Abstract
We have developed an AFLP-based gene expression profiling method called ‘high coverage expression profiling’ (HiCEP) analysis. By making improvements to the selective PCR technique we have reduced the rate of false positive peaks to ∼4% and consequently the number of peaks, including overlapping peaks, has been markedly decreased. As a result we can determine the relationship between peaks and original transcripts unequivocally. This will make it practical to prepare a database of all peaks, allowing gene assignment without having to isolate individual peaks. This precise selection also enables us to easily clone peaks of interest and predict the corresponding gene for each peak in some species. The procedure is highly reproducible and sensitive enough to detect even a 1.2-fold difference in gene expression. Most importantly, the low false positive rate enables us to analyze gene expression with wide coverage by means of four instead of six nucleotide recognition site restriction enzymes for fingerprinting mRNAs. Therefore, the method detects 70–80% of all transcripts, including non-coding transcripts, unknown and known genes. Moreover, the method requires no sequence information and so is applicable even to eukaryotes for which there is no genome information available.
INTRODUCTION
With the vast amount of sequence information now available for genomes, genome-wide expression profiling provides a powerful tool for studying genes involved in various biological phenomena. Several good hybridization-based microarray methods have been developed to date (1–3), but they require sequence information for their analysis and leave room for improvement in the areas of reproducibility, coverage (percentage of expressed genes that are observable) and cost (4–7). On the other hand, there are gene expression profiling methods that do not require cDNA information such as differential display and arbitrarily primed polymerase chain reaction (8,9). However, these methods suffer from a high rate of false positives, up to 50% (10,11). Often false positive peaks overlap with real peaks, making it difficult to do gene expression profiling with wide coverage.
Here we report on the development of an expression profiling method, ‘high coverage gene expression profiling’ (HiCEP). This method was developed through substantial improvement, including improving the false positive rate, of the amplified fragment length polymorphism (AFLP) technique (12).
MATERIALS AND METHODS
Preparation of RNAs
Mouse embryonic fibroblasts (MEF) were prepared and exposed to 7 Gy irradiation (Pantac HF320; Shimadzu) and incubated for 3, 6 and 24 h at 37°C in 5% CO2. After incubation, each cell sample was used for preparation of mRNA. A total of 1.5 µg of mRNA prepared from MEFs, mouse embryonic stem (ES) cells and yeast cells with Fast TrackII (Invitrogen, Carlsbad, CA) was digested with DNase I (1.5 U/ml at 25°C for 15 min) and used for the HiCEP reaction.
Creation of HiCEP template
First strand cDNA was synthesized using a Superscript™ First-Strand Synthesis System (Invitrogen) with 100 pmol of 5′ biotinylated oligo(dT) primer (5′-biotin-TTTTTTTTTTTTTTTTTTV-3′) and the second strand was synthesized according to the protocol of the manufacturer (Invitrogen). As shown in Table 1, HiCEP analyzes two types of cDNA populations, MspI-MseI-poly(A) and MseI-MspI-poly(A). Here we describe the method for MspI-MseI-poly(A) mRNA. The double-stranded cDNA (dscDNA) was digested with 50 U MspI (TaKaRa, Ohtsu, Japan) followed by ligation to 5.0 µg of MspI adapter (5′-AATGGCTACACGAACTCGGTTCATGACA-3′ and 5′-CGTGTCATGAACCGAGTTCGTGTAGCCATT-3′) with 400 U T4 DNA ligase (NEB, Beverly, MA). The ligated products bearing biotin at the 5′-terminus were bound to magnetic beads coated with streptavidin (Dynabeads M-280 Streptavidin; Dynal, Oslo, Norway) and washed twice with 1.0 ml of washing buffer (5 mM Tris–HCl pH 7.5, 0.5 mM EDTA, 1.0 M NaCl). The cDNA fragments on the magnetic beads were digested with 20 U MseI (NEB) and then the supernatant including the digested fragments was collected. Ligation was performed with 10.2 pmol MseI adapter (5′-AAGTATCGTCACGAGGCGTCCTACTGCG-3′ and 5′-TACGCAGTAGGACGCCTCGTGACGATACTT-3′) using 400 U T4 DNA ligase in the presence of 2 U MseI in 15 µl of reaction mixture. To this was added 1485 µl of 0.1× TE (1.0 mM Tris–HCl pH 8.0, 0.1 mM EDTA), and 1.0 µl of the resulting solution was used as a template for selective PCR.
Table 1. Putative HiCEP coverage in three species.
aThe number of ‘RefSeq’ prefixed with ‘NM_’ in latest UniGene of mouse (build 122) and human (build 160).
Selective PCR
The selective PCR step in HiCEP analysis is based on AFLP (12). We used fluorescently labeled primers in PCR for detection. For labeling, 6-carboxyfluorescein (FAM), VIC and NED were used for 6, 5 and 5 of the 16 primers, respectively. We used HPLC-purified grade primers and the synthesis scale was 0.2 mmol (Applied Biosystems, Foster City, CA). The fluorescently labeled primer was designed to match the MspI adapter; 16 sequences of MspI-NN primers (5′-label-ACTCGGTTCATGACACGGNN-3′) and 16 sequences of MseI-NN primers (5′-AGGCGTCCTACTGCGTAANN-3′) were synthesized. The mixture was 4.0 pmol of MspI-NN primer, 10.0 pmol of MseI-NN primer, 40 nmol of dNTPs, 1× Titanium DNA polymerase buffer and 1× Titanium DNA polymerase (Clontech, Palo Alto, CA) in 20 µl solutions. The PCR conditions were 95.0°C for 1 min and 28 cycles of 95.0°C for 20 s, 71.5°C for 30 s and 72.0°C for 1 min, followed by 60.0°C for 30 min.
Electrophoresis and data analysis
The PCR products labeled with three different fluorescent dyes were mixed and 10 µl of formamide, 0.3 µl of GeneScan 500 ROX (Applied Biosystems) and 0.4 µl of GeneScan 1000 Red Dye (Applied Biosystems) were added to 3.0 µl of the mixed solution. The products were denatured and loaded on an ABI Prism 310 (Applied Biosystems) for electrophoresis. The conditions of injection were 15.0 kV for 5 s for a sample and 15.0 kV for 45 min in 310 POP4 for the electrophoresis (Applied Biosystems). The data analysis was performed using GeneScan 3.1.2 (Applied Biosystems).
Fractionation and sequencing of peaks of interest
An aliquot of 1 µl of HiCEP analysis products, 2.7 µl of formamide, 0.3 µl of GeneScan 500 ROX (Applied Biosystems) and 2.0 µl of 10× loading buffer (TaKaRa) were mixed, denatured by incubation at 95.0°C for 2 min and loaded on a denaturing gel: 4, 6 or 10% polyacrylamide containing 7.0 M urea. The conditions of electrophoresis were 1500 V for 4 h. Fluorescence from HiCEP analysis products was detected with a Typhoon 9210 (Amersham Biosciences, Piscataway, NJ) and slices of gel containing the bands of interest were cut out. The gel slices were suspended in 60 µl of TE buffer and 2.0 µl of it was used for PCR with MspI-universal-T7 primer (5′-TAGGTAATACGACTCACTATAGGGCGAATTGGGTACTCGGTTCATGACACGG-3′) and MseI-universal primer (5′-AGGCGTCCTACTGCGTAA-3′). DNA sequencing was carried out using T7 primer (5′-TAATACGACTCACTATAGGG-3′).
Real-time PCR
Samples of 5 µg of total RNA were treated with RNase-free DNase I (Invitrogen) and used with a Superscript™ First-Strand Synthesis System (Invitrogen). The reaction was performed according to the manufacturer’s protocol using an oligo(dT)18 primer (5′-TTTTTTTTTTTTTTTTTT-3′). The first strand cDNA was diluted to 1/10 with 0.1× TE and used as the template for the quantitative PCR. The real-time PCR (SYBR Green PCR Master Mix; Applied Biosystems) was performed and analyzed using an ABI Prism 7700 (Applied Biosystems). The primers for the mouse p21 gene were 5′-TCTCAGGGCCGAAAACGGAG-3′ and 5′-ACACAGAGTGAGGGCTAAGG-3′. The PCR conditions were 95.0°C for 10 min and 50 cycles of 95.0°C for 15 s, 60.0°C for 30 s and 78.0°C for 40 s. The data were normalized in relation to the expression level of the glyceraldehyde phosphate dehydrogenase (GAPDH) gene.
RESULTS
Reaction
A flow chart of the HiCEP method is shown in Figure 1. This method is based on the mRNA fingerprinting technique for detection of restriction fragment length polymorphisms. Poly(A) RNAs were prepared from cultured cells or tissues of interest and first strand cDNA was synthesized using biotinylated oligo(dT) primer. Second strand cDNAs were then synthesized and subjected to digestion with the restriction enzyme MspI or MseI, followed by adapter ligation. After the ligation step, cDNAs were trapped using magnetic beads coated with avidin and excess adapters, including the adapter dimer, were washed off. This process allowed collection of single tag fragments from each mRNA molecule. Selective PCR, used in the AFLP procedure (12), was used for amplification of the cDNA fragments, and the products with fluorescently labeled primer were then analyzed by capillary electrophoresis. The selective PCR procedure reported to date is not capable of precise selection of the corresponding sequences; 50% of its peaks are false positives (10,11). The presence of many overlapping peaks generated by false positive peaks causes two experimental difficulties, one in fragment cloning and the other in assessing quantitative changes in gene expression. Therefore, we strove to improve the accuracy of selective PCR in HiCEP analysis through extensive optimization of the selective PCR conditions. We randomly picked 4000 peaks and determined their sequences. We found that 3836 (95.9%) of the peaks appearing in electrophoresis were amplified with precise selection or, stated conversely, only 4.1% (164 peaks) were false positives. Of these 164, 106 were caused by a mismatched 5′ adapter side nucleotide in a primer and the other 58 by a mismatched 3′ donor side nucleotide. This high level of precise selection was demonstrated in Mus musculus mRNAs.
Figure 1.
Flowchart of the HiCEP method. The HiCEP method consists of seven steps. (A) dscDNA (open box) synthesis with a biotinylated oligo(dT) primer. (B) Digestion of dscDNA by MspI (or MseI). (C) Ligation with MspI (or MseI) adapter (gray box) and purification by biotin–avidin affinity. M, magnetic beads. (D) Digestion of dscDNA by MseI (or MspI). (E) Ligation with MseI (or MspI) adapter (closed box). (F) 256 sets of selective PCR primers. Arrows indicate primers, f indicates fluorescence (FAM, VIC and NED) and arrowheads correspond to the two selective nucleotides. (G) Detection of the fluorescence from selective PCR products. PCR samples were electrophoresesed with a 310 Genetic Analyzer (Applied Biosystems). Data analysis was conducted with GeneScan 3.1.2 software (Applied Biosystems). The x-axis indicates the length of the PCR product and the y-axis indicates the intensity of fluorescence of the PCR product.
Reproducibility and sensitivity
The results of four independently performed HiCEP analyses using the same poly(A) RNA preparation are shown in Figure 2A. Each peak corresponds to a single transcript and the peak area represents the quantity of mRNA molecules. Number, position and intensity of peaks were all highly reproducible, as demonstrated in four independent analyses. The reproducibility as tested with 2000 HiCEP peaks using 1.5 µg of mRNA as starting material is shown in Figure 2B. The peak intensity ratio between two independent experiments was 1.0001, with a standard deviation of 0.0739, indicating that HiCEP analysis can distinguish even 1.15- and 1.22-fold differences with 95 and 99% reliability, respectively. In this paper we focus our discussion on the reproducibility of reactions in the steps involved after the RNA preparation step, because the preparation of cells or tissues is carried out on a case-by-case basis. In actual experiments, however, the reproducibility of the cell and RNA preparation steps is also critical. The high reproducibility of HiCEP analysis was demonstrated with RNAs independently prepared from different cell fractions (Fig. 2C).
Figure 2.
Reproducibility of HiCEP analysis. (A) Reproducibility of separate reaction profiles by HiCEP analysis. In the four upper panels (black, red, green and blue) are the results of four independent reactions. The results of four reactions are superimposed in the lower panel. (B) Reproducibility of intensity of fluorescence of PCR products in separate reactions. Red, green and pink lines indicate a difference of 2.0, 1.5 and 1.2 times, respectively. We randomly selected 2000 peaks for the data points. (C) Fission yeast (S.pombe) cells were grown twice (lots 1 and 2) and mRNA was purified from each cell sample. The results of HiCEP using lot 1 and lot 2 cells are shown in the top and middle, respectively. The results of lots 1 and 2 are overlaid in the bottom.
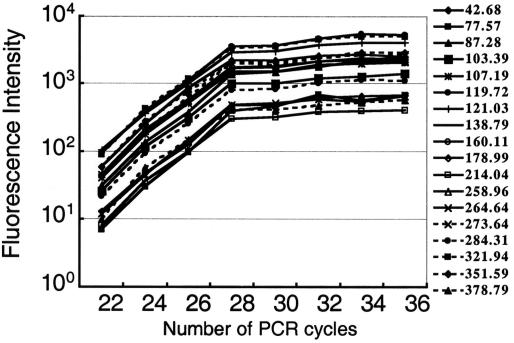
The amplification efficiency of PCR depends on the nucleotide composition and sequence of template DNAs and, therefore, the optimum number of PCR cycles for quantification varies from template to template within the same sample. To achieve proportional amplification of cDNAs from the whole mixture of mRNA molecules, we targeted our PCR to a short region of cDNA close to the poly(A) tail, mostly in the 3′ untranslated region (3′-UTR), which is known to be low in GC content. The use of four nucleotide recognition site restriction enzymes for mRNA fingerprinting enabled us to generate short fragments, almost all less than 700 bp in length (Table 1). Optimizing the number of PCR cycles resulted in the production of fragments with similar amplification efficiencies, independent of the fragment length and nucleotide sequence. All peaks reached a plateau phase of amplification at 28 cycles of PCR under the conditions used (Fig. 3). Similar efficiencies of PCR among different fragments may be due to the fact that almost all cDNA fragments amplified by our system are AT-rich and less than 700 bp in length.
Figure 3.
Relationship between PCR cycle and length of PCR product for the intensity of fluorescence of each peak. The x-axis indicates the number of PCR cycles. The y-axis indicates the intensity of fluorescence of the PCR products. Results shown are representative peaks. The numbers on the right show the peak position that corresponds to the length of each fragment (base pair).
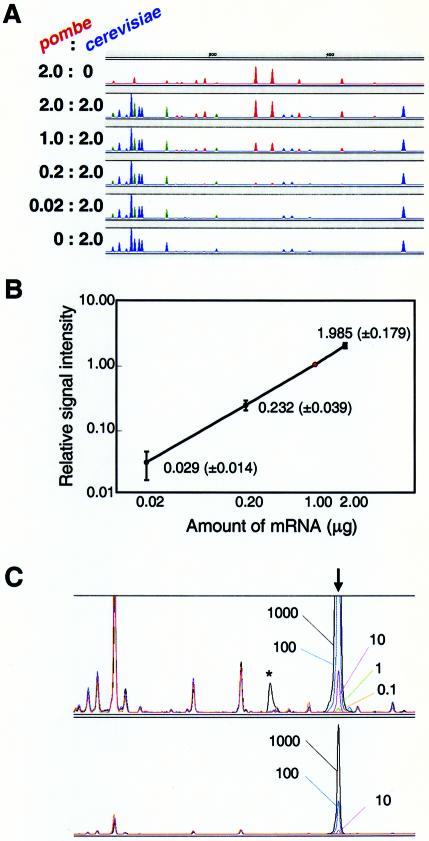
To find out how accurately the intensity of fluorescence reflected the original amount of mRNA in the initial reaction mixture, we carried out HiCEP analysis using mRNA from Saccharomyces cerevisiae and Schizosaccharomyces pombe mixed in various ratios. RNA samples mixing 2.0, 1.0, 0.2 or 0.02 µg of S.pombe mRNA with 2.0 µg of S.cerevisiae mRNA were used as templates. We observed a clear correlation between the signal intensities and the initial amounts of mRNA used. Comparison of the peaks corresponding to 1 and 2 µg of S.pombe RNA showed clear resolution of the 2-fold difference in the amount of RNA, shown by the red peaks in Figure 4A. Analyzing 100 randomly chosen S.pombe HiCEP peaks, we found a linear relationship between the starting amount of mRNA and signal intensity (Fig. 4B).
Figure 4.
Dependency of the signal on the amount of mRNA. (A) Various mixtures of S.pombe and S.cerevisiae mRNAs (2.0:0.0, 2.0:2.0, 1.0:2.0, 0.2:2.0, 0.02:2.0 and 0.0:2.0 µg) were analyzed using HiCEP analysis. The peaks of S.pombe and S.cerevisiae mRNAs are shown in red and blue, respectively. Green indicates mixed peaks of S.pombe and S.cerevisiae. All panels except the top panel feature 2.0 µg of S.cerevisiae mRNA. Blue peaks are consistent with the peaks from 2.0 µg of S.cerevisiae mRNA. (B) Amount-dependent signal demonstrated by 100 randomly picked HiCEP peaks. We analyzed 100 S.pombe peaks (i.e. red peaks) for varying amounts of S.pombe mRNA and plotted their intensities relative to the intensities of the compounding peaks using 1.0 µg of S.pombe mRNA (indicated by the red dot). Average relative intensity and standard deviation are shown. (C) Minimum number of copies detected by HiCEP. Various amounts of poly(A) RNA, synthesized with an in vitro transcription system, were added during the total RNA extraction step for S.cerevisiae. HiCEP provided a peak for the synthetic RNA at the expected position (vertical arrow). Data from five independent experiments are overlaid. The amount of synthetic RNA added to each reaction is indicated by the number of copies per cell. Peak detection with diluted samples (factor of 1/10) is also shown in the lower panel. An asymmetric peak (asterisk) is found in the analysis at 1000 copies/cell. A saturated peak frequently generates a mechanical artifact (ghost peak) downstream.
Experiments with four mRNAs prepared by in vitro transcription showed that HiCEP can quantitatively detect transcripts in a range as low as 1–500 copies/cell (Fig. 4C). This extremely high sensitivity permits detection of mRNA molecules expressed in a few cells of a heterogeneous cell population.
In another experiment, we observed induction of p21 in MEFs by ionizing radiation (IR) using HiCEP. mRNA samples were prepared at 0, 3, 6 and 24 h after exposure and two HiCEP analyses were performed with each sample (Fig. 5A). The magnified p21 peaks demonstrate an ability to reproducibly detect a slight expression change (Fig. 5B), and the expression change was confirmed by real-time PCR (Fig. 5C). Isolation and sequencing of this peak revealed that it was derived from the p21 transcript.
Figure 5.
Induction of mouse p21 transcript in response to IR. (A) Raw data of HiCEP analysis. Blue, red, green and orange lines indicate HiCEP data using mRNAs prepared at 0, 3, 6 and 24 h, respectively, after exposure to IR (7 Gy). (B) The p21 peaks. HiCEP analysis was performed twice. The colors have the same meaning as in (A). (C) Expression of p21 after exposure to IR, analyzed twice by HiCEP (solid line) and real-time PCR (dotted line). Standard deviations are shown. The y-axis indicates the intensity of fluorescence relative to the intensity at 0 h. For real-time PCR, data were normalized relative to the expression level of the GAPDH gene. Both HiCEP and real-time PCR analyses were carried out twice with the same RNA sample. (D) Induction of the CyclinG, Mdm2 and Gadd45 transcripts, whose transcription is controlled by p53, as detected by HiCEP. Black and red lines indicate HiCEP data using mRNAs prepared at 0 and 6 h, respectively, after exposure to IR (7 Gy).
Coverage
PCR-based technologies for mRNA quantification have been successfully used for identification of certain genes showing expression changes (8,13). The low rate of false positive peaks in HiCEP enabled us to use four nucleotide recognition site restriction enzymes for fingerprinting of cDNAs and to detect expressed genes with high coverage. An in silico study revealed that 79.9 and 85.0%, respectively, of the cDNAs converted from mRNA have both recognition sites in mice and humans and 74.5 and 79.8%, respectively, of these are within the range easily resolved using standard capillary electrophoresis (between 40 and 700 bp in length, and thus can be detected as HiCEP peaks (Table 1). Using more enzyme combinations, we can achieve nearly 100% coverage. The relationship between coverage and enzyme combinations is shown in Figure 6.
Figure 6.
Estimation of coverage when using multi-enzyme sets in HiCEP analysis. The y-axis indicates coverage. The x-axis indicates number of enzyme combinations. Open squares and open circles indicate the coverage estimated in human and mouse, respectively. The broken line indicates 99% coverage. The order of enzymes is CCGG/TTAA (MspI/MseI), GATC/CATG (Sau3AI/NlaIII), CTAG/CATG (BfaI/NlaIII), CCGG/ACGT (MspI/HpyCH4IV), TTAA/AATT (MseI/Tsp509I), GCGC/CCGG (HinP1I/MspI), ACGT/AATT (HpyCH4IV/Tsp509I), CATG/TTAA (NlaIII/MseI), CATG/AATT (NlaIII/Tsp509I) and TCGA/TTAA (TaqI/MseI).
We used S.cerevisiae as a test case to experimentally determine the coverage of HiCEP analysis. Saccharomyces cerevisiae is the only eukaryote for which all ORFs (5726 genes) are known (14). Under the culture conditions used, 72.55% of these genes were expressed (15). Our analysis requires sequence information on both the 3′-UTR and ORF of each gene to successfully assign peaks to corresponding genes. Few yeast cDNA sequences, however, have been analyzed and deposited in the public databases, presumably because most yeast genes lack introns and therefore the information on genomic DNA is sufficient for functional analyses. During the course of our study, therefore, we determined the nucleotide sequences of approximately 10 000 ESTs, with particular emphasis on the 3′-UTR to determine non-coding exons and heterogeneity of poly(A) sites. Our studies demonstrated that an average of seven different mRNA transcripts are produced from each ORF, apparently because of polyadenylation at heterogeneous sites, but that only 1.125 HiCEP peaks on average were generated from them (data not shown).
The number of expressed transcripts predicted was 4956 and the number actually detected by HiCEP analysis was 3638, for a calculated coverage rate of 73.41%. Meanwhile, a study using the public EST database and our own EST database revealed that 71.0% of the genes of S.cerevisiae contain both MspI and MseI enzyme recognition sites (Table 1). Thus HiCEP analysis experimentally detected 103.39% of the predicted number of transcripts. To be sure, some of the expressed transcripts detected by HiCEP are non-coding, but the number of these is believed to be small (14). HiCEP analysis is the first PCR-based method for which a wide coverage of expressed genes has been verified experimentally.
Evidence using other species further supports the high coverage rate. We tested whether HiCEP analysis could detect the induction of five randomly chosen IR-inducible genes in mice: p21, Gadd45, CyclinG, Mdm2 and Bax. We successfully detected the induction of the first four in response to IR (Fig. 4A and D), but not Bax, because Bax lacks the recognition sites used in the HiCEP analysis. The predicted fragment lengths from the databases are 212, 626, 1055 and 358 bp for p21, CyclinG, Mdm2 and Gadd45, respectively. In contrast, experiments using capillary electrophoresis estimated the fragment lengths to be 210.42, 628.85 and 355.60 bp for p21, CyclinG and Gadd45, respectively. The size of Mdm2 could not be determined by electrophoresis, because the molecular weight marker was not available. Cloning and sequencing of these peaks were performed to confirm the results. In addition, HiCEP analysis was able to detect almost all of the mating type-specific genes by comparing MATa with MATα yeast strains (data not shown).
Assigning HiCEP peaks to genes
We adopted two methods for assigning HiCEP peaks to their corresponding genes: (i) prediction using the sequence information available in the public database and our ESTs; (ii) actual fractionation and sequencing. At present, prediction is possible and efficient with S.cerevisiae because of the existing body of knowledge about its entire genome sequence. However, it is not efficient for humans or mice because we don’t have enough information about their cDNAs. Capillary electrophoresis can usually discriminate even 1 bp differences in length. Resolution near big peaks sometimes decreases, but even in these cases differences of 2 or 3 bp in length can be discriminated. In addition, overlapping peaks appear in some species that have a large number of genes. Therefore, at present, cloning and sequencing are more effective for humans and mice. Currently, more than 100 HiCEP peaks can be fractionated per day and their corresponding DNA sequences determined. In addition, the false positive rate of ∼4% will enable us to prepare a database of all peaks, allowing gene assignment without having to isolate and sequence peaks of interest.
DISCUSSION
We expected to detect more than 70% of the expressed genes using four nucleotide recognition site restriction enzymes, and this was confirmed experimentally using an RNA sample from S.cerevisiae.
We also further optimized the selective PCR procedure to reduce false positive peaks. Sequencing 4000 randomly chosen peaks showed that 95.9% of the peaks identified by the HiCEP analysis were positives. This enabled us to assign almost all peaks to a corresponding gene or transcript and then to analyze the huge number of peaks generated by the HiCEP reaction using four nucleotide recognition site enzymes with few overlapping peaks. We could even exclude the remaining 4.1% of false positive peaks by comparing the results obtained from two HiCEP analyses performed at different temperatures for the elongation step, 68.0 and 71.5°C. Only false positive peaks showed decreased intensity at 71.5°C.
Most existing PCR-based mRNA fingerprinting methods use oligo(dT) primers and consequently suffer from certain associated problems. One such problem is the complexity of data, including multiple peaks from a single ORF, due to missannealing of primers or the heterogeneity of polyadenylation sites in mRNA (16,17). To overcome these difficulties, we optimized the PCR conditions and adapter sequences. All peaks in each PCR reaction are amplified in a competitive PCR fashion with an adapter-specific primer set, and so the reproducibility of the relative intensity of the peaks in each primer set, usually containing approximately 100 peaks, is extremely high. These improvements paved the way for gene coverage of more than 70% and high reproducibility.
Unlike hybridization-based methods, HiCEP analysis requires the cloning of peaks of interest after fragment separation by capillary electrophoresis. Although this may seem to be a disadvantage, HiCEP does not require any genome-wide information for its analysis and therefore can be used for any eukaryote. The low false positive rate of HiCEP peaks allow us to isolate peaks of interest easily and, furthermore, enables us to obtain information on peaks of interest without having to isolate them.
HiCEP analysis can distinguish even 1.2-fold differences in gene expression, allowing elucidation of the detailed time course of gene expression. Its ability to detect non-coding transcripts will contribute to understanding gene expression regulation at the RNA level (18,19).
Acknowledgments
ACKNOWLEDGEMENTS
We would like to express special thanks to K. Kurachi for critical reading of the manuscript and helpful discussions, M. Ajimura for supplying total RNA of yeast and helpful discussions and N. Sasaki for helpful discussions on statistics. We would also like to thank K. Yokoro and S. Takahashi for encouragement and B. Burke-Gaffney and J.J. Rodrigue for editing the English. We are grateful to Y. Miyamoto, M. Nakahara, T. Asano, T. Ohhata, K. Shingu, M. Nakamura, H. Muto and K. Nishikawa for technical assistance. This work was partly supported by Research Grants from the Special Coordination Funds of the Japan Society for the Promotion of Science.
REFERENCES
- 1.Panda S., Antoch,M.P., Miller,B.H., Su,A.I., Schook,A.B., Straume,M., Schultz,P.G., Kay,S.A., Takahashi,J.S. and Hogenesch,J.B. (2002) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell, 109, 307–320. [DOI] [PubMed] [Google Scholar]
- 2.Cho R.J., Campbell,M.J., Winzeler,E.A., Steinmetz,L., Conway,A., Wodicka,L., Wolfsberg,T.G., Gabrielian,A.E., Landsman,D., Lockhart,D.J. et al. (1998) A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell, 2, 65–73. [DOI] [PubMed] [Google Scholar]
- 3.Lockhart D.J., Dong,H., Byrne,M.C., Follettie,M.T., Gallo,M.V., Chee,M.S., Mittmann,M., Wang,C., Kobayashi,M., Horton,H. et al. (1996) Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol., 14, 1675–1680. [DOI] [PubMed] [Google Scholar]
- 4.Wang X., Ghosh,S. and Guo,S.W. (2001) Quantitative quality control in microarray image processing and data acquisition. Nucleic Acids Res., 29, e75–e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim J.H., Kim,H.Y. and Lee,Y.S. (2001) A novel method using edge detection for signal extraction from cDNA microarray image analysis. Exp. Mol. Med., 33, 83–88. [DOI] [PubMed] [Google Scholar]
- 6.Kurian K.M., Watson,C.J. and Wyllie,A.H. (1999) DNA chip technology. J. Pathol., 187, 267–271. [DOI] [PubMed] [Google Scholar]
- 7.Bowtell D.D. (1999) Options available—from start to finish—for obtaining expression data by microarray. Nature Genet., 21, 25–32. [DOI] [PubMed] [Google Scholar]
- 8.Liang P. and Pardee,A.B. (1992) Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science, 257, 967–971. [DOI] [PubMed] [Google Scholar]
- 9.Welsh J. and McClelland,M. (1990) Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res., 18, 7213–7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green C.D., Simons,J.F., Taillon,B.E. and Lewin,D.A. (2001) Open systems: panoramic views of gene expression. J. Immunol. Methods, 250, 67–79. [DOI] [PubMed] [Google Scholar]
- 11.Martin K.J. and Pardee,A.B. (2000) Identifying expressed genes. Proc. Natl Acad. Sci. USA, 97, 3789–3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vos P., Hogers,R., Bleeker,M., Reijans,M., van de Lee,T., Hornes,M., Frijters,A., Pot,J., Peleman,J., Kuiper,M. et al. (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res., 23, 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato K. (1995) Description of the entire mRNA population by a 3′ end cDNA fragment generated by class IIS restriction enzymes. Nucleic Acids Res., 23, 3685–3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kellis M., Patterson,N., Endrizzi,M., Birren,B. and Lander,E.S. (2003) Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature, 423, 241–254. [DOI] [PubMed] [Google Scholar]
- 15.Velculescu V.E., Zhang,L., Zhou,W., Vogelstein,J., Basrai,M.A., Bassett,D.E.,Jr, Hieter,P., Vogelstein,B. and Kinzler,K.W. (1997) Characterization of the yeast transcriptome. Cell, 88, 243–251. [DOI] [PubMed] [Google Scholar]
- 16.Kato K. (1997) Molecular indexing. Tanpakushitsu Kakusan Koso, 42, 2876–2881. [PubMed] [Google Scholar]
- 17.Pauws E., van Kampen,A.H., van de Graaf,S.A., de Vijlder,J.J. and Ris-Stalpers,C. (2001) Heterogeneity in polyadenylation cleavage sites in mammalian mRNA sequences: implications for SAGE analysis. Nucleic Acids Res., 29, 1690–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eddy S.R. (2001) Non-coding RNA genes and the modern RNA world. Nature Rev. Genet., 2, 919–929. [DOI] [PubMed] [Google Scholar]
- 19.Mattick J.S. (2001) Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep., 2, 986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]