Abstract
Recent developments of single molecule detection techniques and in particular the introduction of fluorescence correlation spectroscopy (FCS) led to a number of important applications in biological research. We present a unique approach for the gene expression analysis using dual-color cross-correlation. The expression assay is based on gene-specific hybridization of two dye-labeled DNA probes to a selected target gene. The counting of the dual-labeled molecules within the solution allows the quantification of the expressed gene copies in absolute numbers. As detection and analysis by FCS can be performed at the level of single molecules, there is no need for any type of amplification. We describe the gene expression assay and present data demonstrating the capacity of this novel technology. In order to prove the gene specificity, we performed experiments with gene-depleted total cDNA. The biological application was demonstrated by quantifying selected high, medium and low abundant genes in cDNA prepared from HL-60 cells.
INTRODUCTION
Gene expression profiling plays an important role in streamlining several critical steps in modern pharmaceutical development: disease diagnostics, new target identification, lead drug identification, and optimization and monitoring of clinical trials (1–4). Several powerful techniques have been developed for analyzing gene expression. The micro-array technique using high-density oligonucleotide chips (5,6) or cDNA arrays (7,8) enables simultaneous analysis of several thousands of genes. These DNA arrays are becoming essential tools for research and drug discovery. Without question, this technology offers an enormous scientific potential, but there are still some limitations with respect to the quality of the data. While both the oligonucleotide and cDNA arrays are reliable at identifying regulated genes, the degree of regulation is not accurate (9). In particular, results obtained upon corresponding measurements performed with either cDNA arrays or oligonucleotide chips showed a poor correlation (10). An accurate, reliable and reproducible determination will be difficult to achieve without signal optimization for each individual probe–target pair (11). So far, RT–PCR is the most sensitive and precise method for detection and quantification of mRNA molecules. However, it is a complex technique, and substantial problems are associated with the reproducibility and specificity (12–15). To address some of these problems, we describe a new method for gene expression analysis using the analysis of single molecule events (16–18). Due to the high sensitivity of this method, it does not require any amplification step. The gene expression assay is performed in solution and is based on the simultaneous hybridization of two dye-labeled DNA probes to a selected gene target. The encounter frequency of double-labeled targets in the detection volume element is determined using dual-color fluorescence cross-correlation (19–22) and event analysis. The detection of non-amplified genomic DNA by hybridization with dye-labeled DNA probes using flow detection and dual-color coincidence analysis has been reported previously (23). We now describe a gene expression assay that allows the absolute quantification of gene copies in a complex biological sample. This determination (copies per µg cDNA and/or mRNA) is calculated from the linear regression of a simultaneously generated calibration curve (standard curve), as gene-specific differences in probe binding efficiency have to be taken into account. The gene-specific DNA probes are designed and selected with respect to their hybridization properties as well as their gene specificity to result in high accuracy and specificity. The quantification assay including all the components required within this process will be described in detail. The expression levels of selected high, medium and low abundant genes were determined in cDNA prepared from HL-60 cells.
MATERIALS AND METHODS
cDNA preparation of biological samples
The two cell lines K562 and HL-60, a human chronic myeloid leukemia and a human acute myeloid leukemia cell line, respectively, were cultured in RPMI 1640 medium with 10% fetal calf serum. Total RNA was extracted using the RNeasy maxi kit (Qiagen, Hilden, Germany). mRNA was isolated using the mRNA purification kit (Amersham Biosciences Europe GmbH, Dübendorf, Germany). Conversion of mRNA to cDNA was accomplished with M-MLV reverse transcriptase (Invitrogen, Groningen, The Netherlands). cDNA was purified over nucleospin extract columns (Macherey-Nagel, Düren, Germany). The quality (integrity) of the total RNA, mRNA and cDNA was analyzed with a Bioanalyzer 2100 using the RNA 6000 Nano Lab-Chip kit (Agilent Technologies, Basel, Switzerland). The cDNA/RNA samples were quantified via a UV spectrophotometer and stored at –80°C (RNA) and –20°C (cDNA), respectively.
Genes to be analyzed
The following genes were quantified in cDNA prepared from HL-60 cells: human β-actin (BC009275, β-actin); human transcription elongation factor 1-α 1 (L41490, EL-1); human S19 ribosomal protein (M81757, S19); human tubulin-α (K00558, tuba); human DNA-binding protein A variant (X95325, DNAbp), mRNA encoding phosphoglycerate kinase (V00572, PGK1), human GTP-binding protein (M28209, RAB1) and human transcription factor NF-κB p65 subunit (L19067, p65). The codes are used as abbreviations for each gene within this manuscript.
Preparation of gene-specific DNA
In general, gene-specific single-stranded (ss) DNA was prepared in two steps. First, a PCR amplification using 0.5 U of Taq DNA polymerase (Roche Diagnostics, Rotkreuz, Switzerland) in K562 total cDNA (1–10 ng) with a gene-specific 5′-biotinylated forward and an unmodified reverse DNA primer was performed. The PCR product was attached to streptavidin-coated superparamagnetic polystyrene beads (Dynal Biotech, Hamburg, Germany) and washed several times. Finally, gene-specific ssDNA was obtained by elution of the non-biotinylated DNA strand from the beads according to the manufacturer’s instructions. The samples were analyzed with the Bioanalyzer 2100 using the RNA 6000 Nano Lab-Chip kit. The primer sequences used were: L19067 (nt 1477–2424), 5′-bio-TTTTGATGGAGTACCCTGAGGCTATAA, 5′-AGCTTGGCAACAGCTTTATTAGTT; M28209 (nt 115– 621), 5′-bio-TTTGGAAAGTCTTGCCTTCTTCTTAGG, 5′-TAACATTGGACTTCTCAGCACCAC; X95325 (nt 580–1057), 5′-bio-TTTCGCAGTGTAGGAGATGGAGAA, 5′-GGTACCTTGGGCGGTAAGTT; V00572 (nt 720–1689), 5′-bio-TTTGAGCTAAAGTTGCAGACAAGATCC, 5′-CATGCTGAGTAGTGAAACAGTGAC; M81757 (nt 24–425), 5′-bio-TTTTGCCTGGAGTTACTGTAAAAGACG, 5′-CGATTCTGTCCAGATCTCTTTGTC; K00558 (nt 607–1553), 5′-bio-TTTGTAGTTGAGCCCTACAACTCCATC, 5′-CAGGTACACATGGAAAAGACATGA; L41490 (nt 932–1962), 5′-bio-TTTGGAAATTGTTAAGGAAGTCAGCAC, 5′-CCTTTCCTTCTGAAAGGTTTTACG; and BC009275 (nt 88–2252), 5′-bio-TCATGTCTTCCCCTCCATCGTGGGG, 5′-CTGGTCTCAAGTCAGTGTACAGG.
Depletion of β-actin cDNA in total cDNA
For the depletion of β-actin cDNA in total cDNA, 5′ biotin-tagged ssDNA complementary to nt 88–413 of β-actin cDNA was prepared. A 1 µM concentration of primer P68 (5′-TCATGTCTTCCCCTCCATCGTGGGG) was added to a 13 nM β-actin DNA fragment (nt 1–413) in 1× Dynazyme reaction buffer, 200 nM dNTP, 2.5% (v/v) dimethylsulfoxide (DMSO) and 0.8 U of Dynazyme EXT (Finnzymes, Espoo, Finland) in a final volume of 50 µl and subjected to 50 reaction cycles: 96°C for 20 s, 60°C for 45 s, 72°C for 2 min. After purification on nucleospin extract columns (Macherey-Nagel, Düren, Germany), this ss cDNA was added to total cDNA (76 ng/µl final concentration) at a final concentration of 5 nM in 6× SSC buffer. Hybridization was performed by heating at 96°C for 2 min, and then by stepwise reduction of the temperature with 10 min for each step: 70, 60, 50, 40 and 30°C, on ice. Finally, the biotin-tagged complex was removed by addition of 1 µg of streptavidin-coated paramagnetic beads (Promega, Wallisellen, Switzerland) and incubation at room temperature for 10 min with mixing. The beads were trapped with a magnet and the supernatant corresponding to β-actin-depleted total cDNA was recovered.
Probe design and probe synthesis
Gene-specific DNA probes (probes) were designed by iSenseIt AG (Bremen, Germany). The designed probes (40–45 nt) were synthesized by Thermohybaid (Ulm, Germany). Each probe contains either a green (Rhodamine green) or a red label (Bodipy 630) at the 5′ end. For some green-labeled probes, low c.p.m. (counts per molecule) values were determined. In order to increase the c.p.m. value, these probes were re-synthesized with an additional rhodamine green label at the 3′ end. The purity of the probes was analyzed by reverse-phase HPLC (column 214 TP1010, Dionex AG, Olten, Switzerland) and determined to be >95%.
96-well microstructures
Measurements were performed in 96-well microstructures (microplates, 24.0 × 60.0 mm) made of PMMA according to specifications by Gnothis AB (Kista, Sweden). The microstructures have a well diameter of 1.4 mm and a total well volume of 1.8 µl (optimized for 1 µl sample volume). The microstructures for the 100 nl hybridization experiments have a well diameter of 0.5 mm and a total well volume of 234 nl. The wells have a thin window (PMMA) of 170 µm that enables optical measurements with confocal microscopy. The plates are sealed with a commercial PCR lid and are thermostable up to 75°C.
Denaturation and hybridization
Hybridization experiments were performed in a buffered system [6× SSC, 0.06% (v/v) NP-40]. For low and medium abundant genes, 0.1 nM, and for high abundant genes, 2 nM final probe concentrations were used for titration and quantification. Gene-specific ss DNA in various concentrations (0.001–1 nM) was used for the generation of the calibration curve. For the quantification, 100 and 200 ng of total cDNA were used for each experiment, respectively. The hybridization mixture was heated at 75°C for 4 min in order to denature the target cDNA, then kept at 60°C for 2 h for high abundant genes and 16 h for medium and low abundant genes.
Definition of controls
All hybridization samples were measured in triplicate (three wells per microstructure). Additionally, the following control samples were included: (i) set-up alignment controls: Cy5-dUTP (Amersham Pharmacia Bioscience Europe GmbH, Dübendorf, Germany), rhodamine green (Molecular Probes, Leiden, The Netherlands), dual-labeled PCR product; (ii) background controls: hybridization buffer, probe pair, gene-specific DNA, total cDNA; (iii) hybridization control: hybridization sample (2 nM probe pair, 1 nM gene-specific DNA).
Measurement
The samples were measured with a ConfoCor2 instrument (Carl Zeiss, Jena, Germany), which allows simultaneous excitation at 488 and 633 nm (Argon and HeNe laser, respectively) and the detection of the emission in the green (band-pass filter 505–550 nm) and red (long-pass filter 650 nm) wavelength range by avalanche photodiodes. Optics Neofluar 40× NA 1.2 water immersion generating a volume element of ∼0.4 fL with the half-axes ωx,y 0.25 µm and ωz = 1 mm for the green laser, and ∼0.8 fL with the half-axes ωx,y 0.32 µm and ωz = 1.3 mm for the red laser. The two excitation intensities, the pinhole sizes and positions have been optimized for the highest c.p.m. using the labeled nucleotides rhodamine green-dUTP and Cy5-dCTP. A double dichroic mirror reflecting and transmitting at 488 and 633 was used. The excitation intensities are ∼100 µW for the argon laser and ∼200 µW for the HeNe laser (at this level, differences in excitation intensity do not influence the correlation amplitude). The pinhole size is 50 µm for the green and 60 µm for the red detection channel. Thereby, the number of molecules occupying the triplet state (non-fluorescent dark states) was minimized and a maximal detection yield obtained. Intensity fluctuation traces were obtained from the freely diffusing molecules. The cross- and auto-correlation curves were calculated on-line and the recorded correlation curves were evaluated with a non-linear least square-fitting algorithm (Marquardt). Measurement times were 60 s for 2 nM probe concentrations and 240 s for 0.1 nM probe concentrations. In order to apply a measurement temperature of 50°C, the microstructures were placed into a temperature-controlled holder, where temperature was applied from the top via a Peltier element. To ensure a stable temperature, the objective was also heated up by winding a plastic tube around it and pumping tempered water through the tube.
Data analysis
Cross-correlation analysis. Fluorescence correlation spectroscopy (FCS) allows the analysis of molecular interactions and molecular complexes at the level of single molecules (16,18). The cross-correlated emission in two intensities gives a positive result for stochastic processes such as Brownian motion only if they are linked in time and space. Therefore, solely DNA sequences with two complementary probes hybridized to it and emitting at two different wavelengths simultaneously contribute to the cross-correlation. The non-hybridized probes cannot be observed since motions of non-hybridized probes are uncorrelated in time (20,21). Let Ir(t) and Ig(t) be the intensities in red and green wavelengths, and δIr(t) and δIg(t) represent the deviations from their means. The normalized auto-correlation and cross-correlation functions of two intensity deviations are then calculated by
and
where the brackets denote time average and τ the time delay. It has been shown [e.g. Rigler et al. (21)] that the average number of red and green molecules in an illuminated volume can be calculated from the auto-correlation as
and
The number of molecules hybridized with both primers can be calculated with cross-correlation function as
Ngr = Ng · Nr · Ggr (0) 6
Since the probe concentration is constant in both the calibration curve experiment and the quantification experiment, the cross-correlation amplitude Ggr (0) is proportional to Ngr. The background in Ggr is caused by cross-talking of photons from the green primer into the red channel, which leads to a false correlation signal. Its magnitude is related to the number of green primer molecules Ng and a cross-talk factor q, which is in the range of 1–2%. Since the correlation time (diffusion time) of the probes is different from the probe–target complex, this false-positive signal can be separated from the real correlation signal and the background can be accounted for (21).
Event analysis. In addition to cross-correlation analysis, event analysis was used. This method allows the detection and counting of each single molecule with either a red or green fluorescence label that passed the observation volume. The encounter frequency relates directly to the concentration of those molecules (17). The use of a low concentration of fluorescent molecules avoids getting more than one fluorescent particle at any instant in the observation volume. A 1 nM solution of fluorescent molecules corresponds to approximately N = 0.1 molecules in the observation volume. The event analysis was performed in two steps. First the events were identified and, secondly, true events were distinguished and counted. Events were identified from the green fluorescence by a threshold. Each time the intensity integrated over a characteristic time frame was 10 times higher than the corresponding level of background noise, an event was identified. The selected time frame of 0.9 ms corresponds to the time it takes for a DNA molecule of interest to traverse the observation volume. The events, which included a short trace from the green as well as the red fluorescence intensity, were then subjected to characterization by pattern recognition. Each event constituted of 18 points since the intensity was sampled with a bin time of 0.1 ms. Principal components analysis was used to distinguish the fluorescence intensity pattern of true events from that of false events (caused by molecules with only green fluorescence). The two components selected for the projection of all events were those that allowed the separation of true from false events at an optimal level. Inside an area that corresponds to true events, all events had intensities that were 10-fold higher than the background intensity for the green as well as the red fluorescence. Thus, only molecules hybridized to both a green and a red fluorescent probe can cause true events.
RESULTS
Principle of the method
The gene expression measurement is performed in 96-well microstructures. Each well (optimized for 1 µl sample volume) of the microstructure contains a hybridization sample comprised of biological sample solution (cDNA) and two gene-specific DNA probes (labeled with either a green or a red dye, respectively), which were pre-assayed. The probes hybridize under controlled conditions to their target gene and the number of double-labeled molecules is determined (Fig. 1). In order to allow for an absolute quantification, a calibration curve (standard curve) is constructed for each selected gene. The calibration curve is generated by plotting the number of double-labeled target molecules against the defined amount of gene-specific ssDNA. Finally, the copy number of the target gene cDNA within the biological sample can be calculated from the linear regression of the calibration curve.
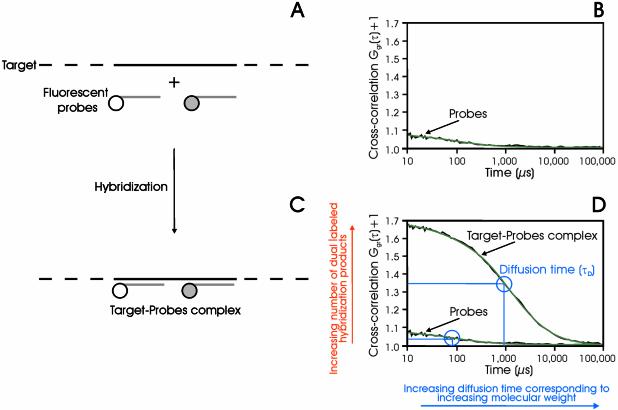
Figure 1.
Schematic diagram of the dual-color cross-correlation assay. Two differently dye-labeled DNA probes hybridize simultaneously to their respective binding side on the same target molecule (A and C). In order to be detected by cross-correlation spectroscopy, the motions of the red and the green dyes have to be linked in time and space. Non-hybridized probes as well as targets with one attached probe only cause a background (B), while the dual-labeled target generates a cross-correlation curve (D, black, measured curve; green, fitted curve). The diffusion time τD corresponds to the occupation time of the labeled molecules in the detection volume. The molecular weight of the target–probe complex is significantly higher than the probe molecule itself. Therefore, the target–probe complex has a slower Brownian motion, which consequently results in a longer diffusion time, τD (the curve is shifted to the right). The amplitude Ggr(τ) of the correlation function is related to the amount of hybridization products. At a constant probe concentration, the amplitude increases with increasing amounts of target concentration. The background signal itself in (C) and (D) is caused by cross-talk (see data analysis).
Probe design
Two main objectives are important for the probe design. In order to achieve an optimal sensitivity of the assay, the probes need to have good hybridization properties; furthermore, to obtain an accurate quantification, the probes have to be gene specific. The hybridization of oligonucleotides to complementary DNA or RNA is complicated by the presence of secondary and tertiary structures within the target, as even at higher temperatures (50–60°C) this folding still exists and therefore has a major impact for the probe design. The probe design is performed by iSenseIt AG (www.isenseit.de) as a service (iQserv). Based on the Vienna RNA package (24,25), a prediction of the secondary structure of RNA and DNA molecules is possible. Including secondary structure effects as well as thermodynamic parameters, the probes were designed with respect to hybridization efficiency and specificity (26,27). The design process already considers the experimental parameters (salt concentration, hybridization temperature, probe concentration). The sequences are located within the first 1000 bases of the cDNA (5′ end), which corresponds to the region close to the poly(A) tract of the mRNA. The distance between the red and green probes was selected to be between 50 and 400 nt. For the initial experiments, the length of the probes was selected to be 25 nt. Later on, due to difficulties in finding gene-specific probes for certain genes, the probe length was increased to 40–45 nt, which led to an adjustment of the hybridization conditions.
Probe pre-assay
For each gene to be quantified, gene-specific probes were synthesized in dye-labeled form. The probe design was based on a prediction of the secondary structure of cDNA. As the two probes had to hybridize simultaneously to the same target, the hybridization properties of each probe within a two-probe target complex had to be evaluated. This test was performed on gene-specific ssDNA. The binding efficiency of each of the two probes depends on experimental conditions (probe/target ratio, hybridization temperature, salt concentration), secondary structure of the cDNA target and its chosen counterpart (second probe). The experimental conditions (see Materials and Methods) were considered in the initial probe design. As the formation of the secondary structure of a selected cDNA molecule is to a certain degree length dependent, the gene-specific ssDNA was designed to have properties similar to the cDNA to be quantified in the biological sample. The ssDNA fragment was prepared by PCR amplification of specific cDNA followed by removal of the complementary DNA strand via attachment to magnetic beads (for details, see Materials and Methods). The length and the integrity of the DNA fragments were confirmed in a quality control by electrophoretic analysis (Fig. 2), and selected samples have been verified by sequencing. Each probe pair combination (one red- and one green-labeled probe at a concentration of 2 nM each) was hybridized to gene-specific ssDNA (at a concentration of 1 nM). The experiments were performed in 6× SSC, 0.06% (v/v) NP-40 buffer for 2 h in a volume of 1 µl. It was very important to heat up the DNA fragment and later on the total cDNA (biological sample) for at least 4 min at 75°C in order to achieve a complete denaturation. After evaluation of the hybridization properties of all the possible probe pair combinations, the best probe pair was selected (highest Ngr value). Figure 1D shows a cross-correlation curve obtained by the hybridization of a probe pair to PGK1 ssDNA. The amplitude (Ngr value; equation 6, see Materials and Methods) correlates with the number of the double-labeled DNA targets, while the diffusion time τD is related to the size of the hybridization product.
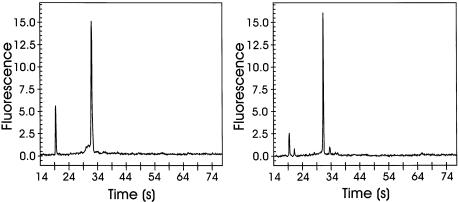
Figure 2.
Electropherograms of gene-specific ssDNA of EL-1 (left panel) and p65 (right panel) were obtained using the Bioanalzyer 2100 (Agilent). The fluorescence signals are generated by dye intercalation into DNA (RNA 6000 Nano LabChip kit, Agilent; for conditions, see Materials and Methods). Theoretical lengths of ssDNA fragments were 1031 nt for EL-1 and 948 nt for p65, respectively. Measured length indicates 1115 nt for EL-1 and 942 nt for p65, respectively.
Sensitivity of the assay
The sensitivity and the accuracy of the assay are determined by several factors, namely the quantum yield of the dyes, the probe concentration, and the binding efficiency and the calibration of the set-up. The quantum yield of a dye is influenced by the sequence of the attached oligonucleotide. Therefore, the c.p.m. values of all probes were determined. Significant differences were observed for the green-labeled probes, where the c.p.m. values varied between 4 and 30 kHz and therefore are lower than the c.p.m. values of the red-labeled probes (70–90 kHz). This reduced quantum yield is caused by quenching of rhodamine green via interaction with DNA (28). In order to increase the quantum yield (>20 kHz), several probes were re-synthesized with an additional rhodamine green label at the 3′ end. The quantification of the target molecules requires the probes to be added in excess in order to obtain a target saturation. As all the fluorescence measured by either one of the detectors contributes to the dominator of correlation function (see equation 3, Materials and Methods), the probe excess has certain limitations and has to be adjusted to the expression level of the selected gene. A sufficient signal upon measurement requires at least 1% of the probe to be present in target-hybridized form (21,29). Taking measurement time, background noise conditions and the given quantum yield into account, simulation studies led to an optimal probe concentration of 50–100 pM, which was confirmed experimentally. Whereas for a 2 nM mono-labeled probe concentration, a sensitivity of 100–200 pM target concentration was observed, the introduction of a second green label and the decrease of the probe concentration to 100 pM increased the sensitivity below 5 pM (Fig. 3). The use of a 5′,3′-bi-labeled red probe did not lead to any further improvement of the sensitivity. While applying event analysis to measurements performed with hybridization samples comprised of 100 pM probe concentrations, the sensitivity could be improved further down to 1 pM.
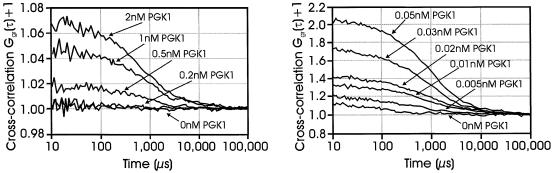
Figure 3.
Comparison of cross-correlation obtained using either mono- or bi-labeled green probes. Left panel: 5′-mono-labeled green and 5′-red-labeled probe at 2.0 nM with PGK1 ssDNA at concentrations of 2, 1, 0.5 and 0.2 nM. Right panel: 5′,3′-bi-labeled green and 5′-red-labeled probe at 0.1 nM with PGK1 ssDNA concentrations of 0.05, 0.03, 0.02, 0.01 and 0.005 nM. For conditions, see Materials and Methods. Although the bi-labeled probe causes a higher background amplitude (compared with the mono-labeled probe), the sensitivity is increased. This background (0 nm PGK1) is caused by cross-talk, which is due to the fact that the emission spectrum of rhodamine green upon excitation at 488 nm is extending into the detection range of the red detector.
Generation of the calibration curve
In order to achieve an absolute and accurate quantification, a calibration curve was measured for each selected gene. At a given probe concentration, different amounts of gene-specific ssDNA were hybridized to the selected probe pair. Figure 3 (right panel) shows cross-correlation curves obtained for PGK1 ssDNA. Since the y-axis value (Ngr) is proportional to the target concentration, these values are plotted resulting in a calibration curve (Fig. 4), which was used as standard curve for the evaluation of the absolute number of gene copies (quantification).
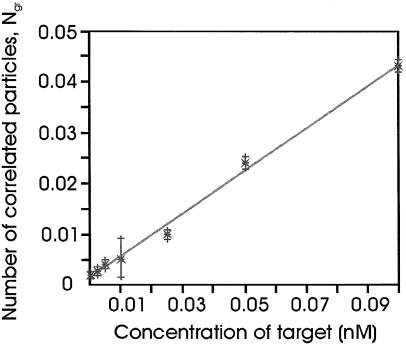
Figure 4.
Calibration curve. The particle numbers (Ngr) are a function of the gene-specific PGK1; ssDNA concentration (0.005–0.1 nM). Hybrid ization conditions: 0.1 nM probe; 16 h at 60°C. Measurement: 240 s at 50°C. Each data point is the average of three independent measurements. The error bars illustrate the deviation in Ngr based on the 95% confidence interval. The slope of the calibration curve is a direct measure for the binding efficiency of the probe pair to the specific gene under the given experimental conditions.
Gene quantification in HL-60 cDNA
For each gene to be quantified, a gene-specific calibration curve was generated. In order to determine the concentration of selected genes within a biological sample, the hybridization experiments were performed under identical conditions to those described for the generation of the calibration curve, whereby the gene-specific DNA was replaced by total cDNA (100 and 200 ng each) of HL-60 cells. Using the particle numbers (Ngr) obtained in these experiments, the concentration of cDNA molecules within the biological sample was calculated from linear regression of the calibration curve. As the quantifications are performed with either 100 or 200 ng biological sample, for each gene two concentration values were obtained. Finally, the mean value was converted into copies/µg of cDNA (Table 1). With the above-described gene expression assay, we quantified various high, medium and low abundant genes. For high abundant genes, typical absolute numbers of (4.3 ± 1.3) × 109 for S19, (0.7 ± 0.3) × 109 for tuba and (5.4 ± 1.3) × 109 for EL-1 were obtained. The calculations of the expression levels are based on a 95% confidence interval. For the medium and low abundant genes, the following results could be achieved: (2.4 ± 0.7) × 108 for DNAbp, (2.4 ± 1.1) × 108 for RAB1, (0.57 ± 0.05) × 108 for PGK1 and (1.3 ± 0.8) × 107 for p65. The uncertainties are caused by systematic as well as measurement errors. In Table 1, the mean values and the corresponding SDs for the selected genes are listed. These data indicate clearly that the dynamic range of detection exceeds three orders of magnitude (107–1010 copies/µg cDNA).
Table 1. Absolute quantification of selected genes in HL-60 cDNA.
| Accession no. | Gene | Gene concentrationa | Gene copies/µg total cDNAb |
|---|---|---|---|
| BC009275 | β-Actin | 1330 ± 260 pM | (8.0 ± 1.6) × 109 |
| L41490 | EL-1 | 810 ± 170 pM | (4.9 ± 1.0) × 109 |
| M81757 | S19 | 660 ± 70 pM | (4.0 ± 0.4) × 109 |
| K00558 | tuba | 115 ± 30 pM | (7.0 ± 1.7) × 108 |
| M28209 | RAB1 | 50 ± 13 pM | (3.0 ± 0.8) × 108 |
| X95325 | DNAbp | 38 ± 3.3 pM | (2.3 ± 0.2) × 108 |
| V00572 | PGK1 | 12.8 ± 3.3 pM | (0.8 ± 0.2) × 108 |
| L19067 | p65 | 2.1 ± 1.3 pM | (1.3 ± 0.8) × 107 |
aMean ± SD in 1 µl assay volume using 100 and 200 ng of HL-60 cDNA, respectively. The concentration obtained for 200 ng was divided by 2.
bMean ± SD.
Gene specificity of assay
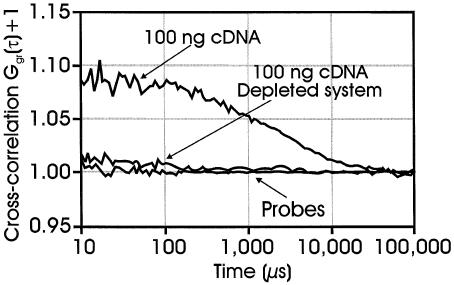
All probes used in our assays were designed with respect to their hybridization properties and their gene specificity (Thermodynamic alignment, iSenseIt AG). Additionally, all measurements were performed under temperature-controlled conditions avoiding unspecific interactions. As the detection technology is based on simultaneous translation of two dye molecules through the detection volume element, which in the case of the presented gene expression analysis only occurs upon hybridization of two probes to the same gene target, a high specificity is inherently given by the method. To obtain a false-positive signal, both probes have to hybridize non-specifically to the same cDNA molecule. We addressed the gene specificity by preparing a β-actin-depleted total cDNA sample. ssDNA complementary to a fragment of the β-actin cDNA was added to the sample cDNA solution. Upon hybridization and attachment of the formed duplex onto streptavidin-coated beads, >95% of the initial amount of β-actin cDNA (determined by PCR analysis, data not shown) was removed from the sample (for details, see Materials and Methods). In the following, we tested the specificity of the interaction using this depleted sample. Figure 5 presents a comparison between the β-actin hybridization measurements of the original, the depleted and the background sample, respectively. The cross-correlation signal for the depleted sample is slightly above the background contributed by the cross-talk of the non-hybridized probes. The signal corresponds to <10% of original sample and therefore confirms the results obtained by PCR analysis. These findings clearly demonstrate the specificity of this method.
Figure 5.
Cross-correlation curves obtained upon hybridization of two gene-specific actin probes to either 100 ng of total cDNA or 100 ng of β-actin-depleted total cDNA. For conditions, see Materials and Methods.
DISCUSSION
We have developed a new method for gene expression analysis and presented data demonstrating that gene quantification in absolute numbers is possible without any amplification of either the sample or the detected signal. The fluctuation analysis allows the direct determination of the number of labeled genes provided they carry both labels (probes) simultaneously. This property is evaluated using the calibration curve, as the measured number of dual-labeled gene molecules can be compared with the real number (defined amount of target). The calibration curve provides a quantitative measure for the efficiency of the probe–target binding which can be below 100% due to a reduced photon emission of one probe and/or a reduced probe binding as the folding of the target nucleic acids imposes substantial penalties on the hybridization. Using the calibration curve, correction factors can be obtained and a quantitative evaluation is possible. The feasibility to detect non-amplified genomic DNA at the level of single genes by double-color DNA probes has been reported previously by Castro and Williamson (23). As there is no information about the binding efficiency of the probes available, a quantitative assessment is not possible. In our assay, the molecular motion of a double-labeled target complex using fluorescence cross-correlation spectroscopy (20,21) is analyzed and a quantification range of at least three orders of magnitude is reached. The analysis is independent of the target size or the probe positioning and, in addition, is providing information on the gene size. Due to stochastic motion, only the double-labeled target is observable but not the uncorrelated probe background even in large excess (21). The RT–PCR also allows the quantification of selected genes in absolute numbers. While the PCR concept is based on determination of Ct values (the fluorescence signal reaches a point where it is statistically significant above background) (30), our assay determines Ngr values (number of double-labeled molecules). In both cases, these numbers are translated into target copies with the help of a measured standard curve. The accurate generation of the calibration curve requires the preparation of pure gene-specific DNA fragments. A high-quality preparation followed by a precise quantification is extremely important. In the case of RT–PCR assays, the generation of a standard curve has to be repeated for every new quantification to ensure accurate reverse transcription and amplifications profiles (15). A clear advantage of the presented new technology can be seen here. In the future, the generation of a calibration curve will be performed only once for each specific gene. Later on, any determination of the copy number per µg of cDNA can be based on this calibration curve by simultaneous measurements of a very limited number of defined controls. The most direct and therefore accurate way would be the determination of the gene copies by counting mRNA molecules within a biological sample. Any additional treatment of the RNA, such as reverse transcription, increases the probability of obtaining incorrect or inaccurate results. Our technology does not require any modifications (labeling, etc.) of the biological targets. Moreover, we are currently developing a direct quantification assay of gene expression levels on mRNA. In the present stage, 106 cells are required in order to detect low abundant genes within a measurement volume of 1 µl. As no amplification is involved in the whole procedure, the next consequence of reducing the sample amount is a decrease in the hybridization volume. For this purpose, microstructures with a well volume of 234 nl (see Materials and Methods) were fabricated. Except for the well volume, these microstructures have identical characteristics (compared with 1 µl microstructures), which allowed us to use the same instrument and measurement set-up. The concentration of all the reagents could be kept at the same highest possible level, which is crucial for the reaction kinetics. Calibration curves using a 100 nl sample volume have already been successfully measured, resulting in a required sample amount of 105 cells per well in order to quantify low abundant genes. By a further reduction of the measurement volume, sample amounts down to 104 cells per well seem to be possible.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank PerkinElmer LifeScience (Boston, USA) for the gift of HL-60 cells, and Klara Båth, Michel Bernard, Jean-François Cajot, Martine Damond, Thomas Hug, Taurai Tasara, Stefan Wennmalm and Nathalie Simon-Vermot for technical assistance.
REFERENCES
- 1.Clarke P.A, te Poele,R., Wooster,R. and Workman,P. (2001) Gene expression microarray analysis in cancer biology, pharmacology and drug development: progress and potential. Biochem. Pharmacol., 62, 1311–1336. [DOI] [PubMed] [Google Scholar]
- 2.Debouck C. and Goodfellow,P.N. (1999) DNA microarrays in drug discovery and development. Nature Genet., 21, 48–50. [DOI] [PubMed] [Google Scholar]
- 3.Mancinelli L., Cronin,M. and Sadée,W. (2000) Pharmacogenomics: the promise of personalized medicine. AAPS PharmSci., 2, E4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dyer M.R., Cohen,D. and Herrling,P.L. (1999) Functional genomics: from genes to new therapies. Drug Discov. Today, 4, 109–114. [DOI] [PubMed] [Google Scholar]
- 5.Lockhart D.J., Dong,H., Byrne,M.C., Follettie,M.T., Gallo,M.V., Chee,M.S., Mittmann,M., Wang,C., Kobayashi,M., Horton,H. and Brown,E.L. (1996) Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol., 14, 1675–1680. [DOI] [PubMed] [Google Scholar]
- 6.Chee M., Yang,R., Hubbell,E., Berno,A., Huang,X.C., Stern,D., Winkler,J., Lockhart,D.J., Morris,M.S. and Fodor,S.P. (1996) Accessing genetic information with high-density DNA arrays. Science, 274, 610–614. [DOI] [PubMed] [Google Scholar]
- 7.Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470. [DOI] [PubMed] [Google Scholar]
- 8.Duggan D.J, Bittner,M., Chen,Y., Meltzer,P. and Trent,J.M. (1999) Expression profiling using cDNA microarrays. Nature Genet., 21, 10–14. [DOI] [PubMed] [Google Scholar]
- 9.Yuen T., Wurmbach,E., Pfeffer,R.L., Ebersole,B.J. and Sealfon,S.C. (2002) Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res., 30, e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuo W.P., Jenssen,T.K., Butte,A.J., Ohno-Machado,L. and Kohane,I.S. (2002) Analysis of matched mRNA measurements from two different microarray technologies. Bioinformatics, 18, 405–412. [DOI] [PubMed] [Google Scholar]
- 11.Medh R.D. (2002) Microarray-based expression profiling of normal and malignant immune cells. Endocr. Rev., 23, 393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freeman W.M., Walker,S.J. and Vrana,K.E. (1999) Quantitative RT–PCR: pitfalls and potential. Biotechniques, 26, 112–115. [DOI] [PubMed] [Google Scholar]
- 13.Keilholz U., Willhauck,M., Rimoldi,D., Brasseur,F., Dummer,W., Rass,K., de Vries,T., Blaheta,J., Voit,C., Lethe,B. and Burchill,S. (1999) Reliability of reverse transcription–polymerase chain reaction (RT–PCR)-based assays for the detection of circulating tumour cells: a quality-assurance initiative of the EORTC Melanoma Cooperative Group. Eur. J. Cancer, 34, 750–753. [DOI] [PubMed] [Google Scholar]
- 14.Bishop G.A, Rokahr,K.L., Lowes,M., McGuinness,P.H., Napoli,J., DeCruz,D.J., Wong,W.Y. and McCaughan,G.W. (1997) Quantitative reverse transcriptase–PCR amplification of cytokine mRNA in liver biopsy specimens using a non-competitive method. Immunol. Cell Biol., 75, 142–147. [DOI] [PubMed] [Google Scholar]
- 15.Bustin S.A. (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol., 25, 169–193. [DOI] [PubMed] [Google Scholar]
- 16.Rigler R., Mets,U., Widengren,J. and Kask,P. (1993) Fluorescence correlation spectroscopy with high count rate and low background: analysis of translational diffusion. J. Eur. Biophys., 22, 169–175. [Google Scholar]
- 17.Eigen M. and Rigler,R. (1994) Sorting single molecules: application to diagnostics and evolutionary biotechnology. Proc. Natl Acad. Sci. USA, 91, 5740–5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rigler R. and Elson,E.L. (2000) Fluorescence Correlation Spectroscopy. Theory and Applications. Springer, Berlin, Germany. [Google Scholar]
- 19.Kinjo M. and Rigler,R. (1995) Ultra sensitive hybridization analysis using fluorescence correlation spectroscopy. Nucleic Acids Res., 10, 1795–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwille P., Meyer-Almes,F.-J. and Rigler,R. (1997) Dual-color fluorescence correlation spectroscopy for multicomponent diffusional analysis in solution. J. Biophys., 72, 1878–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rigler R., Foldes-Papp,Z., Meyer-Almes,F.J., Sammet,C., Volcker,M. and Schnetz,A. (1998) Fluorescence cross-correlation: a new concept for polymerase chain reaction. J. Biotechnol., 63, 97–109. [DOI] [PubMed] [Google Scholar]
- 22.Kettling U., Koltermann,A., Schwille,P. and Eigen,M. (1998) Real-time enzyme kinetics monitored by dual-color fluorescence cross-correlation spectroscopy. Proc. Natl Acad. Sci. USA, 95, 1416–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castro A. and Williams,J.G.K. (1997) Single molecule detection of specific nucleic acid sequences in unamplified genomic DNA. Anal. Chem., 69, 3915–3920. [DOI] [PubMed] [Google Scholar]
- 24.Hofacker I.L., Fekete,M. and Stadler,P.F. (2002) Secondary structure prediction for aligned RNA sequences. J. Mol. Biol., 319, 1059–1066. [DOI] [PubMed] [Google Scholar]
- 25.Fontana W., Stadler,P.F., Bornberg-Bauer,E.G., Griesmacher,T., Hofacker,I.L., Tacker,M., Tarazona,P., Weinberger,E.D. and Schuster,P. (1993) RNA folding and combinatory landscapes. Phys. Rev., 47, 2083–2099. [DOI] [PubMed] [Google Scholar]
- 26.Volkmann G., Wewetzer,D. and Trutnau,H.H. (2003) Integrierte Software-Plattform für Design und Analyse von DNA-Mikroarrays. Transcript, 9, 58–59. [Google Scholar]
- 27.Trutnau H.H., Nölte,M., Volkmann,G., Drutschmann,D. and Blohm,D. (2003) Genotypisierung von Hepatitis-C-Viren mittels DNA-Mikroarray-Technik durch Bio-informatik verbessert. Biospectrum, 9, 93–95. [Google Scholar]
- 28.Seidel C.A.M., Schulz,A. and Sauer,M.H.M. (1996) Nucleobase-specific quenching of fluorescent dyes. 1. Nucleobase one-electron redox potentials and their correlation with static and dynamic quenching efficiencies. J. Phys. Chem., 100, 5541–5553. [Google Scholar]
- 29.Schwille P. (2000) Cross-correlation analysis in FCS. In Rigler,R. and Elson,E.S. (eds), Fluorescence Correlation Spectroscopy, Theory and Applications. Springer, Berlin, pp. 360–378. [Google Scholar]
- 30.Higuchi R., Fockler,C., Dollinger,G. and Watson,R. (1993) Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology, 11, 1026–1030. [DOI] [PubMed] [Google Scholar]