTR)**-εp, the LTR enhancer was surprisingly very active in the presence of the disabled LTR promoter and activated synthesis of GFP from the ε-globin promoter to the highest level among constructs 1–6, at 40-fold that of the GFP plasmid (Fig. 3a). This indicates that optimal LTR enhancer activity was achieved in the absence of a functional LTR promoter, which apparently dampened LTR enhancer activity in these plasmids.
In construct 5, the LTR** enhancer synthesized the long LTR** RNA and GFP mRNA. The LTR** RNA produced a 5′-RACE band of 1100 nt which, as in (LTR)*-εp (construct 3), was initiated from the cryptic promoter in the U5 region; the GFP mRNA produced a 5′-RACE band of 230 nt initiated from the ε-globin promoter (Fig. 3c, lane 5). The results indicate that the LTR** enhancer could initiate synthesis of both long LTR RNAs and GFP mRNA and exhibit elevated enhancer activity independent of the LTR promoter.
In construct 6, (LTR)**-εp*, which contained not only a disabled LTR promoter but also a disabled ε-globin promoter, LTR enhancer activity was not detectable, as indicated by the background level of GFP expression (Fig. 3a). Corres pondingly, the band of 230 nt produced by GFP mRNA was not detected, although the long LTR** RNA producing the band of 1100 nt was present (Fig. 3c, lane 6). These results indicate that the LTR**RNA transcribed through the HS5 site and the ε-globin promoter into the GFP gene was not processed into GFP mRNA or translated into GFP. Furthermore, they showed that the LTR** enhancer could autonomously initiate synthesis of long LTR RNAs from downstream cryptic promoters in U5 and HS5 (see 1100, 900 and 800 bp bands in lanes 5 and 6, Fig. 3c). However, these cryptic promoters produced non-coding RNAs and appeared not to be truly functional promoters, which should produce mRNA that could be translated into the protein product of the gene. Hence, expression of the GFP gene required a functional ε-globin promoter proximal to the GFP gene to synthesize translatable GFP mRNA.
Taken together, the results indicate that (i) the ability of the LTR enhancer to initiate synthesis of long LTR RNAs did not require the LTR and ε-globin promoters, (ii) the long LTR RNAs synthesized autonomously by the LTR enhancer through the HS5 site and the ε-globin promoter into the CAT/GFP gene were not processed into mRNA or translated into protein products, and (iii) manifestation of LTR enhancer activity in recombinant plasmids required a functional ε-globin promoter from which to initiate mRNA synthesis.
Reversing the orientation of the LTR with respect to the ε-globin promoter and GFP gene, thus causing the LTR RNA to be synthesized in the antisense direction away from the ε-globin promoter and gene, diminishes LTR enhancer function
To assess the functional significance of the synthesis of long LTR RNAs in LTR enhancer function, we determined whether interfering with this transcription process affected LTR enhancer function. To this end, we created the plasmid (LTR)rev-HS5-εp-GFP, (LTR)rev-εp (Fig. 4a), in which the orientation of the LTR was reversed with respect to the ε-globin promoter and GFP gene. This plasmid and the reference plasmid (LTR)-HS5-εp-GFP, (LTR) -εp, containing the LTR inserted in the sense orientation, were linearized at a site in the vector upstream of the LTR (see Materials and Methods) to ensure that the LTR enhancer acted only from a location upstream of the HS5 site and the GFP gene but not downstream of the GFP gene by-passing the HS5 site. The linearized plasmids were transiently transfected into K562 cells. The level of GFP expression in the transfected cells was determined by FACS analyses. The sense versus antisense (+ versus –) direction of the RNAs transcribed from the plasmids was determined by directional RT–PCR with primer pairs 1a–1d, 2 and 3, which amplified regions of interest in the plasmids but not the corresponding regions in the K562 genome (see Fig. 4a and Materials and Methods). In directional RT–PCR, the difference in band intensities between each pair of + and – lanes should faithfully reflect the relative abundance of the sense versus antisense RNAs of the region, since the difference in band intensities could not be due to different amplification efficiencies of the primer pair, as in each pair of + and – lanes, the bands were generated by the same primer pair (see Materials and Methods). Furthermore, to show an equal amount of RNA template used in each lane of RT–PCR, an internal control band was generated from the endogenous ε-globin mRNA (see Materials and Methods).
FACS analysis showed that the transfected (LTR)rev-εp expressed GFP at a greatly reduced level at 30% that of the reference (LTR)-εp (Fig. 4a). Directional RT–PCR showed that the LTR enhancer now initiated synthesis of LTR RNA predominantly in the antisense direction (compare + and – lanes amplified by primer pair 1c, Fig. 4b). Interestingly, the HS5 site in (LTR)rev-εp, now located upstream of the LTR, was also transcribed predominantly in the antisense direction (compare + and – lanes amplified by 1d and 2, Fig. 4b). In contrast, in the reference (LTR)-εp plasmid containing the LTR inserted in the sense orientation, the LTR and also the downstream HS5 site were transcribed predominantly in the sense direction, co-linear with synthesis of GFP mRNA (compare + and – lanes amplified by primer pairs 1a, 1b and 2 in (LTR)-εp panel, Fig. 4b). Since the forward primers in 1a and 1c were located in the vector sequence, the LTR transcription machinery could drive transcription from the vector sequence as well as HS5 in either the antisense or sense direction, co-linear with synthesis of the LTR RNAs (Fig. 4a).
In (LTR)rev-εp, in association with transcription of the LTR and the HS5 site in the antisense direction away from the ε-globin promoter and the GFP gene, the level of GFP RNA was reduced by 50% as compared with that from the reference (LTR)-εp plasmid (compare the intensities of the GFP RNA bands in + lanes amplified by primer pair 3, Fig. 4b). The reduction in transcription of GFP mRNA from (LTR)rev-εp could not have been due to the transcription of antisense GFP RNA resulting from elongation of the antisense LTR RNA through the vector DNA into the GFP gene, thus forming GFP RNA duplexes which could trigger the mechanism of RNA interference (42) to degrade GFP mRNA. This RNA interference was ruled out because (i) the transfected plasmid was linearized at a site in the vector such that the antisense LTR RNAs could not elongate through the vector DNA into the GFP gene and (ii) directional RT–PCR confirmed that the GFP gene was not transcribed in the antisense direction (see absence of bands in – lanes amplified by primer pair 3, Fig. 4b).
The 50% reduction in GFP mRNA transcribed from the (LTR)rev-εp plasmid might not have been accurately estimated from the intensities of RT–PCR bands, which produced only semi-quantitative measurements. In addition, since the RNA sample contained both GFP mRNA and LTR RNA that extended into the GFP gene, the RT–PCR band of GFP RNA was amplified not only from GFP mRNA but also the long LTR RNAs. Hence, we performed northern blots in which the GFP mRNA of 1.1 kb and the long LTR RNAs of 2.6 and 2.2 kb, initiated, respectively, from within the LTR enhancer and from the LTR promoter at the 5′ border of the R region, were resolved into separate bands (Fig. 4c), the radioactive intensities of which could be estimated without relying on PCR amplification. Quantification of the GFP mRNA bands showed that GFP mRNA produced by the (LTR)rev-εp test plasmid was 40% of that produced by the reference (LTR)-εp plasmid (see bar graph, Fig. 4c). Thus, the reduction in GFP mRNA transcribed from the (LTR)rev-εp plasmid, as estimated by both RT–PCR and northern blots, was in a similar range of 50–60%.
In (LTR)rev-εp the 50–60% reduction in transcription of GFP mRNA associated with antisense transcription of the LTR and the HS5 site suggests that the LTR transcription machinery, in synthesizing antisense LTR and HS5 RNAs away from the ε-globin promoter, could not reach the ε-globin promoter to activate GFP mRNA synthesis, thus resulting in a lower level of GFP mRNA. On the other hand, in (LTR)-εp the enhanced transcription of GFP mRNA associated with sense transcription of the LTR and the HS5 site suggests that the LTR transcription machinery through synthesizing sense LTR and HS5 RNAs into the ε-globin promoter could reach the ε-globin promoter to activate GFP mRNA synthesis, thus resulting in a higher level of GFP mRNA.
In contrast, (LTR)**rev-εp, containing the LTR enhancer and the disabled LTR promoter inserted in the antisense orientation, activated GFP expression to a very high level, comparable to that exhibited by the reference (LTR)**-εp containing (LTR)** in the sense orientation (Fig. 4d). Correlating with the active LTR enhancer in both (LTR)**rev-εp and (LTR)**-εp, directional RT–PCR showed that the (LTR)** enhancer, regardless of its genomic or reverse genomic orientation, now initiated sense transcription of LTR RNAs through the HS5 site into the ε-globin promoter and the GFP gene [see lanes 1a, 1b, 1d and 2 in the (LTR)**-εp panel and lanes 1d and 2 in the (LTR)**rev-εp panel, Fig. 4e]. In addition, the (LTR)** enhancer in (LTR)**rev-εp also activated synthesis of sense RNAs from within the LTR enhancer (see 1300 and 950 bp bands, Fig. 4f, lane 2) and from HS5, which generated a strong 5′-RACE band of 580 bp (Fig. 4f, lane 2).
Thus, in the presence of a disabled LTR promoter in (LTR)**-εp and (LTR)**rev-εp, the LTR enhancer initiated transcription in the sense direction towards the functional ε-globin promoter and exhibited very high enhancer activity. On the other hand, in the presence of both a functional LTR promoter and a functional ε-globin promoter in (LTR)rev-εp, the LTR enhancer initiated transcription preferentially in the antisense direction towards the LTR promoter and exhibited greatly reduced enhancer activity. Together, the results indicate that the LTR transcriptional machinery, in synthesizing non-coding, sense, long RNAs through the intervening HS5 site, could reach the ε-globin promoter to activate GFP mRNA synthesis and thus mediate enhancer function over a distance.
DISCUSSION
In this study, functional dissection of the 5′ boundary area of the β-LCR shows that the ERV-9 LTR located within 1.5 kb upstream of the HS5 site in the β-LCR exhibited unique enhancer and promoter activities in K562 erythroid cells not shared by other DNA fragments in the 9 kb boundary area. Since the LTR is not linked to immediately downstream retroviral or cellular genes, it is possible that the strong ERV-9 LTR enhancer may interact with the proximal HS5 site in modulating transcription of the β-LCR in erythroid cells. On the other hand, the HS5 site has been reported to exhibit insulator properties, in being able to block the enhancer function of the HS2 and HS3 sites when HS5 was placed downstream of these enhancers (31,33,34). Hence, the HS5 site located naturally downstream of the ERV-9 LTR could similarly block LTR enhancer activity, thus precluding any functional interactions of the LTR enhancer with the β-LCR or the further downstream globin genes. However, transfection experiments in this study showed that in LTR-HS5-εp-GFP/CAT plasmids, the HS5 site located downstream of the LTR enhancer did not block LTR enhancer activity. Furthermore, in integrated plasmid HS5L-HS4-εp-CAT, the HS5 site inserted upstream of HS4 and the ε-globin promoter dramatically stimulated the weak HS4 enhancer to exhibit very high enhancer activity. This is contrary to the reported behavior of the insulator, which does not significantly inhibit or stimulate enhancer activity when it is placed upstream of both the enhancer and the promoter in integrated plasmids (31–33,43). Thus, our results indicate that when the HS5 site was linked in the genomic order to its natural neighbors in the β-LCR, i.e. downstream of the ERV-9 LTR or upstream of the HS4 site, it synergized with the neighboring enhancers and did not exhibit insulator function. Through such synergistic interactions with the neighboring HS5 and HS4 sites, ERV-9 LTR enhancer activity could potentially be transmitted over a distance into the β-LCR to regulate LCR transcription in erythroid cells.
It has been postulated that the β-LCR could act over long intervening DNA of 6–45 kb to activate transcription of the far downstream globin genes by a looping mechanism, in which the LCR complex, the LCR DNA and its associated transcription factors, loops over the intervening DNA to directly interact with the globin promoters (28,29), or by a tracking mechanism, in which the LCR complex or its protein components track along the intervening DNA to reach and activate the far downstream gene (40,44), or by a combination of the two mechanisms (27,45). Recently, the β-LCR has been shown to be in physical proximity to the downstream globin gene that is being transcribed (46,47). These studies indicate that long range LCR function could be mediated by a looping mechanism, although how the LCR complex translocates through the nucleoplasm space to precisely loop with the cis-linked globin genes without encountering, and thus looping with and activating, nearby, unlinked heterologous genes is still not clear.
Viewed within the looping model, the ERV-9 LTR enhancer could apparently loop over the HS5 site with the assembled CTCF complex to reach the ε-globin promoter and activate the CAT/GFP gene. Thus, the LTR enhancer in plasmid (LTR)-HS5-εp-GFP may loop with either the LTR or the ε-globin promoter to activate transcription of the GFP gene through a flip-flop looping mechanism (48). In the (LTR)**-HS5-εp-GFP and (LTR)**rev-HS5-εp-GFP plasmids containing a disabled LTR promoter, the LTR** enhancer, regardless of orientation, interacted exclusively with the functional ε-globin promoter without competition from the disabled LTR promoter and thus exhibited enhancer activity over 3-fold higher than the LTR in (LTR)-HS5-εp-GFP (Fig. 4a and d).
The looping model would thus predict that the (LTR)rev enhancer, inserted in reverse genomic orientation in (LTR)rev-HS5-εp-GFP, should also be able to loop with the ε-globin promoter with similar efficiency and exhibit comparable enhancer activity as the LTR enhancer inserted in the genomic orientation in (LTR)-HS5-εp-GFP. However, transfection results showed that (LTR)rev exhibited an enhancer activity only 30–50% that of (LTR) in the two plasmids. This reduction in enhancer activity could not be attributed to differences in distance or sequence composition of the intervening DNA between the LTR enhancer and the ε-globin promoter, since the intervening DNAs in these two plasmids were identical (Fig. 4a and d). The lack of complete consistency when solely using the looping model to interpret the results of LTR enhancer assays suggests that other mechanisms may also participate in LTR enhancer function in these plasmids.
Indeed, transcriptional analyses by directional RT–PCR showed that the (LTR)rev enhancer in (LTR)rev-HS5-εp-GFP initiated transcription of both the LTR and the HS5 site predominantly in the antisense direction, away from the ε-globin promoter, which correlated with reduced enhancer activity. In contrast, the (LTR) enhancer in (LTR)-HS5-εp-GFP and also the (LTR)** enhancers in (LTR)**rev-HS5-εp-GFP and (LTR)**-HS5-εp-GFP initiated transcription predominantly in the sense direction through the intervening HS5 site into the ε-globin promoter, which correlated with elevated enhancer activities of these plasmids (Fig. 4a–f)
These observations suggest that the transcription machinery assembled by the LTR enhancer contains RNA polymerase, which could track and transcribe through the intervening HS5 site to reach the downstream ε-globin promoter and activate GFP mRNA synthesis. The RNA polymerase associated with the LTR transcriptional machinery appeared to be RNA polymerase II (pol II), since synthesis of LTR RNAs could be initiated from the LTR promoter, a pol II promoter containing a TATA box (Fig. 2e) (30) and their synthesis could be inhibited by a low concentration of α-amenitin (49). Consistent with a transcription mechanism of enhancer function, it has been shown that in yeast the major function of the enhancer is to deliver pol II to the cis-linked promoter (50), since the promoter packaged in nucleosomes is unable to recruit pol II and associated transcription factors to the promoter site (51). Thus, it is possible that pol II associated with the LTR enhancer complex, in transcribing long LTR RNAs through the intervening DNA and the ε-globin promoter, may guide the enhancer complex through this tracking and transcription process to ensure that the enhancer complex interacts and forms a loop not with nearby heterologous genes in trans but only with the cis-linked target gene. In view of the long processivity of pol II which can transcribe genes of over 100 kb, it is possible to envision that the LTR transcription machinery could transcribe long distances to bring the LTR enhancer complex to far downstream target sequences in the β-LCR and the globin genes, thus forming an increasingly enlarging loop between the enhancer complex and the intervening DNA being transcribed and ultimately forming a loop between the enhancer complex and the promoter of the downstream gene. Hence, the LTR enhancer may act through a combination of the tracking and the looping mechanisms, with the tracking and transcription process establishing precise loop formation between the enhancer complex and its downstream target (27,45).
This study also revealed several novel features in the synthesis of LTR RNAs.
Synthesis of the LTR RNAs in the plasmids appeared to be an autonomous transcription process initiated by the LTR enhancer independent of the LTR promoter. In the presence of a disabled LTR promoter (constructs 5 and 6, Figs 3 and 4d–f), the LTR enhancer initiated synthesis of non-coding LTR RNAs from multiple cryptic promoters within the LTR enhancer (see 1300 and 950 bp bands, Fig. 4f, lane 2), in the vector sequence (Fig. 4e, lanes 1a and 1c) and in HS5 (800 and 900 bp bands, Fig. 3c, lanes 5 and 6; 580 bp band, Fig. 4f, lane 2). In contrast, HS5 by itself, correlating with its weak enhancer activity, initiated synthesis of only low levels of HS5 RNAs that were not consistently detectable (Fig. 2b, lanes 1 and 2, and Fig. 3c, lane 1). However, these cryptic promoters did not appear to be functional promoters, which by definition should produce mRNA that could be translated into the protein product of the gene. Providing additional support for this observation, the LTR enhancer containing cryptic promoters, when coupled directly to the GFP gene in an (E)-GFP plasmid, did not produce mRNAs that could be translated into GFP (Fig. 1b). These observations indicate that the cryptic promoters initiated syntheses of non-coding RNAs.
While the LTR enhancer could autonomously initiate synthesis of LTR RNAs from multiple cryptic promoters in the absence of a functional LTR promoter, the LTR promoter when present in the LTR-HS5-εp-GFP plasmid appeared to specify a preferred initiation site for the LTR RNA, at the 5′ border of the R region located 25 bases downstream of the AATAAA (TATA) box of the LTR promoter (Fig. 2e). Although longer LTR RNAs initiated from the vector sequence (Fig. 4a and b) and the LTR enhancer (Fig. 4c) were also synthesized by this plasmid, the shorter LTR RNA initiated from the 5′ border of R by the LTR promoter was the only RNA detectable by 5′-RACE (Figs 2 and 3, lanes 1), apparently because it could more efficiently compete for and was thus preferentially amplified by the common PCR primers used in 5′-RACE (see illustration of 5′-RACE, Fig. 2a). Consistent with this finding, the endogenous ERV-9 LTR in the β-globin gene locus in K562 cells also synthesized endogenous LTR RNA from the same site at the 5′ border of the R region that was detectable by 5′-RACE (30,49). We and others suggest that synthesis of the endogenous ERV-9 LTR RNAs represents the initiating event in transcriptional activation of the β-LCR and may thus regulate the transcriptional status of the globin gene locus (19,30,49).
The LTR promoter could also specify the direction of synthesis of LTR RNAs. Thus, in (LTR)-HS5-εp-GFP the LTR RNAs were synthesized predominantly in the sense direction towards the LTR promoter, whereas in (LTR)rev-HS5-εp-GFP the LTR RNAs were synthesized predominantly in the antisense direction towards the LTR promoter inserted in the antisense orientation (Fig. 4a and b). Earlier, it was reported that the LTRs of murine intracisternal A particles, isolated from different gene loci in the mouse genome and tested in either orientation in reporter gene assays, also show LTR transcription and enhancer activity that are dependent on the orientation of the LTR promoters (52).
The long LTR RNAs transcribed from the cryptic promoters and also the LTR promoter through the HS5 site into the ε-globin promoter and the GFP gene did not serve as mRNA and was not translated into GFP (Fig. 3). In contrast, when the LTR was linked directly to the GFP gene in a (LTR)-GFP plasmid, the LTR-GFP RNA initiated from the LTR promoter and elongated directly into the GFP gene served as mRNA and was translated into GFP (Fig. 1b). Sequence analysis of the long LTR-HS5-εp-GFP RNA revealed that the 0.5 kb HS5 contained eight start codons and an extraordinary 59 stop codons, which is up to 6- to 8-fold higher than those in the U5 and ε-globin promoter, each of which contained one start and 10–11 stop codons. It is possible that in the short LTR-GFP RNA the ribosomes could escape the limited number of start and stop codons in the U5 leader sequence to translate the downstream GFP gene (53). However, in the long LTR-HS5-εp-GFP RNA, the ribosomes might not be able to escape the many AUG codons in the long leader sequence, which could initiate out-of-frame translation, and the many stop codons in HS5, which could prematurely terminate translation, thus resulting in truncated translation products without GFP activity. Other factors contributing to the translation block of the long LTR RNAs may also exist and await further investigation.
Large-scale transcriptional analysis of human chromosomes 21 and 22 indicates that >90% of the transcribed DNA is located in the non-coding regions of the chromosomes (54). Non-coding RNAs have been reported to participate in various cellular functions, including regulation of translation and X chromosome dosage compensation (55). In this study, we showed that synthesis of non-coding LTR RNAs by the enhancer-assembled transcription machinery could yet contribute to a new function in regulating LTR enhancer function in plasmids. The biological significance of the synthesis of LTR RNAs from the ERV-9 LTR in the β-globin gene locus and from other ERV-9 LTRs in the human genome (30) remains to be elucidated.
Acknowledgments
ACKNOWLEDGEMENT
This work was supported in part by NIH grants HL 39948 and 62308.
REFERENCES
- 1.Wilkison D., Mager,D. and Leong,J. (1994) Endogenous human retroviruses. In Levy,J. (ed.), The Retroviridae. Plenum Press, New York, NY, Vol. 3, pp. 465–535. [Google Scholar]
- 2.Smit A.F. (1996) The origin of interspersed repeats in the human genome. Curr. Opin. Genet. Dev., 6, 743–748. [DOI] [PubMed] [Google Scholar]
- 3.Lower R., Lower,J. and Kurth,R. (1996) The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl Acad. Sci. USA, 93, 5177–5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henikoff S., Greene,E., Pietrokovski,S., Bork,P., Attwood,T. and Hood,L. (1997) Gene families: the taxonomy of protein paralogs and chimeras. Science, 278, 609–614. [DOI] [PubMed] [Google Scholar]
- 5.Temin H.M. (1981) Structure, variation and synthesis of retrovirus long terminal repeat. Cell, 27, 1–3. [DOI] [PubMed] [Google Scholar]
- 6.Coffin J., Hughes,S. and Varmus,H. (1997) Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- 7.International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 8.Doolittle W.F. and Sapienza,C. (1980) Selfish genes, the phenotype paradigm and genome evolution. Nature, 284, 601–603. [DOI] [PubMed] [Google Scholar]
- 9.Medstrand P., Landry,J.R. and Mager,D.L. (2001) Long terminal repeats are used as alternative promoters for the endothelin B receptor and apolipoprotein C-I genes in humans. J. Biol. Chem., 276, 1896–1903. [DOI] [PubMed] [Google Scholar]
- 10.Strazzullo M., Parisi,T., Di Cristofano,A., Rocchi,M. and La Mantia,G. (1998) Characterization and genomic mapping of chimeric ERV9 endogenous retroviruses-host gene transcripts. Gene, 5, 77–83. [DOI] [PubMed] [Google Scholar]
- 11.Perez-Stable C., Ayres,T.M. and Shen,C.J. (1984) Distinctive sequence organization and functional programming of an Alu repeat promoter. Proc. Natl Acad. Sci. USA, 81, 5291–5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmid C.W. (1996) Alu: structure, origin, evolution, significance and function of one tenth of human DNA. Prog. Nucleic Acid Res. Mol. Biol., 53, 283–319. [DOI] [PubMed] [Google Scholar]
- 13.Britten R.J. (1996) DNA sequence insertion and evolutionary variation in gene regulation. Proc. Natl Acad. Sci. USA, 93, 9374–9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moran J.V., DeBerardinis,R.J. and Kazazian,H.H. (1999) Exon shuffling by L1 retrotransposition. Science, 283, 1530–1533. [DOI] [PubMed] [Google Scholar]
- 15.Henthorn P.S., Mager,D., Huisman,D. and Smithies,O. (1986) A gene deletion ending within a complex array of repeated sequences 3′ to the human beta-globin gene cluster. Proc. Natl Acad. Sci. USA, 83, 5194–5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.La Mantia G., Maglione,D., Pengue,G., Di Cristofano,A., Simeone,A., Lanfrancone,L. and Lania,L. (1991) Identification and characterization of novel human endogenous retroviral sequences prefentially expressed in undifferentiated embryonal carcinoma cells. Nucleic Acids Res., 19, 1513–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zucchi I. and Schlessinger,D. (1992) Distribution of moderately repetitive sequences pTR5 and LF1 in Xq24-q28 human DNA and their use in assembling YAC contigs. Genomics, 12, 264–275. [DOI] [PubMed] [Google Scholar]
- 18.Di Cristofano A., Strazzullo,M., Parisi,T. and LaMantia,G. (1995) Mobilization of an ERV9 human endogenous retroviral element during primate evolution. Virology, 213, 271–275. [DOI] [PubMed] [Google Scholar]
- 19.Long Q., Bengra,C., Li,C., Kutlar,F. and Tuan,D. (1998) A long terminal repeat of the human endogenous retrovirus ERV-9 is located in the 5′ boundary area of the human β-globin locus control region. Genomics, 54, 542–555. [DOI] [PubMed] [Google Scholar]
- 20.Orkin S.H. (1992) GATA-binding transcription factors in hematopoietic cells. Blood, 80, 575–581. [PubMed] [Google Scholar]
- 21.Tenen D.G., Hromas,R., Licht,J. and Zhang,D. (1997) Transcription factors, normal myeloid development and leukemia. Blood, 90, 489–519. [PubMed] [Google Scholar]
- 22.Miller I. and Bieker,J. (1993) A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol. Cell. Biol., 13, 2776–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuan D., Solomon,W., Li,Q. and London,I. (1985) The “β-like-globin” gene domain in human erythroid cells. Proc. Natl Acad. Sci. USA, 82, 6384–6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forrester W., Takegawa,S. Papayannopoulou,T., Stamatoyannopoulos,G. and Groudine,M. (1987) Evidence for a locus activation region: the formation of developmentally stable hypersensitive sites in globin-expressing hybrids. Nucleic Acids Res., 15, 10159–10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grosveld F., van Assendelft,G., Greaves,D. and Kollias,G. (1987) Position-independent, high-level expression of the human b-globin gene in transgenic mice. Cell, 51, 975–985. [DOI] [PubMed] [Google Scholar]
- 26.Higgs D.R. (1998) Do LCRs open chromatin domains? Cell, 95, 299–302. [DOI] [PubMed] [Google Scholar]
- 27.Li Q. and Peterson,K. (1999) Locus control regions coming of age at a decade plus. Trends Genet., 10, 403–408. [DOI] [PubMed] [Google Scholar]
- 28.Bulger M. and Groudine,M. (1999) Looping versus linking: toward a model for long-distance gene activation. Genes Dev., 13, 2465–2477. [DOI] [PubMed] [Google Scholar]
- 29.Engel J.D. and Tanimoto,K. (2000) Looping, linking and chromatin activity: new insights into β-globin locus regulation. Cell, 100, 499–502. [DOI] [PubMed] [Google Scholar]
- 30.Ling J., Pi,W., Bollag,R., Zeng,S., Keskintepe,M., Saliman,H., Krantz,S., Whitney,B. and Tuan,D. (2002) The solitary long terminal repeats of ERV-9 endogenous retrovirus are conserved during primate evolution and possess enhancer activities in embryonic and hematopoietic cells. J. Virol., 76, 2410–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung J., Whiteley,M. and Felsenfeld,G. (1993) A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell, 74, 505–514. [DOI] [PubMed] [Google Scholar]
- 32.Yu J., Bock,J., Slightom,J. and Villeponteau,B. (1994) A 5′ β-globin matrix-attachment region and the polyoma enhancer together confer position-independent transcription. Gene, 139, 139–145. [DOI] [PubMed] [Google Scholar]
- 33.Li Q. and Stamatoyannopoulos,G. (1995) Hypersensitive site 5 of the human β locus control region functions as a chromatin insulator. Blood, 84, 1399–1401. [PubMed] [Google Scholar]
- 34.Farrell C., West,A. and Felsenfeld,G. (2002) Conserved CTCF insulator elements flank the mouse and human β-globin loci. Mol. Cell. Biol., 22, 3820–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frohman A. (1993) Rapid amplification of complementary DNA ends for generation of full-length complementary DNAs: thermal RACE. Methods Enzymol., 218, 340–356. [DOI] [PubMed] [Google Scholar]
- 36.Tuan D., Solomon,W., London,I. and Lee,D. (1989) An erythroid-specific, developmental-stage-independent enhancer far upstream of the human ‘β-like globin’ genes. Proc. Natl Acad. Sci. USA, 86, 2554–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stamatoyannopoulos J., Goodwin,A., Joyce T. and Lowrey,C. (1995) NF-E2 and GATA binding motifs are required for the formation of DNase I hypersensitive site 4 of the human beta-globin locus control region. EMBO J., 14, 106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J., Fritsch,E. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, Vol. 1, Ch. 7. [Google Scholar]
- 39.Li Q., Zhang,M., Duan,Z. and Stamatoyannopoulos,G. (1999) Structural analysis and mapping of DNase I hypersensitivity of HS5 of the beta-globin locus control region. Genomics, 6, 183–193. [DOI] [PubMed] [Google Scholar]
- 40.Kong S., Bohl,D., Li,C. and Tuan,D. (1997) Transcription of the HS2 enhancer toward a cis-linked gene is independent of the orientation, position and distance of the enhancer relative to the gene. Mol. Cell. Biol., 17, 3955–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nienhuis A., Anagnou,N. and Ley,T. (1984) Advances in thalassemia research. Blood, 63, 738–758. [PubMed] [Google Scholar]
- 42.Hannon G. (2002) RNA interference. Nature, 418, 244–250. [DOI] [PubMed] [Google Scholar]
- 43.Cai H. and Levine,M. (1995) Modulation of enhancer-promoter interactions by insulators in the Drosophila embryo. Nature, 376, 533–536. [DOI] [PubMed] [Google Scholar]
- 44.Hatzis P. and Tallanidis,I. (2002) Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol. Cell, 10, 1467–1477. [DOI] [PubMed] [Google Scholar]
- 45.Tuan D., Kong,S. and Hu,K. (1992). Transcription of the hypersensitive HS2 enhancer in erythroid cells. Proc. Natl Acad. Sci. USA, 89, 11219–11223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carter D., Chakalova,L., Osborne,C., Dai,Y. and Fraser,P. (2002) Long-range chromatin regulatory interactions in vivo. Nature Genet., 32, 1–4. [DOI] [PubMed] [Google Scholar]
- 47.Tolhuis B., Palstra,R., Splinter,E., Grosveld,F. and Laat,W. (2002) Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell, 10, 1453–1465. [DOI] [PubMed] [Google Scholar]
- 48.Wijgerde M., Grosveld,F. and Fraser,P. (1995) Transcriptional complex stability and chromatin dynamics in vivo. Nature, 377, 209–213. [DOI] [PubMed] [Google Scholar]
- 49.Plant K., Routledge,S. and Proudfoot,N. (2001) Intergenic transcription in the human b-globin gene cluster. Mol. Cell. Biol., 21, 6507–6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keaveney M. and Struhl,K. (1998) Activator-mediated recruitment of RNA polymerase II is the predominant mechanism for transcriptional activation in yeast. Mol. Cell, 1, 917–924. [DOI] [PubMed] [Google Scholar]
- 51.Imbalzano A., Kwon,H., Green,M.R. and Kingston,R.E. (1994) Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature, 370, 481–485. [DOI] [PubMed] [Google Scholar]
- 52.Christy R. and Huang,R.C. (1988) Functional analysis of the long terminal repeats of intracisternal A particle genes: sequences within the U3 region determine both the efficiency and direction of a promoter activity. Mol. Cell. Biol., 8, 1093–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kozak M. (1999) Initiation of translation in prokaryotes and eukaryotes. Gene, 234, 187–208. [DOI] [PubMed] [Google Scholar]
- 54.Kapranov P., Cawley,S., Drenkow,J., Bekiranov,S,. Strausberg,R., Fodor,S. and Gingeras,T. (2002) Large-scale transcriptional activity in chromosomes 21 and 22. Science, 296, 916–919. [DOI] [PubMed] [Google Scholar]
- 55.Erdmann V.A., Szymanski,M., Hochberg,A., de Groot,N. and Barciszewski,J. (1999) Collection of mRNA-like non-coding RNAs. Nucleic Acids Res., 27, 192–195. [DOI] [PMC free article] [PubMed] [Google Scholar]