Abstract
We have refined a series of isomorphous crystal structures of the Escherichia coli DNA mismatch repair enzyme MutS in complex with G:T, A:A, C:A and G:G mismatches and also with a single unpaired thymidine. In all these structures, the DNA is kinked by ∼60° upon protein binding. Two residues widely conserved in the MutS family are involved in mismatch recognition. The phenylalanine, Phe 36, is seen stacking on one of the mismatched bases. The same base is also seen forming a hydrogen bond to the glutamate Glu 38. This hydrogen bond involves the N7 if the base stacking on Phe 36 is a purine and the N3 if it is a pyrimidine (thymine). Thus, MutS uses a common binding mode to recognize a wide range of mismatches.
INTRODUCTION
Genomic integrity in organisms is maintained by a number of important DNA repair pathways. The DNA mismatch repair (MMR) pathway repairs mismatches and short insertion or deletion loops (IDLs). In addition, MMR also helps in preventing recombination between homologous but diverged DNA sequences (1–3). The fundamental mechanisms of MMR are similar in all organisms ranging from Escherichia coli to humans. In E.coli, MMR is initiated when the enzyme MutS recognizes and binds to mismatches or IDLs. This is followed by the uptake of ATP by MutS and the formation of a complex between MutS and the enzyme MutL. This complex initiates a number of events, leading to the recognition of the daughter strand, followed by its removal and resynthesis. In humans, the role of MutS is played by its homologs, the heterodimers MSH2/MSH6, which binds mismatches and IDLs, and MSH2/MSH3, which binds longer loops (1,2). The role of MutL in humans is played by the heterodimer MLH1/PMS2. Mutations in the genes that encode MMR proteins lead to mutator phenotypes in bacteria and cause a predisposition to cancer, called hereditary non-polyposis colorectal carcinomas (HNPCC) in humans (4).
Two structures of MutS–DNA complexes have been reported already, the Thermus aquaticus enzyme in complex with a single unpaired thymidine (5), and the E.coli enzyme in complex with a G:T mismatch (6). The striking feature in both these structures is a sharp 60° kink in the DNA at the site of the mismatch–protein interaction. Mismatch recognition by MutS involves a phenylalanine, widely conserved in the MutS family (Phe 36 in E.coli, Phe 39 in Taq), which is seen stacking on one of the mismatched bases. The same base is also seen forming a hydrogen bond to a widely conserved glutamic acid (Glu 38 in E.coli, Glu 41 in Taq). In both these structures, the conserved phenylalanine stacks on the thymidine. To find out how other mismatches are recognized, we have solved the structures of E.coli MutS in complex with A:A, G:G, C:A mismatches and an unpaired thymidine. Our results show that all these different lesions are recognized in a similar way, indicating a common binding mode for all mismatches.
MATERIALS AND METHODS
Protein expression and purification
ΔC800, an 800 residue C-terminal deletion construct of E.coli MutS (853 residues) in a pET3d vector (6), derived from the pMQ372 plasmid (7) was used to transform B834 (DE3) pLysS cells. A colony was picked, inoculated into 10 ml of minimal medium (8) and allowed to grow overnight at 30°C. This culture was diluted into 1 l minimal medium and grown at 30°C till it reached an OD (600 nm) of ∼0.7. The temperature was then lowered to 23°C and the culture induced with IPTG (final concentration 1 mM) for 4 h. The cells were harvested, suspended in 25 ml of lysis buffer [50 mM HEPES pH 7.5, 200 mM NaCl, 10 mM β-mercaptoethanol, 5 mM EDTA, 1 mM PMSF and two protease inhibitor tablets (Roche)] and lysed by sonication. Following centrifugation at 39 000 r.c.f. for 50 min at 4°C, the cleared lysate was subjected to streptomycin precipitation. The volume of the lysate was first measured and streptomycin sulphate solution (25% w/v in water) added drop by drop while stirring on ice (9). The volume of streptomycin solution added was equal to 25% of the initial volume of the lysate. This was further stirred on ice for 15 min and centrifuged at 3000 r.c.f. for 25 min at 4°C. The supernatant from this step was subjected to ammonium sulphate precipitation by adding saturated ammonium sulphate solution, drop by drop, while strirring on ice continuously (9). The volume of the ammonium sulphate solution added was equal to 62% of the volume of the supernatant. This was stirred further on ice for 25 min and cleared by centrifugation (3000 r.c.f. for 25 min at 4°C). The pellet obtained was resuspended in GF1 buffer (25 mM HEPES pH 7.5, 150 mM NaCl, 10 mM β-mercaptoethanol, 5 mM EDTA and 0.1 mM PMSF) and applied on a Superdex 200 gelfiltration column (Pharmacia) pre-equilibrated with the same buffer. The peak corresponding to the dimer (160 kDa) was pooled and applied on a Mono-Q HR 10/10 ion-exchange column (Pharmacia) using buffers A (25 mM HEPES pH 7.5, 10 mM β-mercaptoethanol, 5 mM EDTA and 0.1 mM PMSF) and B (A with 1 M NaCl). The protein was eluted using a gradient running from 10 to 50% (buffer B) over 10 column volumes. The peak eluting between 20 and 42% of buffer B was pooled and its salt concentration was adjusted to ∼150 mM by adding buffer A. This was then applied on a HiTrap Heparin HP column (Pharmacia) (4 × 5 ml), which used the same buffers A and B, as the Mono-Q column. The protein was eluted using a gradient of 16–100% (buffer B) over 9 column volumes with the protein coming off between 52 and 70%. The final purification step was a second gelfiltration using a Superdex 200 column pre-equilibrated with GF2 buffer (25 mM HEPES pH 7.5, 250 mM NaCl, 10 mM β-mercaptoethanol). The fractions corresponding to the peak were pooled and concentrated to ∼14 mg/ml using a Centriprep concentrator (Millipore). Aliquots were then flash frozen in dry ice-ethanol and stored at –80°C.
DNA substrates
The two single strands of DNA purified by the reverse-phase cartridge purification method (Sigma-Genosys), were dissolved in 10 mM Tris–HCl pH 7.5, 1 mM MgCl2 and annealed on a heat block. The purity of the final double stranded product was checked using a 20% native polyacrylamide gel stained with ethidium bromide. The sequence of the top strand was 5′-AGC TGC CAM GCA CCA GTG TCA GCG TCC TAT and that of the lower strand was 5′-ATA GGA CGC TGA CAC TGG TGC MTG GCA GCT. The bold Ms indicate the positions of the mismatched nucleotides. The C:A mismatch had the C on the top and the A on the bottom strand and the G:T mismatch had the G on the top and T on the bottom strand, respectively. The sequence of the bottom strand for the unpaired thymidine substrate was 5′-ATA GGA CGC TGA CAC TGG TGC CTTG GCA GCT while that of the top strand was 5′-AGC TGC CAG GCA CCA GTG TCA GCG TCC TAT. The unpaired thymidine is indicated by the bold T.
Crystallization
The MutS–DNA substrates were mixed in a ratio of 2.8 (MutS monomer) to 1 (double stranded DNA) and crystallized using the hanging drop technique. Microseeding was done to improve the crystal quality. The quality of the crystals improved further upon addition of 0.1 mM ADP to the protein–DNA mixture. All crystals grew in the same space group, from a well solution containing 11–14% PEG 6000, 350–750 mM NaCl, 10 mM MgCl2 and 25 mM HEPES pH 7.5. Prior to data collection, cryobuffer (30% PEG 6000, 15% glycerol, 300 mM NaCl, 10 mM HEPES pH 7.5) was gradually added into the crystallization drop. The crystals were then removed, soaked into a drop of pure cryobuffer and frozen in liquid nitrogen.
Data collection, structure solution and refinement
All data collection was done either at the ESRF in Grenoble, France or at the EMBL outstation at DESY, Hamburg, Germany, and the data processed using the HKL suite (9) (Table 1). The structure of the MutS–G:T complex (6) was used as a model for structure solution. Unless otherwise specified, all refinement jobs were carried out using REFMAC5 (10) in the CCP4 suite (11). The waters, DNA and ligands (ADP-Mg) were first removed and rigid body refinement was carried out using the two protein monomers as rigid domains. This was followed by 20 cycles of rigid body refinement using the individual domains of the protein as rigid bodies. After this, restrained refinement was done and the first electron density maps were generated. The DNA with the corresponding mismatch and the ADP-Mg were then built into the difference density using the program O (12). Torsion angle refinement for the lower resolution structures (MutS–C:A, MutS–unpaired T) using CNS (13) and TLS refinement using REFMAC5 (14) for all the structures were performed which led to improved Rfree values (Table 2). The domains used in the rigid body refinement were used as TLS groups during the TLS refinement. Waters were built into the structures using ARP/wARP (15). All the structures were refined to good stereochemistry (Table 2) with >99% of the residues in the allowed and additionally allowed and none in the disallowed regions of the Ramachandran plot. Stereochemical checks on all structures were carried out using WHATCHECK (16) and the DNAs were analyzed using the program 3DNA (17). All figures except 1C and 1D were generated using MolScript (18) and Raster3D (19).
Table 1. Crystallographic data.
| Complex | MutS–A:A | MutS–C:A | MutS–G:G | MutS–unpaired T |
|---|---|---|---|---|
| Beamline | BW7B (DESY) | X11 (DESY) | ID14EH1(ESRF) | ID14EH2 (ESRF) |
| Resolution range (Å) | 15.0–2.4 (2.5–2.4) | 15.0–2.9 (3.0–2.9) | 15.0–2.6 (2.7–2.6) | 15.0–2.9 (3.0–2.9) |
| Complete (%) | 98.4 (86.1) | 95.4 (93.8) | 91.2 (60.5) | 98.3 (92.4) |
| I/sig(I) | 12.60 (1.60) | 12.21 (1.97) | 8.70 (1.10) | 9.54 (1.57) |
| Rmerge (%) | 8.9 (61.4) | 9.5 (76.0) | 10.4 (62.8) | 12.4 (72.5) |
| Space group | P212121 | P212121 | P212121 | P212121 |
| Cell parameters (Å) | a = 89.48 | a = 89.91 | a = 89.41 | a = 89.46 |
| b = 91.81 | b = 91.88 | b = 91.81 | b = 91.80 | |
| c = 260.04 | c = 261.17 | c = 260.44 | c = 259.70 | |
| Observations | 565 453 | 454 107 | 382 803 | 332 212 |
| Reflections | 84 338 | 48 834 | 75 563 | 48 882 |
Data in parentheses are those of the highest resolution shell.
Table 2. Refinement statistics.
| Complex | MutS–A:A | MutS–C:A | MutS–G:G | MutS–unpaired T |
|---|---|---|---|---|
| Resolution range (Å) | 15.0–2.4 | 15.0–2.9 | 15.0–2.6 | 15.0–2.9 |
| Number of atoms | 13 233 | 12 968 | 13 004 | 12 902 |
| Waters | 376 | 53 | 85 | 133 |
| R (%) | 20.5 | 22.4 | 22.7 | 21.6 |
| Rfree (%) | 25.2 | 29.3 | 27.7 | 29.2 |
| r.m.s.d. bonds (Å) | 0.012 | 0.010 | 0.014 | 0.010 |
| r.m.s.d. angles (°) | 1.362 | 1.293 | 1.488 | 1.261 |
Data collection and refinement statistics for the MutS-G:T complex published by Lamers et al. (6).
RESULTS
Overall structures of the MutS–mismatch complexes
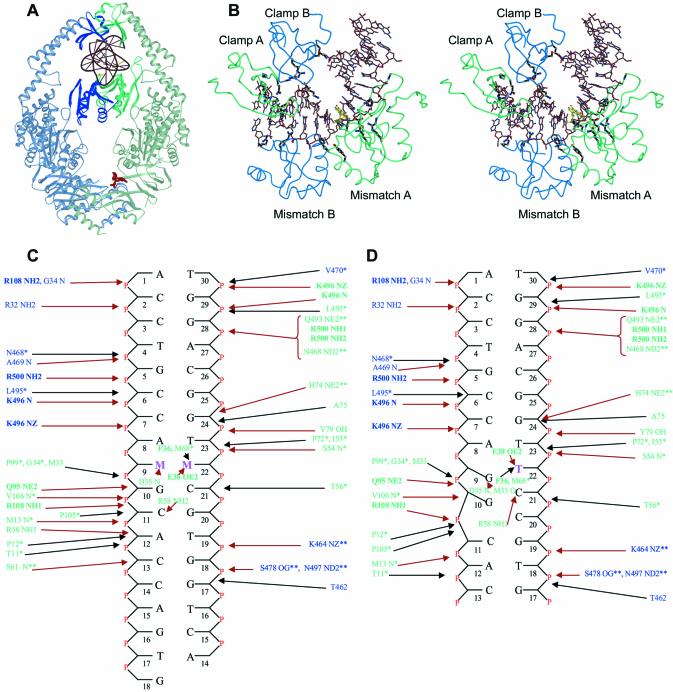
The overall structures of the MutS complexes are very similar to the published structure of the MutS–G:T complex (6) (Fig. 1A). The r.m.s. deviations (on Cαs) of the complexes with the MutS–G:T complex are 0.41 (MutS–C:A), 0.51 (MutS–unpaired T), 0.35 (MutS–G:G) and 0.36 Å (MutS–A:A). The DNA is held in place by the mismatch binding and clamp domains of both the monomers (Fig. 1B). Only one of the two monomers, monomer A, contacts the mismatch directly while the other, monomer B, only makes contacts to the DNA backbone. As in the original MutS–G:T structure, only 15–17 [13 in the MutS-unpaired T (Fig. 1D)] out of the 30 DNA base pairs, starting from the 5′-end of the top strand are visible. The remaining base pairs are untraceable in the density. Since the attempts to crystallize the protein in complex with a 16 bp oligo have been unsuccessful, the remaining bases could play a role in stabilizing the crystal packing or prevent alternative packing modes.
Figure 1.
DNA binding by MutS. (A) View of the MutS–DNA complex showing the DNA and ADP in red. The mismatch binding monomer (A) is coloured green and the other (monomer B) is coloured blue. (B) Stereo view of the protein–DNA interaction interface. The residues forming hydrogen bonds are coloured black. The mismatched bases are coloured yellow. (C) Schematic representation of the interactions between the mismatched DNAs and E.coli MutS. Residues from monomer A are shown in green and those from monomer B in blue. The bases marked M:M indicate the mismatches. Residues conserved in and making contacts to the DNA in Taq MutS (5) and also conserved in the eukaryotic homologs are indicated in bold and underlined. The residues conserved only in E.coli and Taq MutS and interacting with the DNA in the same way are shown with a single asterisk. Residues conserved in E.coli and Taq MutS but interacting with the DNA in a different way are shown with a double asterisk. Hydrogen bonds/salt bridges are shown with red arrows and Van der Waals interactions with black arrows. (D) Schematic representation of the interactions between the E.coli MutS–unpaired T.
Several contacts between the protein and DNA are seen, which are generally conserved in all the structures. The protein–DNA interface in the complexes is extensive, comprising of many hydrogen bonds, salt bridges and Van der Waals interactions (Fig. 1B, C and D). The surface area of the protein–DNA interface is ∼1850 Å2. The mismatch binding domain (residues 1–115) of monomer A accounts for more than half of this area (970 Å2) with several residues from it forming both hydrophobic and hydrophilic contacts to the DNA (Fig. 1B, C and D). The other three domains in contact to the DNA have much smaller interfaces. They are the clamp domain (residues 450–512) of monomer B (525 Å2), clamp domain of monomer A (285 Å2) and the mismatch binding domain of monomer B (105 Å2). The contacts from these domains are predominantly hydrophilic (Fig. 1C and D).
Mismatch binding by MutS
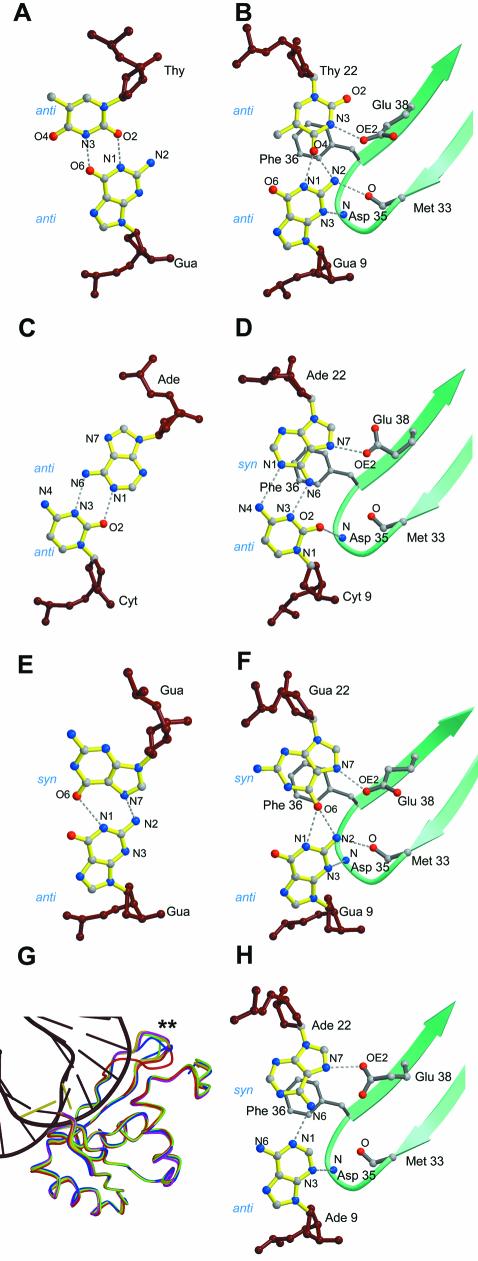
The most striking feature of the DNA in all the complexes is a sharp kink of ∼60° with the mismatched bases located at the vertex of the kink (Fig. 1B). This kinking causes a widening of the minor groove around the mismatch. The distance between the backbone phosphates (P–P distance) of the mismatched base pairs increases to 21–22 Å from an average of 11–12 Å for the rest of the base pairs in the minor groove. Mismatch binding by MutS involves the stacking of a phenylalanine residue, Phe 36 of one of the monomers, onto one of the mismatched bases. The same base is reoriented such that a particular nitrogen on it is brought into proximity to the glutamate, Glu 38. This enables the formation of a hydrogen bond between the carbonyl oxygen (OE2) of the glutamate and the nitrogen of the base. In the structures of the MutS–G:T and the MutS–unpaired T complexes, Phe 36 stacks over the thymidines with their N3s forming the hydrogen bonds to Glu 38 (Figs 2B and 3A). In the structures of the MutS–C:A and MutS–A:A complexes, Phe 36 stacks on the adenosines and their N7s form hydrogen bonds to Glu 38 (Fig. 2D and H). The same is seen in the MutS–G:G complex (Fig. 2F) where the N7 of the guanosine is seen in this conformation. The purine bases on which Phe 36 stacks are in the syn orientation in contrast to the thymidines in the G:T mismatch and the unpaired thymidine complex which are in the anti orientation of the glycosyl bond.
Figure 2.
Comparisons between mismatches bound by MutS and those seen in oligos without the protein. (A) Base pairing in the unbound G:T mismatch (21). (B) G:T mismatch bound to MutS (6). (C) Base pairing in the unbound C:A mismatch (20). (D) C:A mismatch bound to MutS. (E) Base pairing in the unbound G:G mismatch (22). (F) G:G mismatch bound to MutS. (G) Superposition of the mismatch binding domains of all the structures. The variation in the loop between Ala 60 and Gly 63 is indicated by the two asterisks. (H) A:A mismatch bound to MutS.
Figure 3.
Comparison of the E.coli MutS–unpaired T complex with Taq MutS. (A) View of the protein–mismatch interaction site in the MutS–unpaired T complex. The guanosine forms a G:T mismatch with the unpaired thymidine. (B) Side stereoview of the MutS–unpaired T complex showing disruption of the Watson–Crick base pairs downstream to the unpaired T (shown in yellow). (C and D) Front view and side stereoview of the protein–mismatch interaction site in the Taq MutS structure (5).
In the structure of the MutS–unpaired T, more severe unstacking and disruptions in the base pairs adjacent to the unpaired thymidine, Thy 22, are seen (Fig. 3A and B). Phe 36 stacks on the unpaired Thy 22, which is seen forming a G:T base pair with the Gua 9 with significant rearrangements taking place in the Gua 9: Cyt 21 and Gua 10: Cyt 20 Watson–Crick base pairs. The contacts between the protein and the DNA are preserved (Fig. 1C and D) and are identical to those of the MutS–mismatch complexes. A small difference is the conformation of the loop between Ala 60 and Gly 63, which causes a change in the orientation of the side chain of Arg 58 (Fig. 1C and D). Since this loop is not visible in the other monomer due to lack of density, it seems to be highly mobile and takes on a different conformation in each of the structures (Fig. 2G).
DISCUSSION
Mismatch recognition by MutS and the effect of mismatches in DNA have been widely studied, biochemically and structurally. In our structures, MutS binds to DNA containing single G:T, A:A, C:A, G:G mismatches and an unpaired thymidine in a very similar way. Most protein–DNA interactions are conserved (Fig. 1C and D) among the complexes. Although the packing of these molecules is similar, it is clear that rearrangements of loops and side chains would be possible. In fact there is substantial rearrangement of the DNA in the complex with the unpaired thymidine. However, the interface between the DNA and protein is remarkably similar in all the five structures of the E.coli MutS, while the major features are also very well conserved in the Taq MutS structure (5), indicating a common mismatch binding mode by MutS. As the crystal structures of unbound oligos containing G:T, C:A and G:G base pairs (20–22) show no kinking of the DNA, it is clear that this occurs only upon MutS binding. A similar kink in the DNA has also been seen in the structure of Taq MutS in complex with an unpaired thymidine. This seems to be an important requirement as a straight piece of DNA would lead to severe Van der Waals clashes with the mismatch binding domains (Fig. 1B). Phe 36, which is widely conserved in the MutS family, is seen stacking on to one of the mismatched bases. It has been shown that mutating this phenylalanine to an alanine eliminates both DNA binding and MMR by MutS (23).
Comparison of the mismatches bound to MutS to those in crystal structures of free oligos (Fig. 2A and B, C and D, E and F) (20–22) shows rearrangements in the base pairing upon protein binding. This rearrangement exposes either the N3 of the thymidine [in the MutS–G:T and MutS–unpaired T structures (Figs 2A and 3A)] or the N7 of the adenosine/guanosine [in the MutS–C:A, MutS–A:A and the MutS–G:G structures (Fig. 2D, H and F)] to Glu 38 for hydrogen bonding. The widening of the minor groove upon kinking of the DNA probably gives the protein enough room to reorient these bases to achieve this. In the MutS–G:T structure, the protein only has to shift the thymidine from its unbound position (Fig. 2A and B) to expose the N3 to Glu 38. In the MutS–G:G structure, a similar rearrangement of the syn guanosine (Fig. 2E and F) is enough to expose the N7 to Glu 38. In contrast, in the MutS–C:A structure, the adenosine is rotated around its C1′–N9 bond, from its anti orientation in the unbound state (Fig. 2C) to the syn orientation (Fig. 2D). In the MutS–unpaired T structure (Fig. 3A and B) and in the Taq MutS–unpaired T complex (5) (Fig. 3C and D) the N3s of the thymidine stacking on to Phe 36/Phe 39 form this hydrogen bond. These data suggest a scenario where the stacking of Phe 36 on any pyrimidine would lead to the N3 forming a hydrogen bond to Glu 38 while the stacking on a purine would involve its N7 forming the same hydrogen bond.
Glu 38 is a widely conserved residue in the MutS family of proteins. Besides forming the hydrogen bond to the base stacked upon by Phe 36, the role played by Glu 38 in mismatch recognition remains unclear. It has been shown that mutating Glu 38 to an alanine destroys MMR activity in MutS and increases the affinity of the protein towards homoduplex DNA (24). The requirement of a hydrogen bonding donor/acceptor for this residue in the base stacking on Phe 36 has also been demonstrated (24). Removal of the N3 of the thymidine by replacing it with difluorotoluene, which lacks the N3, leads to an 8-fold decrease in mismatch binding affinity by MutS (24). Replacement of the adenosine with 4-methylbenzimidazole, which lacks the N6, N1 and N3, also shows a similar effect. Although in the MutS–C:A and MutS–A:A structures, the N7s of the adenosines form hydrogen bonds with Glu 38, the N3s, N6s and N1 are involved in stabilizing the complex by forming base pairing hydrogen bonds (Fig. 2D and H). Thus, the disruption of any of these sites can affect the complex formation with MutS.
The purine N7–Glu 38 (OE2) hydrogen bond in the MutS–A:A, MutS–C:A and MutS–G:G structures is unexpected since neither of the atoms involved is protonated. Therefore, either the purines are in a tautomeric form where their N7s are protonated or the OE2 of Glu 38 must be protonated. While buried and protonated acids have been seen in crystal structures (25–27), it is unclear how the required pKa shift would occur here. Recently, Junop et al. (28) have shown that mutating Glu 38 into a glutamine improves homoduplex DNA binding relative to mismatch binding, thereby eliminating MMR completely. This glutamine would be able to make the same hydrogen bond and so the role of this residue in mismatch recognition seems complex. It has been suggested that the acidity of the glutamate plays a role in kinking the DNA during mismatch recognition (28). More evidence on the protonation of Glu 38 or the tautomerization of the purines awaits biochemical testing.
The extensive contacts between the protein and DNA play an important role in the stabilization of the protein–DNA complexes as it has been seen that the mismatch binding and the clamp domains are disordered in the absence of the DNA (5). An interesting observation is the involvement in DNA binding, of many other residues widely conserved in the MutS family besides Phe 36 and Glu 38. In the mismatch binding domain, Arg 108 and Gln 95 side chains seem to be important as they are not only conserved in Taq MutS, where they form hydrogen bonds to the DNA (5), but also in eukaryotic MSH3 and MSH6. Since the eukaryotic MSH2–MSH6 complex is involved in the recognition of mismatches and short IDLs and MSH2–MSH3 recognizes longer IDLs (1,2) these residues could play a role in mismatch recognition. Conserved residues are also seen making contacts to the DNA in the clamp domain. Of these, Lys 496 and Arg 500 are conserved in Taq MutS where they are involved in hydrogen bond formation to the DNA. Lys 496 is also conserved in eukaryotic MSH2, MSH3 and MSH6 and so may play an important role in DNA binding, while Arg 500, conserved in MSH3 and MSH6, may play some role in mismatch recognition.
Although several crystallographic studies have shown that the presence of a mismatch in DNA does not change its structure dramatically (20–22,29), mismatches destabilize DNA. This can be seen by the reduction in melting temperatures of DNA upon incorporation of a mismatch (30,31). A mismatch binding enzyme like MutS could be making use of this local weakening to detect the presence of mismatches and unpaired bases. The extensive DNA–protein interface in our complexes is suggestive of a mechanism in which regions of DNA ∼13–14 bp are scanned for the presence of a mismatch. That MutS binds homoduplex DNA with low affinity (24,32,33) and that it remains localized on the chromosomes in cells (34) suggests that it stays on DNA all the time, constantly scanning for mismatches.
A comparison of MutS–mismatch complexes with the MutS–unpaired T complex reveals only a few differences. The protein–DNA contacts are largely the same, with the exception of Arg 58 of the mismatch binding monomer adopting a different conformation in the MutS–unpaired T complex (Figs 1C, D and 2G). The main difference in DNA binding between the Taq MutS–unpaired T (Fig. 3C and D) and our E.coli MutS–unpaired T complex is the significant rearrangement in the Watson–Crick base pairs adjacent to the unpaired T in the E.coli MutS complex (Fig. 3A and B). In fact, the E.coli enzyme appears to recognize this as a G:T base pair with the unpaired T seemingly base paired to Gua 9. An interesting parallel in the difference between two similar enzymes binding the same substrate is seen in the structures of two methyltransferases in complex with DNA. While the structure of HaeIII methyltransferase-DNA complex (35) shows similar rearrangements of adjacent Watson–Crick base pairs upon recognition of the target cytosine, that of the HhaI methyltransferase–DNA complex (36) does not. Apparently such rearrangements are possible but not essential features of substrate binding by these enzymes. However, such rearrangements in DNA have so far only been observed in G:C base pairs (35,37). In both the HaeIII methyltransferase (35), and in our E.coli MutS–unpaired thymidine structures, the rearrangement involves G:C base pairs adjacent to the base being recognized (Fig. 3B). The HaeIII methyltransferase has only one G:C base pair being rearranged while our MutS structure has two (Fig. 3B). An explanation for the rearrangements occurring in G:C base pairs could be that there are more possibilities for creating new hydrogen bonds compared to A:T base pairs. So the energetically unfavourable rearrangement of the base pairs by the protein is at least partially compensated by the formation of new stabilizing hydrogen bonds. This can be seen in our MutS–unpaired T structure where the rearranged Gua 9:Cyt 21 and the Gua 10:Cyt 20 bp form an extensive network of hydrogen bonds, also involving the protein and the unpaired thymidine (Fig. 3B). Further, since mismatch binding by MutS is also known to be influenced by sequence context (32,38) the involvement of neighbouring Watson–Crick base pairs is significant. It suggests that the protein, in addition to kinking the DNA and rearranging the base pairs of the mismatch itself, may use the rearrangement of the adjacent base pairs in order to obtain the common binding mode.
CONCLUSION
We have shown that MutS binds to A:A, C:A and G:G mismatches by stacking Phe 36 over the purine and either keeping it or bringing it into the syn orientation to expose the N7 to Glu 38 for hydrogen bonding. In the G:T and unpaired T, MutS binds in such a way that the N3 of the thymidine forms this hydrogen bond. We have also shown that MutS rearranges the mismatched base pairs from their positions in unbound DNA to achieve this. This is indicative of a common mismatch binding mode for all mismatches.
In our structures, we see the protein interacting with 12 DNA base pairs other than the mismatch itself. This is suggestive of an ability of MutS to scan such regions of DNA, looking for mismatches. Thus, MutS uses the local weakening due to the mismatch to locate it and binds to it by rearranging the base pairs to the conformation defined by the common mismatch binding mode.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Hein te Riele and Niels de Wind for useful comments and Joyce Lebbink for critically reading the manuscript. They also thank the staff at the ESRF, Grenoble and the EMBL outstation at DESY, Hamburg for support during data collection. Funding was provided by NWO–CW and the Dutch Cancer Society (KWF). The coordinates of the structures have been deposited in the PDB with access codes 1OH6 (MutS–A:A), 1OH5 (MutS–C:A), 1OH7 (MutS–G:G) and 1OH8 (MutS–unpaired T).
REFERENCES
- 1.Buermeyer A.B., Deschenes,S.M., Baker,S.M. and Liskay,R.M. (1999) Mammalian DNA mismatch repair. Annu. Rev. Genet., 33, 533–564. [DOI] [PubMed] [Google Scholar]
- 2.Kolodner R.D. and Marsischky,G.T. (1999) Eukaryotic DNA mismatch repair. Curr. Opin. Genet. Dev., 9, 89–96. [DOI] [PubMed] [Google Scholar]
- 3.Modrich P. and Lahue,R. (1996) Mismatch repair in replication fidelity, genetic recombination and cancer biology. Annu. Rev. Biochem., 65, 101–133. [DOI] [PubMed] [Google Scholar]
- 4.Lynch H.T. and de la Chapelle,A. (1999) Genetic susceptibility to non-polyposis colorectal cancer. J. Med. Genet., 36, 801–818. [PMC free article] [PubMed] [Google Scholar]
- 5.Obmolova G., Ban,C., Hsieh,P. and Yang,W. (2000) Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature, 407, 703–710. [DOI] [PubMed] [Google Scholar]
- 6.Lamers M.H., Perrakis,A., Enzlin,J.H., Winterwerp,H.H., de Wind,N. and Sixma,T.K. (2000) The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature, 407, 711–717. [DOI] [PubMed] [Google Scholar]
- 7.Wu T.H. and Marinus,M.G. (1999) Deletion mutation analysis of the mutS gene in Escherichia coli. J. Biol. Chem., 274, 5948–5952. [DOI] [PubMed] [Google Scholar]
- 8.Budisa N., Steipe,B., Demange,P., Eckerskorn,C., Kellermann,J. and Huber,R. (1995) High-level biosynthetic substitution of methionine in proteins by its analogs 2-aminohexanoic acid, selenomethionine, telluromethionine and ethionine in Escherichia coli. Eur. J. Biochem., 230, 788–796. [DOI] [PubMed] [Google Scholar]
- 9.Otwinowski Z. and Minor,W. (1997) Processing of x-ray data collected in oscillation mode. In Carter,C.W. and Sweet,R.M. (eds), Methods in Enzymology, Macromolecular Crystallography Part A. Academic Press, New York, Vol. 276, pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 10.Murshudov G.N., Vagin,A.A, and Dodson,E.J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D, 53, 240–255. [DOI] [PubMed] [Google Scholar]
- 11.CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- 12.Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 13.Brunger A.T., Adams,P.D., Clore,G.M., DeLano,W.L., Gros,P., Grosse-Kunstleve,R.W., Jiang,J.S., Kuszewski,J., Nilges,M., Pannu,N.S., Read,R.J., Rice,L.M., Simonson,T. and Warren,G.L. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 14.Winn M.D., Isupov,M.N. and Murshudov,G.N. (2001) Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D, 57, 122–133. [DOI] [PubMed] [Google Scholar]
- 15.Lamzin V.S. and Wilson,K.S. (1993) Automated refinement of protein models. Acta Crystallogr. D, 49, 129–149. [DOI] [PubMed] [Google Scholar]
- 16.Hooft R.W., Vriend,G., Sander,C. and Abola,E.E. (1996) Errors in protein structures. Nature, 381, 272. [DOI] [PubMed] [Google Scholar]
- 17.Lu X.J., Shakked,Z. and Olson,W.K. (2000) A-DNA conformational motifs in ligand-bound double helices. J. Mol. Biol., 300, 819–840. [DOI] [PubMed] [Google Scholar]
- 18.Kraulis P.J. (1991) MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- 19.Merritt E.A. and Bacon,D.J. (1997) Raster3D photorealistic molecular graphics. In Carter,C.W. and Sweet,R.M. (eds), Methods in Enzymology, Macromolecular Crystallography Part B. Academic Press, New York, Vol. 277, pp. 505–524. [DOI] [PubMed] [Google Scholar]
- 20.Hunter W.N., Brown,T. and Kennard,O. (1987) Structural features and hydration of a dodecamer duplex containing two C:A mismatches. Nucleic Acids Res., 15, 6589–6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunter W.N., Brown,T., Kneale,G., Anand,N.N., Rabinovich,D. and Kennard,O. (1987) The structure of guanosine-thymidine mismatches in B-DNA at 2.5-A resolution. J. Biol. Chem., 262, 9962–9970. [DOI] [PubMed] [Google Scholar]
- 22.Skelly J.V., Edwards,K.J., Jenkins,T.C. and Neidle,S. (1993) Crystal structure of an oligonucleotide duplex containing G.G base pairs: influence of mispairing on DNA backbone conformation. Proc. Natl Acad. Sci. USA, 90, 804–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto A., Schofield,M.J., Biswas,I. and Hsieh,P. (2000) Requirement for Phe36 for DNA binding and mismatch repair by Escherichia coli MutS protein. Nucleic Acids Res., 28, 3564–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schofield M.J., Brownewell,F.E., Nayak,S., Du,C., Kool,E.T. and Hsieh,P. (2001) The Phe-X-Glu DNA binding motif of MutS. The role of hydrogen bonding in mismatch recognition. J. Biol. Chem., 276, 45505–45508. [DOI] [PubMed] [Google Scholar]
- 25.Borgstahl G.E., Williams,D.R. and Getzoff,E.D. (1995) 1.4 A structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site and chromophore. Biochemistry, 34, 6278–6287. [DOI] [PubMed] [Google Scholar]
- 26.Genick U.K., Soltis,S.M., Kuhn,P., Canestrelli,I.L. and Getzoff,E.D. (1998) Structure at 0.85 A resolution of an early protein photocycle intermediate. Nature, 392, 206–209. [DOI] [PubMed] [Google Scholar]
- 27.Luecke H., Schobert,B., Richter,H.T., Cartailler,J.P. and Lanyi,J.K. (1999) Structure of bacteriorhodopsin at 1.55 A resolution. J. Mol. Biol., 291, 899–911. [DOI] [PubMed] [Google Scholar]
- 28.Junop M.S., Yang,W., Funchain,P., Clendenin,W. and Miller,J.H. (2003) In vitro and in vivo studies of MutS, MutL and MutH mutants: correlation of mismatch repair and DNA recombination. DNA Repair (Amst), 2, 387–405. [DOI] [PubMed] [Google Scholar]
- 29.Hunter W.N., Brown,T., Anand,N.N. and Kennard,O. (1986) Structure of an adenine-cytosine base pair in DNA and its implications for mismatch repair. Nature, 320, 552–555. [DOI] [PubMed] [Google Scholar]
- 30.Peyret N., Seneviratne,P.A., Allawi,H.T. and SantaLucia,J.,Jr (1999) Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A,C.C,G.G and T.T mismatches. Biochemistry, 38, 3468–3477. [DOI] [PubMed] [Google Scholar]
- 31.Werntges H., Steger,G., Riesner,D. and Fritz,H.J. (1986) Mismatches in DNA double strands: thermodynamic parameters and their correlation to repair efficiencies. Nucleic Acids Res., 14, 3773–3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown J., Brown,T. and Fox,K.R. (2001) Affinity of mismatch-binding protein MutS for heteroduplexes containing different mismatches. Biochem. J., 354, 627–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su S.S., Lahue,R.S., Au,K.G. and Modrich,P. (1988) Mispair specificity of methyl-directed DNA mismatch correction in vitro. J. Biol. Chem., 263, 6829–6835. [PubMed] [Google Scholar]
- 34.Smith B.T., Grossman,A.D. and Walker,G.C. (2001) Visualization of mismatch repair in bacterial cells. Mol. Cell, 8, 1197–1206. [DOI] [PubMed] [Google Scholar]
- 35.Reinisch K.M., Chen,L., Verdine,G.L. and Lipscomb,W.N. (1995) The crystal structure of HaeIII methyltransferase convalently complexed to DNA: an extrahelical cytosine and rearranged base pairing. Cell, 82, 143–153. [DOI] [PubMed] [Google Scholar]
- 36.Klimasauskas S., Kumar,S., Roberts,R.J. and Cheng,X. (1994) HhaI methyltransferase flips its target base out of the DNA helix. Cell, 76, 357–369. [DOI] [PubMed] [Google Scholar]
- 37.Timsit Y., Vilbois,E. and Moras,D. (1991) Base-pairing shift in the major groove of (CA)n tracts by B-DNA crystal structures. Nature, 354, 167–170. [DOI] [PubMed] [Google Scholar]
- 38.Joshi A. and Rao,B.J. (2001) MutS recognition: multiple mismatches and sequence context effects. J. Biosci., 26, 595–606. [DOI] [PubMed] [Google Scholar]